Proper segregation of genetic material is a universal requirement for all living organisms. In eukaryotic organisms, the mitotic spindle, a specialized tubulin-based cytoskeletal structure, actively separates the chromosomes into two new nuclei during cell division (1). For prokaryotes, the story is not much different. Although they contain only one chromosome, plasmids, and no spindle apparatus, the genetic information has to be replicated and segregated during cell division. Decades of research have shown that chromosomal segregation in prokaryotes requires the action of proteins that actively move chromosomes to their intended place. Plasmids have their own way of ensuring proper distribution during cell division. For high-copy plasmids, numbers are the strength; chance alone ensures that each daughter cell receives some copies guaranteeing genetic transmission. Low-copy plasmids, however, are actively partitioned and code for their own segregation machinery. They carry a tripartite system, composed of two proteins and a centromere-like recognition sequence in the plasmid itself. One of the proteins creates the force involved in plasmid movement. The second acts as an adaptor; it binds the DNA at the recognition sequence and links it to the force-generating protein (2). Interestingly, while all low-copy plasmids share this mechanism, different plasmids use completely unrelated sets of proteins. The force-generating proteins are particularly interesting, with three kinds already known. Two belong to the tubulin (3) and actin families (4), respectively. The third corresponds to a P-loop ATPase, similar to those involved in the segregation of the chromosome (5). All of them couple nucleotide hydrolysis and filament polymerization, which itself is coupled to force generation. In PNAS, Szewczak-Harris and Löwe (6) and Usluer et al. (7) present two back-to-back papers describing the near-atomic resolution structures of AlfA, the protein in charge of the segregation of the pLS32 plasmid from Bacillus subtilis subsp. natto.
AlfA belongs to the actin superfamily. Although it shares only ∼20% sequence identity with its eukaryotic relative, its sequence has the signature motifs of actin (8). The actin fold is composed of two clamped domains (I and II), each of which can be further divided into two subdomains (Ia, Ib, IIa, and IIb). What sets AlfA apart from the rest of the actin family is that it completely lacks subdomain IIb, making it the most divergent member characterized so far. This extreme divergence is typical of bacterial actins. A comparison between different groups of bacterial actin-like proteins shows that they share as much sequence identity with each other as they do with eukaryotic actin (9). Interestingly, eukaryotic actin is one of the most conserved proteins and its versatility stems from countless adaptor proteins that mediate different functions. In contrast, bacterial actin-like proteins seem to have created a version of actin for each purpose. Many of them have only recently been discovered (9).
The closest homolog of AlfA—with ∼20% sequence identity—is ParM, one of the best-characterized force-generating proteins involved in low-copy plasmid segregation (Fig. 1). Like all known actin superfamily members, AlfA and ParM form double-stranded filaments. Contrary to the right-handed eukaryotic actin, AlfA and ParM are left-handed helical filaments, which bundle under physiological conditions. Usluer et al. (7) and Szewczak-Harris and Löwe (6) demonstrate that while AlfA lacks a complete subdomain, a large rearrangement of the filament’s lattice ensures that the polymer structure is mostly preserved. Moreover, bundling seems to be key for their function. Szewczak-Harris and Löwe (6) show that AlfA filaments form antiparallel bundles, which would be necessary to push the plasmids to opposite poles of the cell.
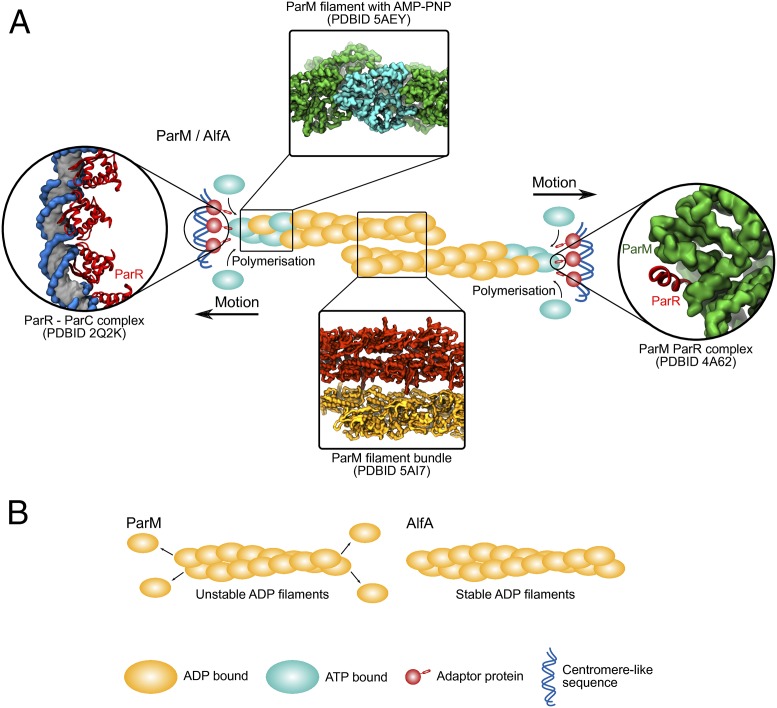
Fig. 1.
Schematic representation of the plasmid segregation mechanism involving actin-like bacterial proteins. (A) Overview of the key macromolecular complexes involved in the plasmid segregation process. Bundling and growth of the filaments toward opposite directions push the plasmids toward different ends of the cell. The filaments are coupled to the plasmid with an adaptor protein that binds the filament end and the centromere-like sequence. For the ParMRC system a structure is available for each key step. This includes the complex between the centromere-like sequence (ParC) and the adaptor (ParR) (14), the complex between ParM and ParR (15), filament with AMP-PNP (10), and filament bundles (10). (B) Polymerization is connected to nucleotide hydrolysis. For ParM, hydrolysis destabilizes the filament, which ends in depolymerization in the absence of ParR/ParC. In contrast, AlfA filaments are stable in their ADP form.
In the case of ParM, bundling is also part of the segregation process. For this protein polymerization can occur at both sides of the filaments, and their stability is strongly linked to the nucleotide bound to their active site (Fig. 1). Soon after polymerization, the ATP at the active site is hydrolyzed to ADP. In the absence of the adaptor protein (ParR) bound to the centromere sequence (ParC), nucleotide hydrolysis changes the helical lattice, which results in catastrophic depolymerization (10) (Fig. 1B). This process of repeated growth and shrinkage is known since dynamic instability and is key for ParM activity. Interestingly, this has been invented at least once more during cytoskeletal evolution, as dynamic instability is one of the key features of microtubule dynamics (11). For AlfA, the picture is very different. Polymerization occurs primarily on one end, and nucleotide hydrolysis does not affect filament formation (Fig. 1B). Fortunately, while Usluer et al. (7) determined the structure of AlfA in complex with the nonhydrolysable ATP analog AMP-PNP, Szewczak-Harris and Löwe (6) solved the structure with ADP. The structures clearly show that there is little difference in helical structure, arguing that the mechanism of plasmid segregation cannot be completely equivalent to ParM. One way of explaining this comes from the most prominent feature of the protein. The missing part, subdomain IIb, contains part of the nucleotide-binding site in the active fold. Both groups report that its absence has led to a new binding mode for the adenosine moiety, something that could explain the different nucleotide sensitivity of these filaments. Szewczak-Harris and Löwe (6) go on to speculate that the lack of this subdomain has an adaptive advantage, as plasmids might prefer coding for simpler proteins to replicate faster.
Why are these new AlfA structures important? Obviously, they contribute to our understanding of how plasmids have adapted to ensure their transmission despite their numbers. But for molecular evolution, the implications are much deeper. The fact that the segregation of many low-copy plasmids, or even chromosomes, occurs using the same basic mechanism but with completely unrelated proteins means that evolution has reinvented this push/pull strategy of nucleic acid segregation multiple times, a wonderful example of convergent evolution. For the understanding of the evolution of the actin superfamily, they are also key. While all known members share the same fold, prokaryotes have evolved a large array of actin-like filaments with different helical lattices: left- or right-handed, staggered, unstaggered, or even apolar (12). This observation raises the question as to whether the actin filament has been invented more than once in evolution. The answer will probably require solving a much larger array of actin-like filament structures than we have so far. From the functional side, rather than being an inert rail on which motor proteins can travel, the actin cytoskeleton is a dynamic structure which allows force to be generated directly from filament growth. For ParM and AlfA, this results in plasmid segregation; for eukaryotic actin polymerization can, for example, create the motion responsible for cellular migration (13). It is tempting to speculate then, that the combination of nucleotide hydrolysis and pushing/pulling activity represents the original function of the actin cytoskeleton.
All in all, these structures help us to understand the evolution of the cytoskeleton and push forward our ever-increasing understanding of the complexity of microbial life. Moreover, they give us clues as to how the dynamics of the cytoskeleton are controlled—on a molecular level—to generate force. Hopefully, new structures of other divergent actin-like filaments will help us to fully understand the evolution of the microbial cytoskeleton.
Acknowledgments
Our work is supported by the Max Planck Society and the European Council under the European Union’s Seventh Framework Programme (FP7/ 2007–2013) (Grant 615984).
Footnotes
References
- 1.Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- 2.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 3.Larsen RA, et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salje J, Gayathri P, Löwe J. The ParMRC system: Molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol. 2010;8:683–692. doi: 10.1038/nrmicro2425. [DOI] [PubMed] [Google Scholar]
- 5.Gerdes K, Møller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: Surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 6.Szewczak-Harris A, Löwe J. Cryo-EM reconstruction of AlfA from Bacillus subtilis reveals the structure of a simplified actin-like filament at 3.4-Å resolution. Proc Natl Acad Sci USA. 2018;115:3458–3463. doi: 10.1073/pnas.1716424115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usluer GD, et al. Cryo-EM structure of the bacterial actin AlfA reveals unique assembly and ATP-binding interactions and the absence of a conserved subdomain. Proc Natl Acad Sci USA. 2018;115:3356–3361. doi: 10.1073/pnas.1715836115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker E, et al. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25:5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derman AI, et al. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: Regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bharat TA, Murshudov GN, Sachse C, Löwe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015;523:106–110. doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 12.Merino F, Raunser S. The mother of all actins? eLife. 2016;5:e23354. doi: 10.7554/eLife.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 14.Schumacher MA, et al. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature. 2007;450:1268–1271. doi: 10.1038/nature06392. [DOI] [PubMed] [Google Scholar]
- 15.Gayathri P, et al. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science. 2012;338:1334–1337. doi: 10.1126/science.1229091. [DOI] [PMC free article] [PubMed] [Google Scholar]