Abstract
Background
Antibiotic resistance of environmental Escherichia coli in hospitals could be increased due to extensive use of biocides resulting in serious infections. In this study, the prevalence of antibiotic resistance of environmental isolates of E. coli from hospitals and household settings were evaluated and compared. In addition, the association between biocide minimum inhibitory concentration (MIC) and multiple drug resistance (MDR) was investigated.
Methods
Environmental samples were collected from different homes and hospitals in Amman, Jordan. The isolates were identified phenotypically and by PCR. Antibiotic susceptibility tests and MIC of selected biocides were performed on the isolates. Screening for blaCTX-M group 1 was also performed.
Results
Of 21 E. coli strains isolated, 47.6% were MDR and 67.9% were phenotypically identified as extended spectrum beta-lactamase (ESBL) producers. The occurrence of these ESBL isolates was comparable between household and hospital settings (P > 0.05). The MIC values of the biocides tested against all isolates were well below the in-use concentration of biocides. Moreover, the MICs of biocides were comparable between isolates from households and those from hospitals (P > 0.05). No association was found between MDR and biocide MIC (P > 0.05). Most of ESBL isolates harboured blaCTX-M 1.
Conclusions
The extensive use of biocides in hospitals is not associated with MDR nor does it affect the MIC of biocides against E.coli.
Keywords: Escherichia coli, Biocide, Environment, ESBL, Hospital, Multiple drug resistance
Background
Health care associated infections (HAIs) are known to contribute to morbidity and mortality among patients affected by them. In addition, they cause significant medical and financial consequences accompanied by emotional devastation [1]. Among the factors contributing to increased risk of HAIS are poor facilities cleaning and inadequate disinfection of health care settings [1]. These issues have led to the extensive use of biocides (including antiseptics and disinfectants) in hospitals. Since biocides have some common properties with antibiotics regarding their activity, mechanism of action and development of resistance [2], there is a possibility that resistance to biocides can contribute to resistance to antibiotics [3]. However, the contribution of biocide use to antibiotic resistance is still controversial and, despite some evidence, remains largely unproven. Several studies have shown that certain disinfectants have increased the expression of specific multiple drug resistant (MDR) efflux pumps which eventually resulted in resistance to some antibiotics [4]. Other studies have shown that over-exposure of bacteria to disinfectants results in reduced susceptibility towards some antibiotics [2, 5]. On the other hand, other studies have failed to show any cross-resistance between biocides and antibiotics [6, 7].
E. coli is a highly diverse species with respect to its virulence and pathogenicity. It is widely distributed in open systems and can easily spread in the environment causing risks to human health [8]. E. coli is one of the most common bacteria causing nosocomial infections. Its presence on inanimate surfaces in hospitals is one of the major infection control challenges facing hospitals [9]. In fact, contaminated surfaces and inanimate objects (fomites) are considered reservoirs for pathogen transmission to the patients. Therefore, the use of biocides in hospitals is of a paramount importance to control infections and transmission of pathogens.
In this study the biocides were selected to represent different chemical classes that are used extensively in Jordan in hospital and/ or household settings. Ethanol is an alcohol, which is used as antiseptic in hospitals and community settings. 4-Chloro-3,5-xylenol (known as chloroxylenol) is a phenolic compound that is used as a general antiseptic and disinfectant in the community. Iodine is a halogen, which is used in the form of povidone-iodine as a preoperative antiseptic in hospitals and for wound disinfection in household and hospital settings. Cetrimide is a member of the quaternary ammonium compounds that are incorporated in many biocidal preparations in combination with other biocides that are used in hospitals as antiseptics or disinfectants.
The aim of this research is to evaluate and compare the prevalence of antibiotic resistance in E. coli isolated from two distinct environments; hospital settings, where biocides are extensively used, and household settings, where biocide use is limited. E. coli was selected since it is recognized as an indicator for the presence of other Enterobacteriaceae and is a common cause of nosocomial infections. The collected isolates were assessed for the presence of potential association between antibiotic resistance and resistance to biocides, which was measured by an increase in their minimum inhibitory concentration (MIC).
Materials and methods
Sample collection
The environmental samples included in the study were collected from two hospitals, Prince Hamzeh (PH) Hospital and Jordan University (JU) Hospital and ten resident homes located in Amman-Jordan. Sample collection was performed from March to October 2016. The samples were collected using sterile swabs (Amies Transport media, Max Protect, China) that were pre-moistened with Amies medium present in the tube. The swab was rolled and moved over the surface to be sampled. After sampling, the swabs were transferred to the laboratory within one to two hours to be processed. In the laboratory, the swabs were cut off aseptically and placed in Lauryl sulphate tryptose broth (LSB) for enrichment and incubated overnight at 35o ± 2oĊ.
The two hospitals included in the study are among the largest in Amman. Each hospital treats on average more than 500,000 patients annually. Ethical permission to undertake sampling was obtained from both hospitals. Biocides used within the hospitals were recorded. The two hospitals use ethanol based gels and solutions as antiseptic for healthcare personnel, pre-injection disinfection and for visitor use. Different quaternary ammonium compounds based products are used for general disinfection and antisepsis within the two hospitals. Iodine in the form of povidone-iodine is used for wound disinfection and preoperative skin treatment. The hospitals apply strict disinfection policies that are monitored by infection control teams. Samples from hospital environments were collected from the floors, elevators, curtains, patient beds, windows, door knobs, nursing cabinets, bathroom sinks, drains, pressure devices, magnetic resonance device, operation equipment, dialysis device, trolleys, and any device that is transferrable among patients and medical staff. The samples were collected from different hospital wards. In total 344 swab samples were collected from both hospitals.
The homes included in the study apply routine cleaning to the premises using detergents. These homes occasionally use hypochlorite (a halogen) or chloroxylenol (e.g. Dettol®) based preparations for general disinfection. Povidone iodine and ethanol (70%) were the most common antiseptics used in these homes to treat bruises or cuts if occurred. Samples from household environment were collected from the floors, door knobs, bathroom sinks and kitchen sinks. The number of samples collected from the 10 homes was 86.
E. coli Isolation and identification
A loopful of LSB culture was streaked on MacConkey agar medium and incubated overnight at 35o ± 2oĊ. Morphologically distinctive pink colonies were isolated and identified biochemically for oxidase production, Kligler’s iron agar, urease, gas, indole production and then identified using API 20 E kit (Biomerieux, France). The potential E. coli isolates were confirmed genetically using PCR method.
Antibiotic susceptibility test
Antibiotic susceptibility test was performed using disc diffusion test according to Clinical and Laboratory Standard Institute (CLSI, 2016) guideline [10] using the following antibiotics: Amoxicillin, Amoxicillin-clavulanic acid, Cefaclor, Cefixime, Nitrofurantoin, Cefuroxime, Amikacin, Ciprofloxacin, Imipenem, Trimethoprim-Sulfamethoxazole, Doxycycline. These antibiotics represent the major antibiotic classes which have known activity against E. coli and are used clinically. E. coli ATCC 25922 was used as a quality control strain to validate the method [10].
The isolates were further tested to phenotypically detect extended spectrum beta lactamase (ESBL) producing bacteria using double disc diffusion test [10]. In this test, cefotaxime and ceftazidime discs alone and in combination of clavulanate were used. The isolate is considered ESBL producing if there is ≥5-mm increase in the zone diameter for antibiotic tested in combination with clavulanate vs the zone diameter of the antibiotic when tested alone [10].
MIC determination of biocides
Minimum inhibitory concentrations of ethanol, chloroxylenol, cetrimide and iodine were determined using a broth microdilution method according to CLSI, but with slight modification. Stock solutions of ethanol, cetrimide and chloroxylenol were prepared in Mueller Hinton broth (MHB) to get final concentration of 200 mg/ml, 400 μg/ml and 300 μg/ml respectively. In order to enhance the solubility of chloroxylenol in MHB, it was dissolved first in dimethylsulfoxide (DMSO) and the final volume completed by the addition of MHB. The lowest concentration of DMSO needed to ensure complete solubility of chloroxyleneol in MHB was 5%. To ensure that DMSO at 5% concentration has no inhibitory effect on the isolates, positive control containing 5% DMSO in MHB was prepared for each isolate. Since MHB medium contains starch as an ingredient, TSB was used for MIC determination of iodine. Iodine was solubilized with potassium iodide at 1:2 ratio (I2: KI) in TSB to get a stock solution of iodine (1300 μg/ml). Aliquots (200 μl) of each stock solution were dispensed into the wells of a microtitre plate. Double serial dilutions were performed using broth. Each trial was performed in five replicates. Aliquots (20 μl) of each bacterial culture adjusted to 5 × 106 CFU/ ml were used to inoculate the microtitre plate wells to yield a final concentration of ca 5 × 105 CFU/ ml. The microtitre plate was incubated for 20 h at 35o ± 2oĊ. MIC was determined by visual inspection. For ethanol, since the difference between consecutive concentrations is large, linear serial dilutions were performed after determining its MIC by double serial dilutions. E. coli Nissle 1917 was used as a control for MIC testing. This strain is a kind gift from Ardeypharm GmbH, Germany. It is a probiotic non-pathogenic microorganism utilized clinically to treat many gastrointestinal disorders including diarrhoea, ulcerative colitis and uncomplicated diverticular disease [11].
DNA extraction
The Wizard® Genomic DNA Purification Kit (Promega, England) was used to isolate DNA from the isolated E. coli strains. The kit was used according to manufacturer’s instructions.
PCR primers and conditions
blaCTX-M group 1 gene and E. coli16s rRNA gene: PCR reaction was performed using 3 μl of the extracted DNA (2 μl for E. coli 16S rRNA gene), and 0.4 μM of each of the blaCTX-M group 1 gene forward and reverse primer and the (16 E1, 16 E2 and 16 E3) primers of E. coli 16S rRNA gene (Table 1). The gene was amplified using 12.5 μl of PCR Master Mix 2× (GoTaq® Green Master Mix, Promega, USA). The volume was made up to 25 μl using nuclease free water. Cycling conditions for E. coli 16S rRNA gene were applied according to Tsen et al., [12]. Cycling conditions for blaCTX-M 1 gene were applied according to Mirzaee et al., [13]. The amplified gene products were analyzed using 2% agarose gel electrophoresis and visualized by (UVP) system (Alpha Imager®, Japan) using Redsafe™ (Intron biotechnology, Korea).
Table 1.
The targeted genes, primer sequence and product size
| Target | Detection primer | Primer (sequence 5′ to 3′) | Product size (bp) | Reference |
|---|---|---|---|---|
| E. coli 16S rRNA | 16 E1 (F) | GGGAGTAAAGTTAATACCTTTGCTC | 584 | 12 |
| 16 E2 (R) | TTCCCGAAGGCACATTCT | |||
| 16 E3 (R) | TTCCCGAAGGCACCAATC | |||
| blaCTX-M group 1 | CTX-M-7 (F) | GCGTGATACCACTTAACCTC | 260 | 13 |
| CTX-M-8 (R) | TGAAGTAAGTGACCAGAATC |
Statistical analysis
The results were statistically analysed using the non-parametric Mann Whitney U test and the Chi square test as relevant. Analyses at 95% confidence level were performed. The analysis was performed using IBM SPSS Statistics version 23.
Results
The total number of samples collected was 430; 175 samples were taken from PH hospital, 169 samples from JU hospital and 86 samples from household settings. The potential number of E. coli isolates identified by API kit was 21. These isolates were confirmed to be E. coli by PCR followed by agarose gel electrophoresis. Accordingly, the prevalence rate of E. coli in all the settings was 4.9%; seven isolates from PH hospital, four isolates from JU hospital and 10 isolates from households (Table 2).
Table 2.
The antibiogram of E coli isolates from the environment of the two hospitals and 10 homes
| Facility | Isolate | Amx | Amc | Cfc | Cfu | Cfx | Ctz | Ctx | Imp | Nit | Amk | Cip | Tms | Dox | MDR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Household settings | A126 | R | S | R | R | R | R | R | S | S | S | S | S | S | No |
| A409-c | I | I | R | R | R | R | R | S | S | S | S | R | I | No | |
| A410-a | R | R | S | S | S | R | R | S | S | S | S | R | S | Yes | |
| A410-b | R | R | R | R | R | R | R | S | I | R | S | R | S | Yes | |
| A411 | R | S | R | R | R | R | R | S | S | S | S | R | I | Yes | |
| A413 | R | R | R | R | R | R | R | S | S | S | S | R | S | Yes | |
| A414 | R | S | R | R | R | S | S | S | S | S | R | R | S | Yes | |
| A801 | R | I | S | I | S | S | S | S | S | S | S | R | I | No | |
| A814-a | I | S | S | I | S | S | S | S | S | S | S | S | S | No | |
| A824-a | S | S | S | S | S | S | S | S | S | S | S | S | S | No | |
| PH Hospital | B107-a | R | S | R | R | R | S | S | S | R | S | R | R | R | Yes |
| B425-a | S | S | R | R | R | R | R | S | S | S | S | R | I | No | |
| B425-b | S | S | R | R | R | S | R | S | S | S | S | R | S | No | |
| B426-a | R | R | R | R | R | R | I | S | S | S | R | R | R | Yes | |
| B475-a | R | I | R | R | R | R | R | S | S | S | R | R | R | Yes | |
| B506-a | S | S | S | S | S | S | S | S | S | S | S | R | R | No | |
| B705-b | R | S | R | R | R | R | R | S | S | S | S | R | S | Yes | |
| JU Hospital | C604 | R | S | S | I | S | S | S | S | S | S | S | R | I | No |
| C705 | S | S | R | I | S | R | R | S | S | S | S | S | S | No | |
| C715-b | I | S | S | I | S | S | S | S | S | S | S | S | S | No | |
| C906 | R | S | R | R | R | R | R | S | S | S | R | S | I | Yes | |
| E. coli ATCC | S | S | S | S | S | S | S | S | S | S | S | S | S | No |
Amx: amoxicillin, Amc: amoxicillin-clavulanate, Cfc: cefaclor, Cfu: cefuroxime, Cfx: cefixime, Ctz: ceftazidime, Ctx: cefotaxime, Imp: imipenem, Nit: nitrofurantoin, Amk: amikacin, Cip: ciprofloxacin, Tms: trimethoprim-sulfamethoxazole, Dox: doxycycline. MDR: multiple drug resistant
Antibiotic susceptibility tests have shown that the majority of the isolates (71.4%) exhibited resistance to Trimethoprim-sulfamethoxazole (Fig. 1). Resistance to amoxicillin was seen in 61.9% of the isolates. When clavulanic acid was combined with amoxicillin (amoxicillin-clavulanic acid), the resistance was seen in only 19% of the isolates. Resistance to cephalosporins (second and third generation) was considerable and ranged from 57% to 66.7% of the isolates. However, no resistance was detected against imipenem. Multiple drug resistance (MDR), that is resistance to three antibiotics or more from different classes, was observed in 10 isolates from the 21 E. coli strains (47.6%, Table 2). MDR E. coli isolates were found to comprise 50% (5 isolates), 57.1% (4 isolates) and 25% (1 isolate) of the isolated E. coli strains from households, PH hospital and JU hospital respectively (Table 2). The prevalence of MDR E. coli in the household environment was comparable to that in hospitals (p = 0.59). Moreover, 13 strains out of 21 were phenotypically identified as ESBL-producers (61.9%). The prevalence of the phenotypically identified ESBL E. coli in households was 60% and in hospital settings were 71.4% and 50% for PH & JU Hospital respectively. blaCTXM group 1 was found in 11 strains (52%) out of the 21 strains.
Fig. 1.
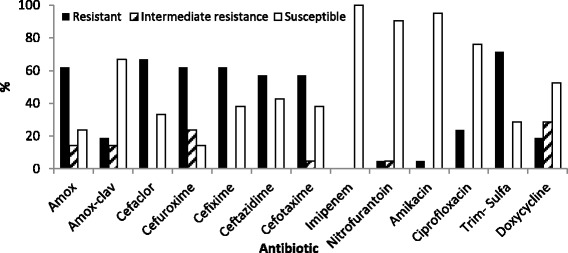
Percentage of resistant, intermediate resistant and susceptible E. coli strains versus the antibiotics tested
The results of biocides MIC against E. coli isolates are given in Table 3. All MIC values measured are well below the in-use concentrations of these biocides (Table 3). It is noteworthy that the quality control strain (ATCC 25922) and the probiotic strain (Nissle 1917) have MIC values that overlap with MIC values of hospital and household isolates. Moreover, the MIC values of the isolates collected from hospitals were found to be comparable to those collected from households (p = 0.23).
Table 3.
MIC values of the biocides tested (mg/ ml) against different isolates compared to the in-use concentration (mg/ ml). Each result is the average of five replicates ± SD
| Environment | Strain NO. | Cetrimide MIC±SD |
Chloroxylenol MIC±SD |
Ethanol MIC±SD |
Iodine MIC±SD |
|---|---|---|---|---|---|
| Household settings | A126 | 0.2 ± 0 | 0.1 ± 0 | 55 ± 12 | 0.4 ± 0.2 |
| A409-c | 0.2 ± 0 | 0.2 ± 0 | 75 ± 0 | 0.4 ± 0.1 | |
| A410-a | 0.4 ± 0.1 | 0.2 ± 0 | 55 ± 12 | 0.4 ± 0.2 | |
| A410-b | 0.4 ± 0.1 | 0.1 ± 0 | 55 ± 12 | 0.4 ± 0.1 | |
| A411 | 0.2 ± 0 | 0.1 ± 0 | 65 ± 14 | 0.4 ± 0.1 | |
| A413 | 0.2 ± 0 | 0.1 ± 0 | 75 ± 0 | 0.4 ± 0.1 | |
| A414 | 0.2 ± 0 | 0.1 ± 0 | 55 ± 12 | 0.2 ± 0.2 | |
| A801 | 0.2 ± 0 | 0.2 ± 0.1 | 85 ± 14 | 0.2 ± 0.2 | |
| A814-a | 0.2 ± 0 | 0.2 ± 0 | 50 ± 0 | 0.4 ± 0.2 | |
| A824-a | 0.2 ± 0 | 0.1 ± 0 | 55 ± 12 | 0.4 ± 0.2 | |
| PH Hospital | B107-a | 0.4 ± 0.1 | 0.2 ± 0.1 | 50 ± 0 | 0.4 ± 0.2 |
| B425-a | 0.4 ± 0.1 | 0.2 ± 0.1 | 65 ± 14 | 0.4 ± 0.1 | |
| B425-b | 0.4 ± 0.1 | 0.1 ± 0 | 50 ± 0 | 0.4 ± 0.1 | |
| B426-a | 0.2 ± 0.1 | 0.1 ± 0 | 50 ± 0 | 0.4 ± 0.2 | |
| B475-a | 0.4 ± 0.1 | 0.2 ± 0 | 65 ± 14 | 0.4 ± 0.1 | |
| B506-a | 0.2 ± 0 | 0.2 ± 0 | 55 ± 12 | 0.2 ± 0.1 | |
| B705-b | 0.2 ± 0 | 0.2 ± 0 | 60 ± 14 | 0.2 ± 0.1 | |
| JU Hospital | C604 | 0.2 ± 0 | 0.2 ± 0 | 55 ± 12 | 0.4 ± 0.1 |
| C705 | 0.4 ± 0.1 | 0.1 ± 0 | 50 ± 0 | 0.4 ± 0.1 | |
| C715-b | 0.2 ± 0 | 0.2 ± 0 | 75 ± 0 | 0.4 ± 0.1 | |
| C906 | 0.2 ± 0 | 0.1 ± 0 | 55 ± 12 | 0.2 ± 0.1 | |
| E. coli ATCC | 0.2 ± 0 | 0.1 ± 0 | 25 ± 0 | 0.4 ± 0.1 | |
| E. coli Nissle | 0.2 ± 0.1 | 0.2 ± 0.1 | 50 ± 0 | 0.4 ± 0.1 | |
| In- use concentration of the biocides | 6[22] | 4–4.8* | 390–710[22] | 2.5–5[22] | |
*Calculated from manufacturer’s instructions for use
Discussion
The pattern of antibiotic resistance detected in the studied isolates is in line with the resistance encountered in different strains of E. coli isolated from various clinical or environmental sources worldwide [14, 15]. In the last few years it has been observed that E. coli is exhibiting resistance to more antibiotic classes, hence rendering these drugs ineffective in treating its infections. On the other hand, the high prevalence of blaCTXM 1 gene is expected since this gene was found to be the most prevalent ESBL enzyme producing genes in Jordan, particularly in E. coli [16, 17].
The results showing the prevalence of MDR and ESBL producing E. coli in the environment are alarming since they reveal the dissemination of ESBL bacteria not only in hospital environment, but also in the community. The spread of ESBL-producing microorganisms is of major concern to health organizations worldwide. In 2013, the CDC published a report listing the top 18 drug resistant threats to the United States [18]. ESBL-producing Enterobacteriaceae were within the group categorized as “serious threat”. Moreover, the spread of MDR organisms has caused the WHO to issue a recent report classifying ESBL-producing Enterobacteriaceae as a “critical priority”, where effective treatment is urgently required [19]. Infections with ESBL-producing bacteria leave limited choices in antibiotic treatment, where carbapenems are the only approved drugs of choice. This effectiveness was observed in this study where all isolates were susceptible to imipenem (Fig. 1).
The biocides investigated in this study were chosen from different classes; alcohols (ethanol), quaternary ammonium compounds (cetrimide), phenolics (chloroxylenol: 4-Chloro-3,5-xylenol,) and halogens (iodine). These classes are used extensively by health care sectors and /or household settings.
A large difference between the MIC and the in-use concentrations of the studied biocides was seen. This suggests that these biocides are effective against the isolated E. coli strains, whether MDR or non-MDR. Moreover, the MICs of household isolates and hospital isolates were comparable for all the biocides tested. They were also comparable to the quality control strain and the non pathogenic strain. These findings indicate that the extensive use of biocides in hospitals didn’t increase the MIC values of biocides, i.e. they didn’t have an impact on the resistance of E. coli isolates. Furthermore, we investigated the possible association of MDR with the MIC values of the isolates. No significant difference or association was found (p > 0.05).
The occurrence of potential cross-resistance between antimicrobial agents and antibiotics is still not well understood. Some reports have shown a relationship between biocide resistance and antibiotic resistance whilst others have failed to do so [2, 20, 21]. Cole et al. [20], performed a study on 1238 (Gram-positive and Gram-negative) bacterial isolates taken from different surfaces and locations from 60 houses. They didn’t observe any cross resistance between antibiotics and biocides. On the contrary, Moken et al. [21] reported cross-resistance between pine oil disinfectant and MDR. In their study, the cross resistance was thought to be through over-expression of multiple antibiotic resistance (marA) gene. Other studies have reported the induction of some resistant mechanisms, such as over-expression of efflux pumps or a decrease in growth rates and alteration in gene expression [2]. These are believed to be part of the bacterial stress response. The scientific controversy about the presence of cross-resistance with antibiotics has led the Scientific Committee on Emerging and Newly Identified Health Risks / Directorate General for Health and Consumers in the European Commission (Directorate General for Health and Consumers, 2009) to adopt and issue an opinion about “Assessment of the Antibiotic Resistance Effects of Biocides” in 2009 [2]. In this report, they state that “there is convincing evidence that common mechanisms that confer resistance to biocides and antibiotics are present in bacteria”. However, due to the limitations in identifying and characterizing cross-resistance in the targeted environment (in situ), the report concluded that more research is needed in this field.
Conclusion
E. coli isolates from household and hospital environments showed high resistance rates to different classes of antibiotics without any significant differences between the two environments. For both groups, many E. coli isolates showed antibiotic multiple resistant patterns. ESBL-producing isolates were detected in both environments. E. coli isolates from both environments showed comparable MIC values of four of the widely used biocides, although the use of biocides in hospitals is more extensive than in households. Data generated from this study failed to show an association between antibiotic resistance and biocide resistance in E. coli isolates.
Acknowledgements
The authors would like to thank Dr. Phillip Collier for revising the manuscript and providing valuable comments.
Funding
This research was funded by the Deanship of Academic Research and Quality Assurance, The University of Jordan.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
RH supervised the work, directed the research, interpreted the results and wrote the manuscript. BG performed the work and the literature survey. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Health Care-Associated Infections. Office of disease prevention and health promotion. U.S. Department of health and human services. 2017 https://health.gov/hcq/prevent-hai.asp. Accessed 16 Sep 2017.
- 2.Assessment of the antibiotic resistance effects of biocides. Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR). Directorate general for health and consumers. European commission. 2009.
- 3.D’Costa M, King C, Kalan L, Morar M, Sung W, Schwarz C, Froese D, Zazula, G, Calmels F, Debruyne R, Golding G, Poinar H, Wright G. Antibiotic resistance is ancient. Nature 2011; 477,457–461. [DOI] [PubMed]
- 4.Hansen L, Sorensen S, Jorgensen H, Jensen L. The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb Drug Resist. 2005;11:378–382. doi: 10.1089/mdr.2005.11.378. [DOI] [PubMed] [Google Scholar]
- 5.Karatzas K, Webber M, Jorgensen F, Woodward M, Piddock L, Humphrey T. Prolonged treatment of Salmonella enteric serovar typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J Antimicrob Chemother. 2007;60:947–955. doi: 10.1093/jac/dkm314. [DOI] [PubMed] [Google Scholar]
- 6.McBain A, Ledder R, Sreenivasan P, Gilbert P. Selection for high-level resistance by chronic triclosan exposure is not universal. J Antimicrob Chemother. 2004;53:772–777. doi: 10.1093/jac/dkh168. [DOI] [PubMed] [Google Scholar]
- 7.Ledder R, Gilbert P, Willis C, McBain A. Effects of chronic triclosan exposure upon the antimicrobial susceptibility of 40 ex-situ environmental and human isolates. J Appl Microbiol. 2006;100:1132–1140. doi: 10.1111/j.1365-2672.2006.02811.x. [DOI] [PubMed] [Google Scholar]
- 8.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nature Rev Microbiol. 2010;8(3):207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 9.Ekrami A, Kayedani A, Jahangir M, Kalantar E, Jalali M. Isolation of common aerobic bacterial pathogens from the environment of seven hospitals, Ahvaz, Iran. Jundishapur J Microbiol. 2011;4(2):75–82. [Google Scholar]
- 10.Clinical and Laboratory Standard Institute (CLSI) M 100S performance standards for antimicrobial susceptibility testing. 26 2016. [Google Scholar]
- 11.Scaldaferri F, Gerardi V, Mangiola F, Lopetuso L, Pizzoferrato M, Petito V, et al. Role and mechanisms of action of Escherichia coli Nissle 1917 in the maintenance of remission in ulcerative colitis patients: an update. World J Gastroenterol. 2016;22(24):5505–5511. doi: 10.3748/wjg.v22.i24.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsen H, Lin C, Chi W. Development and use of 16S rRNA gene targeted PCR primersfor the identification of Escherichia coli cells in water. J Appl Microbiol. 1998;85:554–560. doi: 10.1046/j.1365-2672.1998.853535.x. [DOI] [PubMed] [Google Scholar]
- 13.Mirzaee M, Owlia P, Mansouri S. Distribution of CTX-M β-lactamase genes among Escherichia coli strains isolated from patients in Iran. Lab Medicine. 2009;40(12):724–727. doi: 10.1309/LMUUWBHMZEDYTBW5. [DOI] [Google Scholar]
- 14.van der Donk CF, van de Bovenkamp JH, De Brauwer E, De Mol P, Feldhoff K, Kalka-Moll W, et al. Antimicrobial resistance and spread of multi drug resistant Escherichia coli isolates collected from nine urology services in the Euregion Meuse-Rhine. PLoS One. 2012; 10.1371/journal.pone.0047707. [DOI] [PMC free article] [PubMed]
- 15.Poonia S, Singh T, Tsering D. Antibiotic susceptibility profile of bacteria isolated from natural sources of water from rural areas of East Sikkim. Indian J Community Med. 2014;39(3):156–160. doi: 10.4103/0970-0218.137152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abu Salah M, Badran E, Shehabi A. High incidence of multidrug resistant Escherichia coli producing CTX-M-type ESBLs colonizing the intestine of Jordanian infants. Int Arab J Antimicrob Agents. 2013;3(4:3)
- 17.Badran EF, Qamer Din RA, Shehabi AA. Low intestinal colonization of Escherichia coli clone ST131 producing CTX-M-15 in Jordanian infants. J Med Microbiol. 2016;65(2):137–141. doi: 10.1099/jmm.0.000210. [DOI] [PubMed] [Google Scholar]
- 18.Biggest Threats, Antibiotic / Antimicrobial Resistance. Centers for Disease Control and Prevention (CDC- USA). 2017; https://www.cdc.gov/drugresistance/biggest_threats.html (access date1/9/2017).
- 19.Lawe-Davies O, Bennett S. WHO publishes list of bacteria for which new antibiotics are urgently needed, World Health Organization, Geneva. 2017; http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ (access date, 1/9/2017).
- 20.Cole E, Addison R, Rubino J, Lese K, Dulaney P, Newell M, et al. Investigation of antibiotic and antibacterial agent cross-resistance in target bacteria from homes of antibacterial product users and nonusers. J Appl Microbiol. 2003;95:664–676. doi: 10.1046/j.1365-2672.2003.02022.x. [DOI] [PubMed] [Google Scholar]
- 21.Moken M, McMurry L, Levy S. Selection of multiple-antibiotic-resistant (mar) mutants of Escherichia coli by using the disinfectant pine oil: roles of the mar and acrAB loci. Antimicrob Agents Chemother. 1997;41(12):2770–2772. doi: 10.1128/aac.41.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Adham I, Haddadin R, Collier P. Types of microbicidal and microbistatic agents. In: Fraise AP, Maillard J, Satter SA, editors. Russell, Hugo & Ayliffe's Principles and practice of disinfection, preservation and sterilization 5th edition UK: Wiley Blackwell. 2013. pp. 5–57. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


