Abstract
Type III interferons (IFN-λs) are the most recently found members of the IFN cytokine family and engage IFNLR1 and IL10R2 receptor subunits to activate innate responses against viruses. We have identified IFN-λs as critical instructors of antifungal neutrophil responses. Using Aspergillus fumigatus (Af) as a model to study antifungal immune responses, we found that depletion of CCR2+ monocytes compromised the ability of neutrophils to control invasive fungal growth. Using an unbiased approach, we identified type I and III IFNs as critical regulators of the interplay between monocytes and neutrophils responding to Af. We found that CCR2+ monocytes are an important early source of type I IFNs that prime optimal expression of IFN-λ. Type III IFNs act directly on neutrophils to activate their antifungal response, and mice with neutrophil-specific deletion of IFNLR1 succumb to invasive aspergillosis. Dysfunctional neutrophil responses in CCR2-depleted mice were rescued by adoptive transfer of pulmonary CCR2+ monocytes or by exogenous administration of IFN-α and IFN-λ. Thus, CCR2+ monocytes promote optimal activation of antifungal neutrophils by initiating a coordinated IFN response. We have identified type III IFNs as critical regulators of neutrophil activation and type I IFNs as early stimulators of IFN-λ expression.
Introduction
Cytokines are essential for the activation of host immunity against multiple classes of pathogens including fungi. Type II interferon (IFN-γ) is the prototypical cytokine known to activate the effector mechanisms of myeloid cells for pathogen eradication (1). The type I interferon family is critical for defense against viruses, and is comprised of multiple subtypes including a single IFN-β gene and various IFN-α genes that signal via a shared heterodimeric receptor, IFNAR1/IFNAR2, to activate essential antiviral responses (2). In addition to activating antiviral immunity, type I and II IFNs have been found to promote host defense against non-viral pathogens including fungi (3, 4). The more recently discovered family of type III IFNs is comprised of four lambda genes in humans and two functional genes (IFN-λ2 and IFN-λ3) in mice that signal via a distinct heterodimeric receptor to activate largely overlapping antiviral transcriptional responses as IFNAR (5–7). The evolution of two distinct cytokine families and receptor systems to induce overlapping antiviral immunity has prompted investigators to search for potentially unique functions for IFN-λs. Recent studies suggest that indeed IFN-λs have distinct contributions to antiviral immunity at mucosal sites (8–13). Thus, although type III IFNs were originally thought to act in parallel to type I IFNs, it is increasingly appreciated that they cooperate to activate compartmentalized antiviral responses (14). The potential involvement of type III IFNs in the activation of immunity against pathogens other than viruses remains to be fully explored.
Invasive fungal infections (IFI) are a significant cause of morbidity and mortality to diverse populations throughout the world, and currently available antifungal drugs are often ineffective in preventing the high mortality associated with these infections (15). Aspergillus fumigatus (Af), is the etiological agent of over 90% of invasive aspergillosis (IA) cases, one of the primary IFI of global concern (15, 16). The increased level of drug resistance observed in Aspergillus clinical isolates further highlights the urgent need for the development of new therapeutic options (17). A better understanding of immune mechanisms of host defense against Af will facilitate the identification of potential new pathways that could be targeted by immune-based therapies to work alongside antifungal drugs to improve patient outcomes. Defense against Af is critically dependent on myeloid cells, and low numbers of leukocytes renders patients at high risk to develop IA (18). Neutrophils are particularly important for Af elimination and employ reactive oxygen species (ROS) as one of their primary effector mechanisms (19). The importance of ROS in defense against Af is further underscored by the susceptibility to fungal infections in patients and mice with genetic defects in NADPH oxidase (20, 21).
We previously reported that in addition to neutrophils, CCR2+monocytes are also essential innate cells in pulmonary innate defense against Af (22). Depletion of CCR2+monocytes and their derivative cells (CCR2-depleted mice) impaired the activation of a protective antifungal inflammatory response, and affected the production of multiple cytokines (22, 23). We found that neutrophils from CCR2-depleted mice were less capable of killing Af suggesting that neutrophil antifungal activity might be regulated by exogenous factors that require an intact CCR2+ compartment (22). Here, we employed an unbiased, systems biology approach to uncover the basis of neutrophil dysfunction in CCR2-depleted mice. With this approach, we identified an unexpected essential role for type III IFN in the regulation of antifungal neutrophil function.
Results
Neutrophils from CCR2-depleted mice have an impaired transcriptional response of IFN-inducible genes
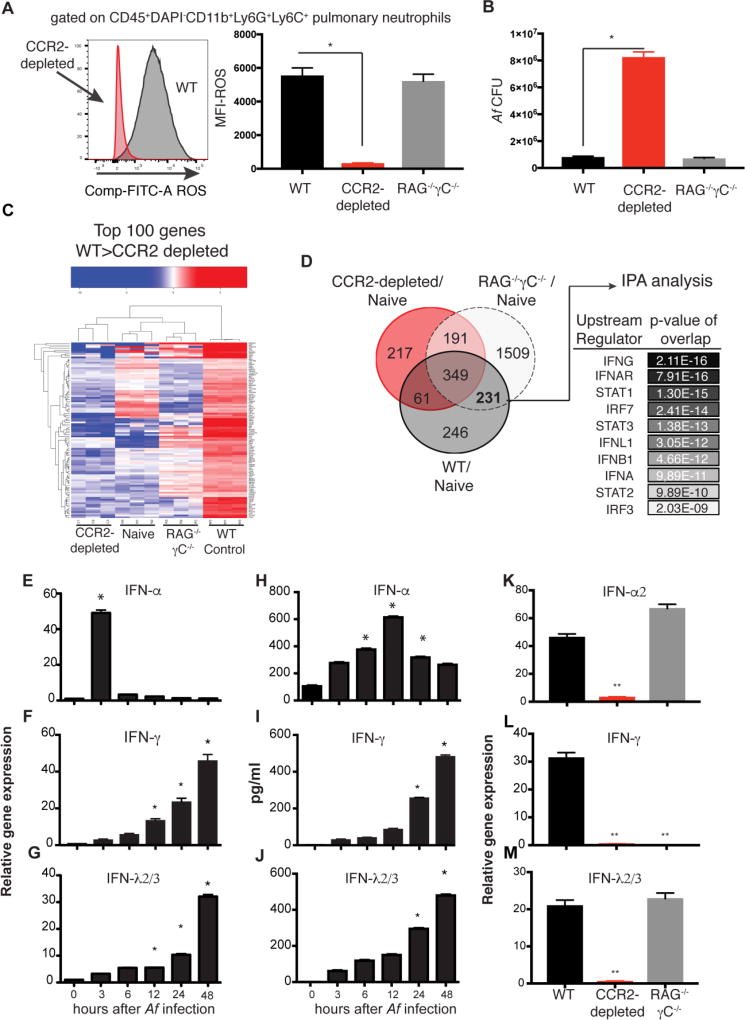
In previous studies, we determined that neutrophils from CCR2-depleted mice had a diminished capacity to inactivate fungal conidia. Consistent with their diminished capacity to kill Af (22), we find that neutrophils isolated from CCR2-depleted mice have an impaired capacity to generate ROS (Fig 1A). In contrast, neutrophils isolated from mice lacking the entire lymphoid lineage (RAG−/−γC−/−) retained their capacity to generate ROS (Fig 1A). Normal ROS generation in RAG−/−γC−/− mice was accompanied by normal control of Af infection in the lung (Fig 1B and Fig S1A–S1B). Thus, signals derived from non-lymphoid cells are sufficient to activate antifungal neutrophils, but require an intact CCR2+ compartment. In order to identify possible signals that mediate antifungal neutrophil activation, we analyzed the transcriptional response of pulmonary neutrophils after Af infection, and examined how CCR2+ cell depletion affected that response. As reference populations, we included pulmonary neutrophils from uninfected mice and neutrophils from lymphoid-deficient mice. Neutrophils from Af-infected mice upregulated the expression of 887 genes as compared to pulmonary naïve neutrophils (Fig S2A). A portion of this transcriptional response to Af was depressed in neutrophils isolated from CCR2-depleted mice (Fig 1C). We further looked for genes that were commonly upregulated in neutrophils from WT and RAG−/−γc−/− mice, but not in CCR2-depleted mice, and identified 231 genes that met this criteria (Fig 1D). We reasoned that this core response was critical to preserved antifungal effector function of WT and RAG−/−γC−/− neutrophils, but was altered in dysfunctional neutrophils from CCR2-depleted mice (Fig 1C). Ingenuity Pathway Analysis of upstream regulators of the common 231 genes expressed in functional neutrophils predicted multiple factors involved in the induction of an interferon response as potential regulators (Fig 1D). Consistently, multiple IFN-inducible genes had lower expression in neutrophils from CCR2-depleted mice than in other neutrophil populations examined (Fig S2B). Altogether, our transcriptional analysis suggested an impaired interferon-inducible response as the possible basis for the dysfunction of neutrophils in CCR2-depleted mice. To further explore the possible involvement of IFNs in the antifungal response to Af, we performed a kinetic analysis of IFN expression at different times after infection. IFNs α, γ and λ2/3 were all upregulated in the lung after Af infection, but with different kinetics (Fig 1E–1J). Type I IFNs were rapidly induced within the first few hours of infection (Fig 1E and 1H) while IFN-γ and IFN-λ steadily increased over the next 48 hours after infection (Fig 1F–1G and 1I–1J). Depletion of CCR2+ cells affected the expression of all IFNs tested while lymphoid-deficient mice had undetectable expression of IFN-γ, but retained intact expression of type I and III IFNs (Fig 1K–1M). Thus, impaired IFN-γ expression did not correlate with susceptibility to IA. Consistently, IFNGR−/− mice were not more susceptible to IA as compared to control mice (Fig S1C–S1D). In aggregate, these findings suggest that impaired type I and III IFN production in CCR2-depleted could be a relevant, underlying reason for dysfunctional neutrophil responses in these mice.
Figure 1. Dysfunctional antifungal neutrophils have impaired expression of IFN-inducible genes.
(A) Reactive oxygen species (ROS) production by airway infiltrating neutrophils isolated from CCR2-depleted mice (red histogram) or control littermates (gray histogram) 48 hours after infection. Graph shows mean ± SEM of ROS MFI for 5 mice per group assayed by FACS as shown. (B)Af pulmonary fungal burden 48 hour after infection. (C) Differential gene expression as assessed by RNA-seq of pulmonary neutrophils isolated from uninfected controls (naïve) or mice infected with Af for 48 hours. Heat map depicts the top 100 genes expressed at FC>2.5 in control neutrophils > neutrophils from CCR2-depleted mice. (D) Venn diagram of differentially expressed genes. Predicted upstream regulators of the 231 genes as determined with Ingenuity Pathway Analysis software. (E–J) Kinetics of IFN expression in the lung of mice at different times after Af infection. E–G shows gene expression as determined by qRT-PCR using TaqMan probes. Data shown is mean ± SEM of 5 mice per time point. H–J graphs show mean ± SEM of each cytokine as measured by ELISA in 5 mice per time point. (K–M) Pulmonary qRT-PCR of IFN gene expression at 3 (K) or 48 hours after Af infection (L–M). Throughout the figure, *p<0.05, **p<0.01 as calculated by Mann-Whitney non-parametric test of experimental group relative to control using Prism software.
Both Type I and III IFN signaling are essential for protection against IA
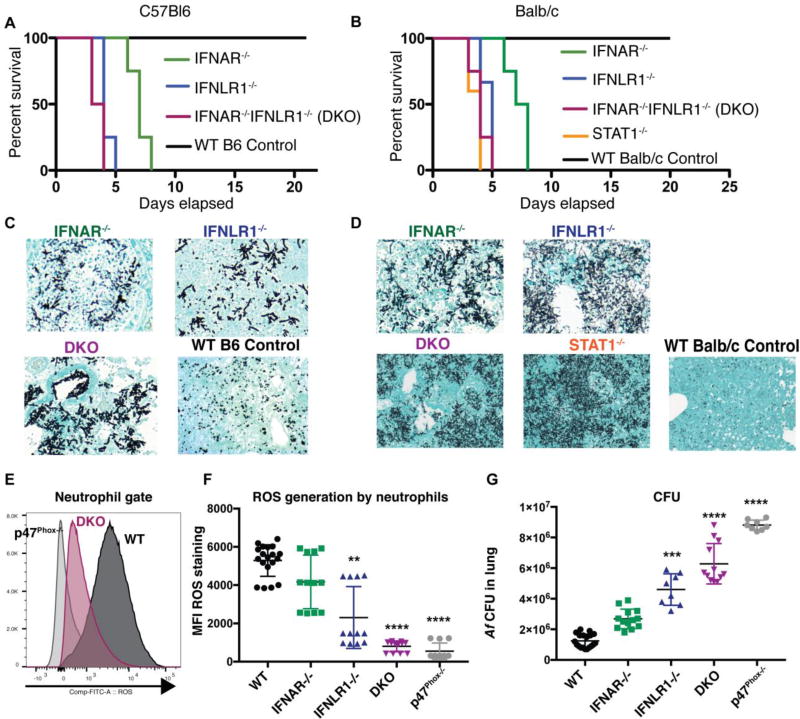
On the basis of our observations, we hypothesized that type I and III IFNs could be acting as critical activators of antifungal neutrophils. To test this hypothesis, we challenged mice with genetic deficiencies in IFNAR, IFNLR1, STAT1 or double deficient in IFNAR and IFNLR1 with Af. Mice with defective expression of type I, type III IFN receptor, or doubly deficient were susceptible to infection with Af (Fig 2A and 2B), and were unable to control fungal growth in the lung (Fig 2C–2D). We examined various parameters of inflammation in IFN receptor deficient mice, including cell recruitment and neutrophil function (Fig S3A–S3F). We observed a cumulative effect of IFN receptor deficiency on the global recruitment of CD45+ cells to the lung of Af infected mice (Fig S3A) with mice with double IFNR deficiency (IFNAR−/−IFNLR1−/−; DKO) displaying more significant reductions in the number of recruited neutrophils (Fig S3B) and monocytes (Fig S3C). Functional measurements of inflammation showed some reductions in TNF production, lower levels of MPO, and diminished generation of NETs (Fig S3D–S3F). Thus, defective IFN signaling had effects on several aspects of what are considered protective parameters in antifungal immunity. DKO mice had a decrease in ROS that was similar in magnitude to mice genetically deficient in NADPH oxidase (p47Phox−/−) (Fig 2E–2F). Defective ROS generation in IFNR deficient mice correlated with a rapid failure to control overall fungal growth in the lung (Fig 2G). Similarly, ROS-deficient mice (p47Phox−/−) developed IA and rapidly succumbed to infection (Fig S4A–S4B). Mortality in p47Phox−/− mice was evident even in the presence of normal inflammatory cytokine production (Fig S4E–S4F) and normal recruitment of monocytes (Fig S4C) and neutrophils (Fig S4D). These findings are consistent with previous work demonstrating the essential role of ROS in defense against IA(24–31). We thus hypothesize that the most likely mechanism of susceptibility to IA in IFNR deficient mice is impaired control of fungal growth due to defective antifungal neutrophil responses, particularly the generation of ROS.
Figure 2. Type I and type III IFN receptor signaling are both essential for protection against invasive aspergillosis.
IFNAR−/− (green lines or symbols), IFNLR1−/− (blue lines or symbols), STAT1−/− (orange line), double deficient IFNAR−/−IFNLR1−/− (purple line or symbols) and wild type control (black lines or symbols) B6 (A) and WT Balb/c (B) animals were challenged with 8 × 107 CEA10 Af conidia and monitored for survival. (A–B) Kaplan-Meier survival plot for 10 mice per group analyzed in two independent experiments. (C–D) Representative images of Gomori ammoniacal silver-stained lung sections captured at 40X magnification. (E) Representative FACS plot of ROS detection in neutrophils derived from WT (dark gray histogram), IFNAR−/−IFNLR1−/−(purple) and p47Phox−/− (light gray histogram) mice. (F) Mean fluorescence intensity of ROS generation by airway infiltrating neutrophils at 48 hours after Af infection as analyzed in E. (G) Pulmonary fungal burden at 48 hours after Af infection (F–G) Each symbol represents one mouse. Data shown is cumulative of three independent experiments. **p< 0.01, **** p<0.0001 values shown in F and G were calculated by Kruskall-Wallis non-parametric test for multiple comparisons for each knockout group compared to WT control. Statistical analysis was done with Prism software.
Optimal type III IFN production requires intact type I and III IFNR expression
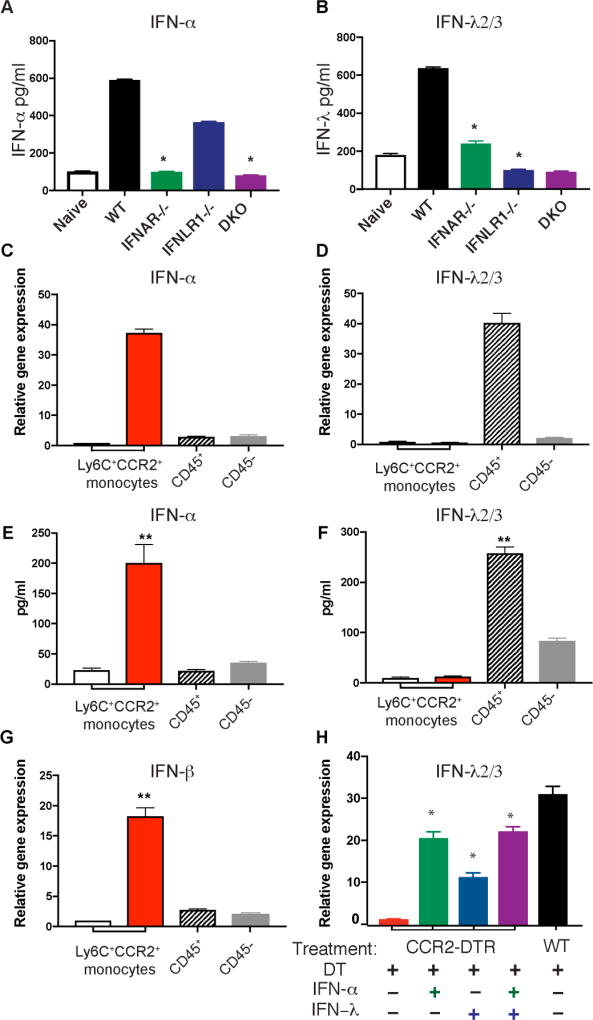
Because mice deficient in either type I or III IFN receptor expression were susceptible to IA, we set out to define the impact of these deficiencies on the production of their ligands. IFNAR−/− mice had diminished production of type I and III IFNs (Fig 3A–3B). IFNLR1−/− mice also had defective production of IFN-λ (Fig 3B). Thus, in the context of Af infection, IFN-λ production is primed by type I IFN and optimal expression requires intact type I and III IFN signaling. These findings are consistent with previous work in viral infections where IFN-λ expression can be induced by stimulation with IFN-α or -λ (32). Together, our findings suggest that type I and III IFNs are both required for antifungal defense, and act at distinct times after infection in a coordinated fashion to induce optimal production of ROS by neutrophils.
Figure 3. CCR2+ monocytes are a relevant source of type I IFN in response to Af.
(A–B) IFNAR−/− (green), IFNLR1−/− (blue), double deficient IFNAR−/−IFNLR1−/− (DKO; purple), and wild type control (black) B6 animals were challenged with 4 × 107 CEA10 Af conidia, and examined for type I IFN expression at 12 hours after infection (A) or type III IFN at 48 hours after infection (B) by ELISA. (C–G) CCR2+Ly6C+ monocytes (red bars), CD45+ cells depleted of monocytes (hatched bars), and CD45 pulmonary cells (gray bars) were isolated form the lung of CCR2-GFP reporter mice at 3 hours (C, E and G) or 48 hours (D and F) after infection with Af. Pulmonary, CCR2+Ly6C+ monocytes were also isolated from naïve CCR2-GFP reporter mice (white bars). All samples were examined for IFN transcription (C, D and G) or secretion of IFN proteins after overnight ex vivo culture (E and F). (H) Endogenous transcription of type III IFN was examined by qRT-PCR in RNA samples isolated from the lungs of CCR2-depleted mice that were left untreated (red bars) and in CCR2-depleted mice that were treated with 1 µg of IFN-α (green bars), 1 µg of IFN-λ (blue bars), or 1 µg of IFN-α and 1 µg of IFN-λ (purple bars). Responses in CCR2+ competent littermates (black bars) were used as positive controls. Data shown is mean ± SEM for four mice per group and is for one experiment representative of two. *p<0.05 as calculated by Mann-Whitney non-parametric test of experimental group relative to WT control mice using Prism software.
CCR2+ Monocytes are an important source of Type I IFN
Given that CCR2-depleted mice showed impaired production of type I and III IFNs (Fig 1K and 1M), we hypothesized that CCR2+monocytes were acting as a direct source of these cytokines. To test this hypothesis, we isolated CCR2+monocytes from the lung of naïve, control mice and from Af-infected animals at the peak of type I and III IFN production (3 and 48 hours after infection respectively, Fig 1E and 1G). Purified cells were examined for IFN transcription at the time of isolation as well as for their capacity to secrete IFN proteins ex vivo. As additional control populations, we sorted CD45+cells depleted of CCR2+Ly6C+ cells and CD45− cells. We also purified CCR2+Ly6C+ monocytes from the lung of naïve mice. We observed that CCR2+monocytes were a significant source of type I IFNs after Af infection (Fig 3C, 3E, and 3G). In contrast, we did not detect much IFN-λ production by CCR2+monocytes (Fig 3D and 3F). We observed that IFN-λ was produced by CD45+ cells depleted of CCR2+monocytes, and to a lesser extent by CD45− pulmonary cells (Fig 3D ad 3F). In aggregate, our observations suggest that CCR2+monocytes are an important source of type I IFN (Fig 3C,3E, and 3G). In turn, type I IFNs are required for optimal production of IFN-λ (Fig 3B) by CD45+ and CD45− pulmonary cells (Fig 3D and 3F). On the basis of our findings, we hypothesize that defective production of type I and III IFNs in CCR2-depleted mice (Fig 1K and 1M) is likely due to a direct contribution of CCR2+monocytes as source of type I IFN (Fig 3C–3G) that is then necessary for optimal induction of type III IFN by other cells (Fig 3B,3D, and 3F). Consistent with this idea, we found that treatment of CCR2-depleted mice with recombinant IFN-α induced significant transcription of IFN-λ in the lung of recipient mice (Fig 3H). Treatment with recombinant IFN-λ protein also induced higher transcription of IFN-λ in CCR2-depleted recipients. These findings further support the idea that in the context of Af, type III IFN expression is primed by early production of type I IFN. Type III IFN further amplifies its own production. Thus, in response to Af, optimal type III IFN production requires both type I and III signaling.
Neutrophils are an important target population for the effects of both type I and III IFNs
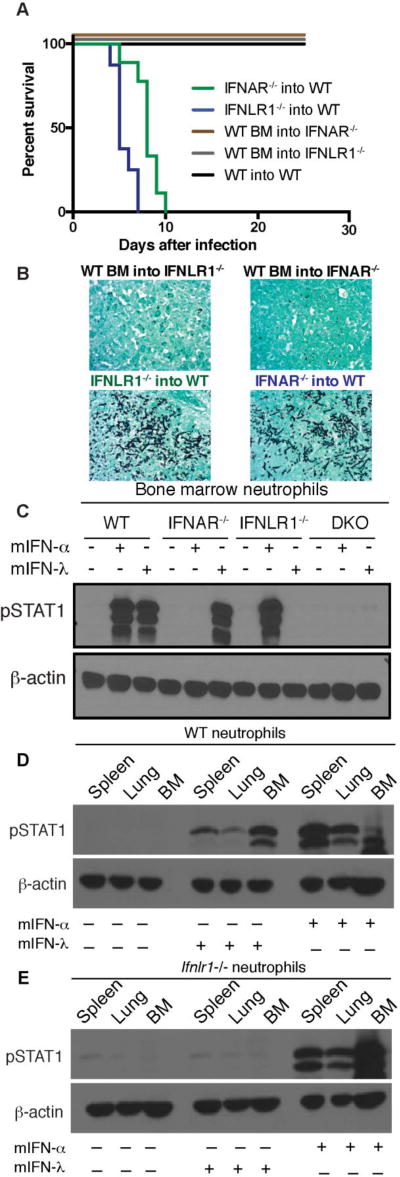
Previous studies in models of viral infection have demonstrated that intestinal and pulmonary epithelial cells are important targets of the effects of IFN-λ while type I IFN is broadly expressed by hematopoietic cells. Thus, in order to decipher the potentially distinct activities of IFN receptors on hematopoietic and non-hematopoietic cells during Af infection, we generated bone marrow chimeric mice to limit deficient receptor expression to host, non-hematopoietic, irradiation-resistant cells or to donor, bone marrow-derived cells. Mice with defective IFNAR or IFNLR1 expression on hematopoietic cells succumbed to infection with Af due to the development of IA (Fig 4A–4B). In contrast, control BM chimeric mice or mice with defective IFNAR or IFNLR1 expression selectively on non-hematopoietic cells were protected from infection (Fig 4A–4B). Our findings thus suggest that defense against IA requires IFNAR and IFNLR1 expression on hematopoietic cells.
Figure 4. Type I and type III IFN receptor expression on hematopoietic cells is required for protection against invasive aspergillosis.
(A–B) Lethally irradiated recipients were reconstituted with donor bone marrow for 8 weeks before infection with 8 × 107 CEA10 Af conidia. (A) Kaplan-Meier survival plot for 8 mice per group analyzed in two independent experiments. (B) Representative images of Gomori ammoniacal silver-stained lung sections captured at 40X magnification. (C) Western Blot analysis of STAT1 phosphorylation at 15 min after treatment with 100 ng of recombinant murine IFN-α2 or mIFN-λ2 as indicated. Bone marrow neutrophils from each experimental group were FACS sorted >99% purity prior to in vitro treatment with IFNs. (D–E) Neutrophils from bone marrow, spleen, or lung of WT mice (D) and IFNLR1−/− mice (E) were FACS sorted >99% purity prior to in vitro treatment with IFNs and examined for responsiveness by Western Blot analysis of STAT1 phosphorylation. Neutrophils were sorted as Live CD45+CD11b+Ly6CintLy6G+.
IFNAR is broadly expressed on hematopoietic cells, but expression of IFNLR1 is more restricted. Although IFNLR1 has been found to be expressed primarily by epithelial cells (33), recent studies have indicated that neutrophils can also respond to IFN-λ (10, 12, 34). Consistently, purified bone marrow neutrophils were equally responsive to treatment with IFN-α or IFN-λ (Fig 4C). As expected, neutrophils defective in IFNAR expression lost their response to IFN-α but retained responsiveness to IFN-λ, IFNLR1−/− cells lost their response to IFN-λ but retained responsiveness to IFN-α while IFNAR−/−IFNLR1−/− (DKO) cells lost responsiveness to treatment with either IFN (Fig 4C). Neutrophils isolated from the spleen and lung continued to respond to both type I and III IFNs (Fig 4D) while neutrophils from IFNLR1−/− mice did not respond to IFN-λ as expected (Fig 4E). Together, these findings suggest that hematopoietic cells are the relevant targets of IFN-α and IFN-λ function in order to protect mice against IA, and that, neutrophils are particularly important targets of both IFNs within hematopoietic cells.
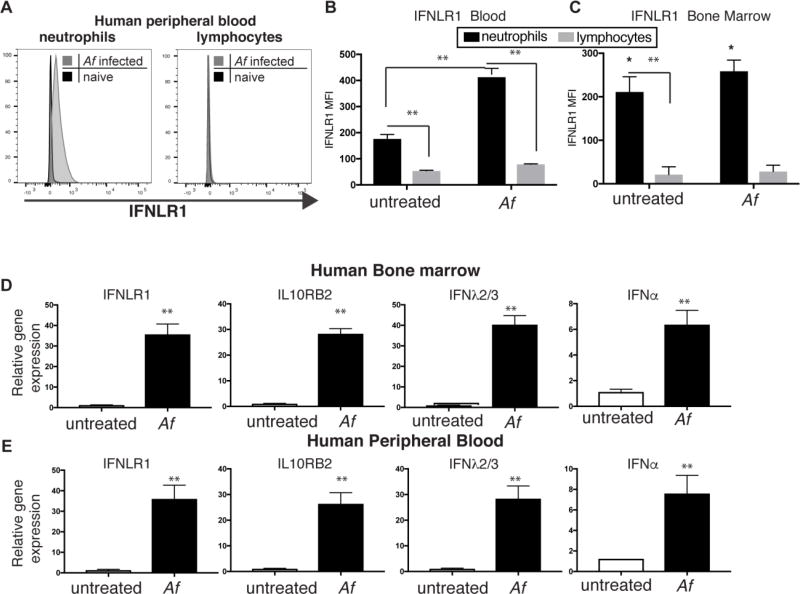
Human neutrophils also express IFNLR1
Our findings thus far indicate that type I and III IFNs are critical cytokines in defense against IA, and that neutrophils are important targets of their effects. We thus wanted to determine whether human neutrophils also expressed IFNLR1 receptor. We tested the expression of the unique IFNLR1 chain in peripheral blood and bone marrow cells by flow cytometry. The antibody used was selected after extensive screening of multiple antibody clones using cell lines with enforced expression of hIFNLR1 (Fig S5). We observed that human neutrophils from peripheral blood or bone marrow expressed IFNLR1 (Fig 5A–5C), and did so at higher levels than lymphocytes (Fig 5B–5C). Stimulation with Af induced further upregulation of IFNLR1 expression in human neutrophils but did not affect the minimal expression seen in lymphocytes (Fig 5B–5C). Expression of IFN-α and IFN-λ was also stimulated by Af infection in human bone marrow and peripheral blood cells (Fig 5D and 5E). Altogether, these findings suggest that human and murine neutrophils might be important targets of the biological effects of both type I and type III IFNs, and that these cytokines are produced in response to Af stimulation.
Figure 5. Human cells produce type I and III IFN upon Af stimulation.
(A) Representative FACS plot of IFNLR1 expression in gated neutrophils or lymphocytes in human peripheral blood cells without treatment or 6 hours after culture with Af. (B–C) IFNLR1 expression as measured by MFI in each gated population in human peripheral blood (B) or bone marrow samples (C). (D–E) qRT-PCR of relative gene expression of IFN genes and type III IFN receptor in RNA isolated from human peripheral blood (E) or bone marrow samples (D). Data shown is mean ± SEM of 5 (B and E) or 4 (C and D) individual donors. **p<0.01 as calculated by paired t-test using Prism software.
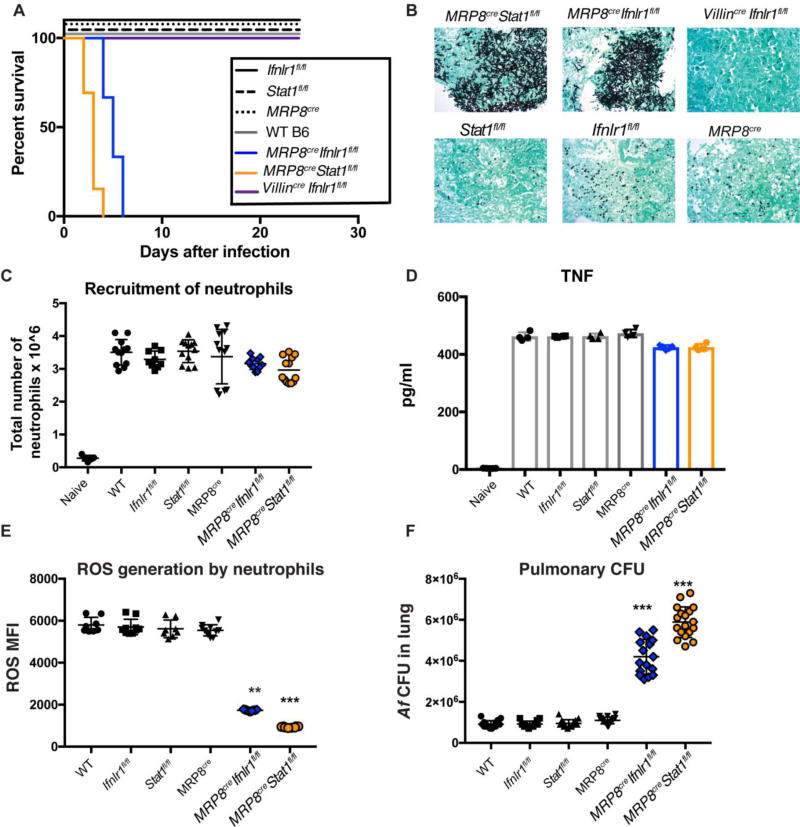
Neutrophil-specific deletion of IFNLR1 and STAT1 renders mice susceptible to IA
In aggregate, our observations suggest that type I and III IFNs are important regulators of antifungal responses (Fig 2), and that in their absence, neutrophils show dysfunctional antifungal responses and impaired ability to generate ROS (Fig 1 and Fig 2). We also find that neutrophils are directly responsive to type III IFN, and that hematopoietic restricted expression of these receptors is sufficient to protect mice against IA (Fig 4). Based on our aggregate observations, we thus hypothesized that neutrophils are the critical hematopoietic cell (Fig 4) that require expression of IFNLR1 and overall IFN responsiveness (STAT1 expression) in order to mount an effective defense against IA. To further examine the potential direct effect of type I and III IFNs on neutrophils, we targeted IFNLR1 or STAT1 gene expression on granulocytes by crossing MRP8cre mice with Ifnlr1fl/fl or Stat1fl/fl mice. Previous studies have shown that MRP8cre targets gene excision efficiently in neutrophils (35, 36). Consistently, IFNLR1 and STAT1 were efficiently targeted in neutrophils of MRP8cre Ifnlr1fl/fl and MRP8cre Stat1fl/fl mice, respectively (Fig S6). MRP8cre Ifnlr1fl/fl and MRP8cre Stat1fl/fl mice were susceptible to Af infection and developed IA while control MRP8cre, Ifnlr1fl/fl, and Stat1fl/fl were fully resistant to infection similar to control wild type mice (Fig 6A–6B). Selective removal of IFNLR1 on intestinal epithelial cells in Villincre Ifnlr1fl/fl mice also had no effect in susceptibility to IA (Fig 6A–6B). Impaired expression of IFNLR1 or STAT1 on neutrophils did not affect their recruitment to the lung (Fig 6C) or the global production of TNF (Fig 6D). In contrast, defective expression of IFNLR1 or STAT1 on neutrophils was accompanied by diminished capacity to produce ROS by these cells (Fig 6E), and overall defective control of Af growth in the lung (Fig 6B and 6F). These findings are reminiscent of the phenotype of p47Phox−/− mice (Fig S4). Altogether, our findings suggest that optimal neutrophil ROS generation and antifungal activity requires expression of IFNLR1 and STAT1 by these cells.
Fig. 6. Mice with neutrophil-specific deletion of IFNLR1 or STAT1 succumb to invasive aspergillosis.
Conditional gene targeting in neutrophils was achieved by crossing Ifnlr1fl/fl and Stat1fl/fl mice with MRP8cre. Gene targeted animals and control mice were infected with 8 × 107 CEA10 Af conidia. Control groups were WT B6 mice, Ifnlr1fl/fl, Stat1fl/fl, and MRP8cre(A) Kaplan-Meier survival plot for 10 mice per group analyzed in two independent experiments. (B) Representative images of Gomori ammoniacal silver-stained lung sections captured at 40X magnification. (C) Neutrophil recruitment to the lung of infected mice at 48 hours after infection with 4 × 107 CEA10 Af conidia. Each symbol represents one mouse. Data is cumulative of two independent experiments. (D) Pulmonary levels of TNF as measured by ELISA. (E–F) ROS generation by airway infiltrating neutrophils (E) and pulmonary fungal burden (F) 48 hours after Af infection. Each symbol represents one mouse. Data shown is cumulative of two independent experiments. Statistical analysis was done with Kruskall-Wallis, non-parametric test for multiple comparisons using Prism software. *p<0.05, ***p<0.001 for each labeled sample as compared to WT mice.
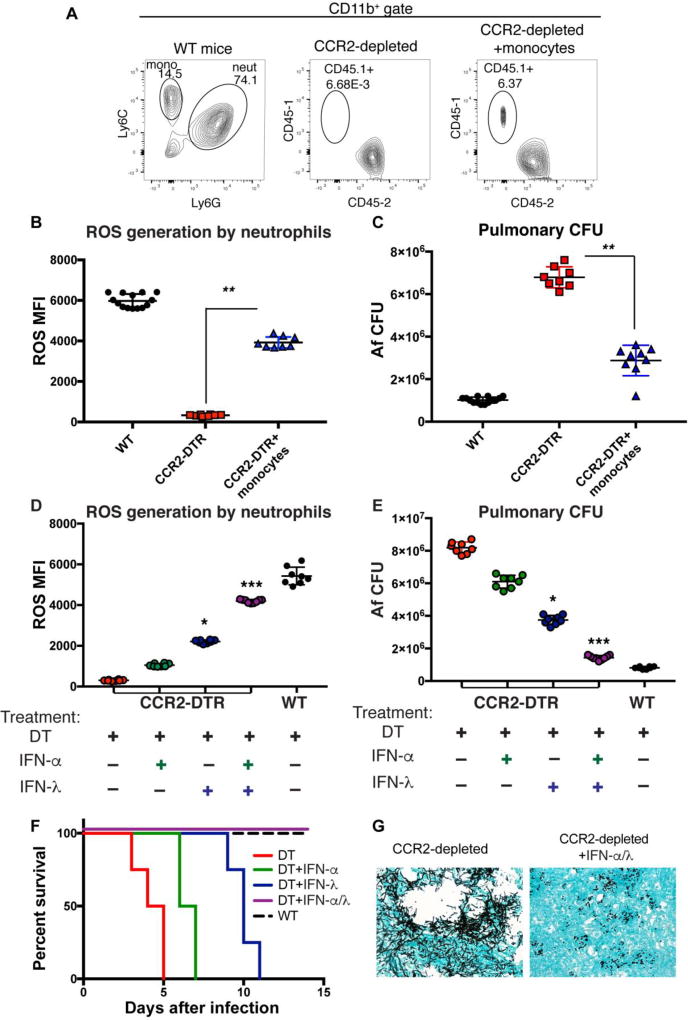
Dysfunctional neutrophil responses in CCR2-depleted mice can be rescued by adoptive transfer of CCR2+ monocytes or treatment with recombinant type I and III IFNs
On the basis of our aggregate findings, we hypothesized that limited availability of type I and type III IFNs in CCR2-depleted animals (Fig 1K and 1M) underlies the dysfunction of antifungal neutrophils in these mice (Fig 1A–1B). Impaired IFN responses in CCR2-depleted mice are likely due to the important contribution of CCR2+monocytes as a source of type I IFN (Fig 3C, 3E, and 3G), which acts on other cells for optimal induction of IFN-λ (Fig 3B). To test this hypothesis, we employed two distinct experimental approaches: 1) we performed adoptive transfer of pulmonary CCR2+monocytes isolated from the lung of mice infected with Af and 2) we tested the capacity of recombinant IFNs to rescue CCR2-depleted mice. Adoptively transferred CD45.1+CCR2-GFP+monocytes were able to effectively migrate to the lung of CD45.2+CCR2-depleted mice (Fig 7A). We isolated pulmonary monocytes from donor CCR2-GFP+CD45.1+ congenic mice that were infected for 3 hours with Af; a time during which CCR2+monocytes are actively transcribing type I IFN (Fig 3C and 3G). Importantly, we observed that ROS production by neutrophils from CCR2-depleted mice was significantly improved by adoptive transfer of monocytes (Fig 7B), and this correlated with improved control of fungal growth (Fig 7C). These findings suggest that the impaired antifungal response of neutrophils in CCR2-depleted mice is due to a direct role for CCR2+monocytes, and not due to other CCR2+ populations that could be targeted in CCR2-depleted mice. IFN-λ production in CCR2-depleted mice was significantly improved by adoptive transfer of CCR2+monocytes (Fig. S7). This is a similar finding to what we previously observed in CCR2-depleted mice that were treated with recombinant IFN-α (Fig. 3H). Based on our aggregate observations, we hypothesize that in the context of IA, CCR2+monocytes mediate optimal activation of antifungal neutrophils by serving as an early source of type I IFN, which is required for optimal IFN-λ production. Consistent with this idea, systemic administration of recombinant IFN-α and/or IFN-λ progressively restored optimal ROS generation by antifungal neutrophils in CCR2-depleted mice (Fig 7D), and improved control of fungal growth in the lung (Fig 7E). Importantly, sustained treatment of CCR2-depleted mice with IFN-α and IFN-λ was able to protect CCR2-depleted mice from mortality and IA development (Fig 7F–7G). Altogether, our study has identified a novel role for type III IFN as a critical regulator of ROS generation and antifungal activity of neutrophils, and that this important response is coordinated by CCR2+monocytes.
Fig. 7. Neutrophil antifungal response in CCR2-depleted mice is rescued by adoptive transfer of CCR2+monocytes or by treatment with recombinant IFNs.
CD45.2+CCR2-depleted-mice were infected with Af and either left untreated or treated by the adoptive transfer of CD45.1+CCR2-GFP+ monocytes at 3 hours after infection. Monocytes (~850,000) were isolated from the lung of Af-infected CD45.1+CCR2-GFP+ donor mice 3 hours post-infection. (A) Representative FACS plot of control, CCR2-depleted mice, and CCR2-depleted mice that received CD45.1+CCR2+monocytes. (B–C) WT control (black symbols), CCR2-depleted (red symbols), or CCR2-depleted mice adoptively transferred with CCR2+monocytes (blue symbols) were examined at 48 hours after infection for ROS generation by neutrophils (B) or Af fungal burden in the lung (C). Each symbol represents one mouse. Data shown is cumulative of two independent experiments. (D–E) CCR2-depleted mice were left untreated or were treated with recombinant IFN-α2 and/or IFN-λ3 as indicated. Mice were treated with 1 µg of individual cytokines, except for the double treated which received 500 ng of IFN-α2 and 500 ng of IFN-λ3. To recapitulate the kinetics of IFN expression in control mice, CCR2-depleted mice were treated shortly after infection with type I IFN followed by type III IFN. Mice were treated with IFNs the same day of infection and then every other day. (D) Analysis of ROS generation by airway infiltrating neutrophils isolated from controls (WT littermates treated with DT) or from untreated and treated CCR2-depleted mice. (E) Pulmonary fungal burden 48 hours after infection. Each symbol represents one mouse. Data shown is for one experiment representative of two independent ones. (F) Kaplan-Meier survival plot of CCR2-depleted mice treated with each IFN and mock-treated controls. (G) Representative images of Gomori ammoniacal silver-stained lung sections captured at 40X magnification. Data shown is cumulative of two-three independent experiments. Statistical analysis was done with Kruskall-Wallis, non-parametric test for multiple comparisons using Prism software. *p<0.05, ***p<0.001 for each labeled sample as compared to CCR2-depleted mice.
Discussion
Our findings identify a critical and non-redundant role for type III IFNs in the activation of innate antifungal neutrophil responses. We find that type I and III IFNs are expressed with distinct kinetics during fungal infection with type I IFN being produced within hours after infection, and type III IFN peaking at later times. The optimal expression of IFN-λ requires early priming by type I IFN. Thus, both IFN families are essential for the activation of antifungal neutrophils by acting in a coordinated manner. In this model, type I IFN acts as an early signal that promotes the expression of type III IFN, which further amplifies its own production. Together, type I and III IFNs act as critical activators of ROS generation by neutrophils, an essential effector killing mechanism in control of fungal infection. The importance of ROS in control of fungal infection is supported by studies in mouse models of disease and by clinical observations in NADPH-deficient patients (21, 24–31). We find that mouse and human neutrophils both express type III IFN receptor thus suggesting a possible conserved role for IFN-λs in regulating neutrophil activation. We observed enhanced expression of IFNLR1 in human neutrophils upon stimulation with Af. Stimulation of human neutrophils with LPS has also been reported to induce increased expression of IFNLR1 by neutrophils (34). It is thus possible that the activation of innate receptors and/or the production of inflammatory cytokines in response to fungi further stimulates IFNLR1 expression on neutrophils. Future studies on the regulation of IFNLR1 expression will likely provide further insight on the pathways that can dynamically regulate IFNLR1 expression in neutrophils and potentially other innate cells. The conserved role of ROS in antifungal defense in mice and humans suggest the possible conserved importance of type I and III IFNs in activating human antifungal responses. Further studies will be necessary to decipher this intriguing possibility. Additional studies might also uncover defective type I and/or type III IFN signaling as potential genetic basis for currently undiagnosed susceptibility to fungal infections.
Our study also identified CCR2+Ly6C+ pulmonary monocytes as important regulators of the IFN response, and depletion of these cells impaired production of both type I and III IFNs during fungal infection. We find that CCR2+Ly6C+ monocytes act as an important, direct source of early type I IFNs. The contribution of CCR2+monocytes to IFN-λ production appears to be indirect in that we did not observe significant production of type III IFN by purified CCR2+Ly6C+monocytes. The majority of measurable IFN-λ during Af infection appears to be produced by CCR2−CD45+ and CD45− pulmonary cells. We find that induction of IFN-λ production during Af appears to be largely dependent on type I IFN expression. Thus, the requisite role for CCR2+ monocytes for the optimal expression of type I and III IFNs appears to be via direct production of type I IFNs and indirect induction of type III IFN in other pulmonary cells. Future studies will likely provide exciting answers about the identity of relevant cellular sources of IFN-λ in response to fungal infection as well as the sensing pathways required for the expression of these important cytokines. An alternate interpretation to CCR2-dependent regulation of IFN-λ expression is that CCR2+Ly6C+ monocyte precursors could differentiate into CCR2−Ly6C− derivatives that then serve as an important source of type III IFN at later times after infection. CCR2-derivative cells can be part of the CCR2−CD45+ population we have analyzed since our current analysis was restricted to cells actively expressing CCR2 and Ly6C. Future studies with CCR2-cre driven expression of fluorescent marker genes in fate mapping studies will allow further examination of possible direct contributions of CCR2+monocytes to IFN-λ production by acting as potential precursors of IFN-λ-producing derivative cells.
The responsiveness of neutrophils to type III IFNs and their importance in control of fungal infection that we have identified in this study, suggest an expanded range of relevant cellular targets for the biological activities of type III IFNs. Specific gene ablation of IFNLR1 or STAT1 in murine neutrophils had a striking effect on the antifungal response of these mice. Restricted genetic deficiency to neutrophils had a similar effect in susceptibility to IA as mice with global defects in IFNLR1 or STAT1 expression. Our findings thus provide strong genetic evidence for a critical and non-redundant role for type III IFN in the activation of antifungal neutrophils. In the context of viral infection, neutrophil-specific targeting of IFNLR1 also affected the host response but in contrast to our findings, defense against influenza required expression of IFNLR1 by epithelial cells (12). In the context of Af infection, our findings suggest that type III IFN activates antifungal responses via effects on hematopoietic cells (neutrophils), and does not depend on targeting on non-hematopoietic cells (epithelial cells). The distinct dependency of type III in hematopoietic cells in the context of Af infection is likely a reflection of the unique biology of viral and fungal pathogens. Viruses are able to efficiently infect epithelial cells, and in many cases, epithelial cells are the first to respond to the presence of an invading virus and to trigger a protective IFN response. In contrast, upon infection with fungal conidia, phagocytes are rapidly engaged in the active uptake of fungal spores, and are essential in containment of fungal infection. Thus, although it is likely that epithelial cells contribute to overall defense against fungal infection, our data suggests that those defense mechanisms operate independently of IFNLR1. Instead, the critical contributions of IFNLR1 to antifungal defense are restricted to hematopoietic cells, especially neutrophils. Reported effects of IFN-λ on neutrophils include inhibition of their recruitment (10), activation of antiviral responses (12), and attenuation of their activation by inflammatory signals (34). The IFN-λ-mediated suppression of neutrophil function was found to be STAT1- and translation-independent (34). In contrast, our results revealed that STAT1-dependent action of IFN-λ on neutrophils is critical for the activation of their antifungal functions in vivo. The diverse activities of IFN-λ on neutrophil function are likely modulated by the presence of other inflammatory cytokines and shaped by the type of infectious insult and tissue microenvironment.
The contributions of IFNLR1 to antifungal control in mice with global defects in IFNLR1 expression are likely multifactorial, and include effects on cellular recruitment and activation of killing mechanisms. Conditional targeting of IFNLR1 deficiency to neutrophils had a more selective effect on ROS generation with a lesser impact on cellular recruitment or inflammatory cytokine production. Thus, a significant contribution of IFN-λ in control of fungal infection appears to be as an important activator of ROS generation in neutrophils. The cellular mechanisms by which IFN-λ influences ROS production remain to be elucidated, but could potentially include STAT1-dependent induction of optimal expression of NADPH enzyme components and/or optimal assembly of enzyme subunits in the phagosome. Altogether, IFN-λ emerges as an important regulator of neutrophil functions, which appear to change in a disease state-dependent manner (10, 12, 34). Additional studies are required to understand how neutrophils integrate specific stimuli to adjust their function according to the task to be accomplished at a given time.
In aggregate, our study revealed an essential function for type III IFN in the regulation of antifungal neutrophils. Our study further suggests that dysfunctional innate antifungal responses can be boosted by systemic administration of recombinant type I and III IFN, which together mediated optimal control of fungal infection. These observations provide proof-of-principle evidence for the possible therapeutic benefit of these important cytokines in activating antifungal responses on myeloid cells.
Materials and Methods
Study Design
Animal studies were performed in an SPF facility in accordance with Institutional guidelines for the humane use and care of mice. Human cell samples were obtained from healthy volunteers in accordance with guidelines of Rutgers Institutional Review Board. Samples from female and male donors were used. All mouse strains were bred in house. Wild type control mice were transgene negative (CCR2-DTR-negative or Cre-negative) littermates of genetically modified cohorts. Age-matched, male and female mice were used. For all studies with CCR2-depleted mice CCR2-DTR+/− and CCR2-DTR −/− littermate controls were treated with diphtheria toxin. All mouse studies were performed independently at least two times with 4–5 mice per group per time point of analysis.
Mice
C57BL/6J and Balb/c mice were purchased from Jackson Laboratory or bred in-house. CCR2-depleter (CCR2-DTR) and CCR2-GFP mice were generated on the C57BL/6 background as previously described (37, 38). p47Phox−/, IFNγR−/−, and MRP8cre mice were purchased from Jackson Laboratory. RAG−/−γC−/− (RAG-2−/−IL2rg−/−) lymphopenic mice were purchased from Taconic. IFNAR−/−, IFNLR1−/−, IFNAR−/− IFNLR1−/− (DKO), STAT1−/−, Ifnlr1fl/fl, and Villincre Ifnlr1fl/fl were obtained from the laboratories of Dr. Sergei Kotenko and Dr. Joan Durbin. Ifnlr1fl/fl and Stat1fl/fl mice were generated as previously described (8, 39)
Bone marrow chimeric mice were generated by transferring 2–3 × 106 of CD45.1+wild-type, CD45.2+IFNAR−/−, or CD45.2+IFNLR1−/− bone marrow cells intravenously using lethally irradiated (10.5 Gy) CD45.1+wild-type, CD45.2+IFNAR−/−, or CD45.2+IFNLR1−/− recipients. Chimerism was verified via flow cytometric analysis.
All strains were maintained and bred in the Rutgers-NJMS Cancer Center Research Animal Facility under specific pathogen-free conditions. Animal studies were performed following biosafety level 2 (BSL-2) protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Rutgers University.
Infection, culture, and histology
Pulmonary fungal infections were done with live Aspergillus fumigatus CEA10 strain (13). cultured on Glucose minimal media (GMM) slants for 7–10 days prior to infection. Mice were challenged with 4–8 × 107 live conidia per mouse using a non-invasive intratracheal (i.t.) infection procedure (40). Fungal burden was assessed by plating serial dilutions of single-cell lung suspensions on SDA. For histological examination, lungs were perfused with 10 mL of PBS to remove blood and fixed in 10% buffered formalin. Fixed lung tissue was paraffin embedded and stained with modified GMS stain at the Histology Core Facility (Rutgers-NJMS). Images were taken at 40X magnification using a Leica confocal microscope and Surveyor software.
CCR2 Depletion, CCR2+Monocyte transfer, and IFN Treatment
For depletion of CCR2+ cells, CCR2-DTR mice and control CCR2-DTR negative littermates received 250 ng of diphtheria toxin i.p. one day prior to infection and every other day thereafter in order to maintain depletion. Mice were injected i.p. with 1.0 µg of either IFNα2 (Novoprotein), IFNλ3 (PBL), or both on D0 and D+1 for ROS analysis 48 hours post-infection. For survival, mice were injected i.p. with 1.0 µg of either IFNα2, IFNλ3, or 500 ng of both for a total of 1.0 µg on D0 and every other day. Mice were euthanized as they developed severe symptoms of IA, and survival was terminated at D+14.
For CCR2+monocyte transfer experiments, CD45.1+CCR2-GFP+ monocytes were sorted from mice that were infected with 4 × 107 Af conidia for 3 hours (peak of IFN-α transcription). CD45.1+CCR2-GFP+ monocytes (~850,000) were transferred via retro-orbital injection into CD45.2+CCR2-DTR mice that have been infected with 4 × 107 Af conidia for 3 hours. CD45.2+CCR2-DTR mice were treated with DT on D-1, D0, and D+1. Congenic markers were used to track transferred cells in recipients by flow cytometry. ROS and fungal burden was assessed 48 hours after infection. Monocytes were sorted as (Live CD45.1+CD11b+NK1.1−CCR2-GFP+Ly6C+)
ROS Measurement Assay
BAL cells from infected mice were cultured with 1.0 µM of CM-H2DCFA (General Oxidative Stress Indicator; Life Technologies) in Hank’s balanced salt solution for 60 minutes at 37°C. After incubation, cells were analyzed using flow cytometry. DAPI was included to exclude dead cells.
Murine Lung Cell Isolation and Flow Cytometry
Lung samples were minced in PBS with 3 mg/mL of collagenase type IV (Worthington), and were incubated at 37°C for 45 min to obtain single cell suspensions. After digestion, lung suspensions were lysed of red blood cells. The staining protocols included combinations of the following antibodies from BD Biosciences: Gr-1 (RB6–8C5), Ly6C (AL-21), Ly6G (1A8), CD11b (M1/70), CD11c (N418), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), MHC Class II I-A/I-E (M5/11.415.2), NK1-1 (PK136), and Siglec-F (E50–2440). F4/80 (BM8) was purchased from Biolegend, and was also included in staining panel. DAPI (Life Technologies) was used as a viability control Samples were collected on a BD LSRFortessa X-20, and analyzed using FlowJo software. Cell counts were calculated via flow cytometric analysis of whole lung tissue.
Measurement of Myeloperoxidase (MPO) and NET-associated Elastase
Neutrophils were isolated from the BAL of different IFNR−/− mice 48 hours post-infection. Neutrophils were treated ex vivo with 20 nM PMA in order to stimulate MPO release and NET formation. MPO was measured using a neutrophil myeloperoxidase activity assay kit (Cayman Chemical) per the manufacturer’s instructions. As a control for the specificity of the assay, we used the MPO inhibitor, 4-aminobenzhydrozide (4-ABH), and treated neutrophils using a 1:100 dilution of the 25 mM stock. NET-associated elastase was measured using a NETosis assay kit (Cayman Chemical) per the manufacturer’s instructions.
Quantitative RT-PCR and ELISA in Murine Lung Tissue
Total RNA from lungs was extracted with Trizol (Invitrogen). One microgram of total RNA was reverse transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). TaqMan Fast Universal Master Mix (2X) No AmpErase UNG and TaqMan probes (Applied Biosystems) for each gene were used, and normalized to GAPDH. Gene expression was calculated using ΔΔCT method relative to naïve sample.
Lungs homogenates were made by passing the tissue through a 70 µm cell strainer for protein analysis. ELISAs were performed according to the manufacturer’s instructions. IFNα and IFNγ ELISA kits were purchased from eBioscience, and the IFNλ2–3 ELISA kit was obtained from R&D.
Characterization of IFN Production by Different Cell Populations
CCR2+monocytes (Live CD45+CD11b+NK1.1−CCR2-GFP+Ly6C+), CD45− cells, and CD45+ cells excluding CCR2+monocytes were FACS sorted from the lungs of CCR2-GFP mice infected for 3 and 48 hours (peak of type I and III IFN expression) as well as naïve. These populations were sorted to >99% purity. In order to assess protein production, approximately 100,000 cells of each of the sorted populations were plated onto a tissue culture treated 96- well plate and left overnight. Supernatants were collected next day, and assessed for IFN production by ELISA using the same kits used for lung homogenates. RNA was also isolated from each of the populations by using the Qiagen RNeasy kit and Qiashredder. RNA reverse transcription and QRT-PCR was performed the same as the lung.
Western blots and Genotyping
Neutrophils from the lung, spleen, and bone marrow as well as bone marrow-derived T and B cells were isolated from naïve wild-type, IFNAR−/−, IFNLR1−/−, IFNAR−/−IFNLR1−/−(DKO), MRP8cre Ifnlr1fl/fl, MRP8cre Stat1fl/fl, MRP8cre, Ifnlr1fl/fl, and Stat1fl/fl mice. Neutrophils and B cells were stimulated with 100 ng of IFNα2 (Novoprotein) or IFNλ2 (PBL). B cells were sorted as Live CD45+B220+, T cells were sorted as Live CD45+Thy1.2+, and neutrophils were sorted as Live CD45+CD11b+Ly6CintLy6G+. These populations were sorted to >99% purity. Cell lysates were collected in lysis buffer containing protease and phosphatase inhibitors. Equal amounts of total protein was separated on 7.5% SDS-PAGE gels, transferred to Nitrocellulose 0.45 µm membrane (BIO-RAD), and subsequently probed with antibody against phosphorylated STAT1 (BD #612133) and β-actin (Sigma #A5441). For PCR genotyping, DNA from sorted cells was isolated using Qiagen DNeasy kit, and run on a 4% agarose gel. Control DNA was extracted from tail snips.
Human Blood and Bone Marrow Analysis
Bone marrow (BM) and peripheral blood samples were obtained from the Whitehead laboratory. Samples were obtained under Rutgers Institutional Review Board approved protocols. The samples were lysed 2–3 times to remove RBCs and resuspended in RPMI containing FCS. For BM samples, 1×106 cells were plated on flat bottom tissue culture treated 96-well plates and allowed to rest for 30–60 min before culturing with pre-germinated CEA10 conidia at an MOI of 1 for 6 hours. For peripheral blood samples, 3×106 cells were plated on 24-well plates and rested for 30–60 min. Pre-germinated CEA10 conidia were added at an MOI of 1 for 12 hours.
For analysis by flow cytometry, cells were stained with the following anti-human antibodies: anti-lambda receptor (PBL); goat anti-mouse (Jackson ImmunoResearch); biotinylated CD15 (HI98, eBioscience), CD16 (eBioCB16, eBioscience); streptavidin AlexaFluor 633 (Invitrogen); CD49d (gF10, BDPharmingen), CD14 (M5E2, DPharmingen), and CD19 (HIB19, BDPharmingen). RNA from human samples was isolated using the QIAshredder cell homogenizer and RNeasy mini kits from Qiagen. QRT-PCR was performed as described above. Human TaqMan probes were purchased from Applied Biosystems.
Cell Sorting for RNA Sequencing and Analysis
Live CCR2GFP+CD45+CD11b+NK1.1−Ly6C+Ly6G− (CCR2+mo) and Live CCR2GFP−CD45+CD11b+NK1.1−Ly6G+ (neutrophils) populations were isolated to >98.9% purity using a BD FACS ARIA II cell sorter dedicated to the processing of BSL-2 samples (Flow Cytometry Core facility). Cells were sorted from lung single cell suspensions obtained from A. fumigatus infected CCR2-GFP mice that were challenged 2 days prior. RNA was immediately extracted from sorted cells using Qiagen RNeasy kit. The transcriptional profile of neutrophils was examined by RNA-seq. RNA processing for library generation and sequencing on an Illumina HiSeq instrument was done by the Genomic Research Core facility as described (22). Trimming of raw reads was performed using Trimmomatic-0.33 with leading and trailing Q score of 25, minimum length of 25 bp, and by removing adaptors. The cleaned reads were mapped to Mus musculus genome GRCm38 using Tophat v.2.0.13. The reference genome sequence and annotation files were downloaded from ENSEMBLE, release.79 (Mus_musculus.GRCm38.79.fa, and Mus_musculus.GRCm38.79.gtf). The aligned read counts were obtained using htseq-count (intersection-nonempty), part of the package HTSeq-0.6.1. 74–79% of the reads were mapped to the mouse genome. A total of 22,289 genes were used for further analysis. The bioconductor package edgeR_3.8.6 with limma_3.22.7 was used to perform the differential gene expression analysis, under R environment, R version 3.1.2. Heatmap was plotted using the log2 transformed fpkm expression values within R heatmap_2 function. Multiple testing correction of p values was performed by limma with the Benjamini-Hochberg method. Pathway analysis of gene expression profiles was done using Ingenuity Pathway Analysis software (Qiagen). RNA-seq data is being deposited to NCBI BioProject under accession number PRJNA406963.
Statistics
Statistical analysis of in vivo and in vitro parameters of antifungal immunity was performed using non-parametric Mann-Whitney test or paired t-test (for human samples) in GraphPad Prism version 7 software. For multiple comparison analysis of 4 groups or more, non-parametric, Kruskall-Wallis test was employed using Prism software. For analysis of RNA-seq data, multiple testing correction of p values was performed by Limma with the Benjamini-Hochberg method.
Supplementary Material
Material and Methods
Fig S1. Lymphoid-deficient and IFN-γ-deficient mice control fungal infection normally.
Fig S2. Global transcriptional response of antifungal neutrophils.
Fig S3. Inflammatory responses in IFNR−/− mice 48 hours post-infection.
Fig S4. Analysis of p47Phox−/− mice susceptibility to Af infection.
Fig S5. Validation of hIFNLR1 antibody.
Fig S6. Analysis of efficient neutrophil-specific gene excision in Mrp8-Cre mice.
Fig S7. Type III IFN transcription in CCR2-depleted mice is rescued by adoptive transfer of CCR2+monocytes.
Excel sheet with differentially expressed gene lists
Acknowledgments
We thank our colleagues Drs. Mark Siracusa and George Yap for helpful comments and discussions. We also thank the staff of the Histology and Flow Cytometry Cores at NJMS for outstanding technical support. Funding: A.R. is supported by NIH grant R01AI114647-01A1 and a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease award. Studies in the Kotenko lab were supported by R01AI104669. Ifnlr1fl/fl mice were generated in the Kotenko lab and are available to the scientific community upon standard material transfer agreement between Rutgers University and the requesting institution. SVK is an inventor on patents and patent applications related to IFN-λs, which have been licensed for commercial development.
Footnotes
Author contributions: AR and VE conceived the project and designed the experiments. VE, OD, CM, BC and AP performed the experiments. AR and VE analyzed the data. PD and Y-JC performed analysis of RNA-seq data. I.W., JJO, JD and SK provided important reagents and mice. AR, SK and VE wrote the manuscript.
Competing interests: The authors declare no competing interests. SVK is an inventor on patents and patent applications related to IFN-λs, which have been licensed for commercial development.
Data deposition and material availability: RNA-seq data is being deposited to NCBI Bioproject under accession PRJNA406963.
References
- 1.Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. 1983;158:670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 3.Boxx GM, Cheng G. The Roles of Type I Interferon in Bacterial Infection. Cell Host Microbe. 2016;19:760–769. doi: 10.1016/j.chom.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez-Ortiz ZG, et al. A nonredundant role for plasmacytoid dendritic cells in host defense against the human fungal pathogen Aspergillus fumigatus. Cell Host Microbe. 2011;9:415–424. doi: 10.1016/j.chom.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotenko SV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 6.Sheppard P, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 7.Crotta S, et al. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 2013;9:e1003773. doi: 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin JD, et al. Distinct Roles of Type I and Type III Interferons in Intestinal Immunity to Homologous and Heterologous Rotavirus Infections. PLoS Pathog. 2016;12:e1005600. doi: 10.1371/journal.ppat.1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nice TJ, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blazek K, et al. IFN-lambda resolves inflammation via suppression of neutrophil infiltration and IL-1beta production. J Exp Med. 2015;212:845–853. doi: 10.1084/jem.20140995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pott J, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galani IE, et al. Interferon-lambda Mediates Non-redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity. 2017;46:875–890. e876. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Willger SD, et al. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotenko SV, Durbin JE. Contribution of Type III Interferons to Antiviral Immunity; Location, Location, Location. J Biol Chem. 2017 doi: 10.1074/jbc.R117.777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown GD, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 16.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verweij PE, Chowdhary A, Melchers WJ, Meis JF. Azole Resistance in Aspergillus fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin Infect Dis. 2016;62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal BH. Aspergillosis. N Engl J Med. 2009;360:1870–1884. doi: 10.1056/NEJMra0808853. [DOI] [PubMed] [Google Scholar]
- 19.Espinosa V, Rivera A. First Line of Defense: Innate Cell-Mediated Control of Pulmonary Aspergillosis. Front Microbiol. 2016;7:272. doi: 10.3389/fmicb.2016.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanternier F, et al. Primary immunodeficiencies underlying fungal infections. Curr Opin Pediatr. 2013;25:736–747. doi: 10.1097/MOP.0000000000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollock JD, et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet. 1995;9:202–209. doi: 10.1038/ng0295-202. [DOI] [PubMed] [Google Scholar]
- 22.Espinosa V, et al. Inflammatory monocytes orchestrate innate antifungal immunity in the lung. PLoS Pathog. 2014;10:e1003940. doi: 10.1371/journal.ppat.1003940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caffrey AK, et al. IL-1alpha signaling is critical for leukocyte recruitment after pulmonary Aspergillus fumigatus challenge. PLoS Pathog. 2015;11:e1004625. doi: 10.1371/journal.ppat.1004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahlin A, Elinder G, Palmblad J. Dose-dependent enhancements by interferon-gamma on functional responses of neutrophils from chronic granulomatous disease patients. Blood. 1997;89:3396–3401. [PubMed] [Google Scholar]
- 25.Boyle KB, Stephens LR, Hawkins PT. Activation of the neutrophil NADPH oxidase by Aspergillus fumigatus. Ann N Y Acad Sci. 2012;1273:68–73. doi: 10.1111/j.1749-6632.2012.06821.x. [DOI] [PubMed] [Google Scholar]
- 26.Chusid MJ, Sohnle PG, Fink JN, Shea ML. A genetic defect of granulocyte oxidative metabolism in a man with disseminated aspergillosis. J Lab Clin Med. 1981;97:730–738. [PubMed] [Google Scholar]
- 27.Falcone EL, Holland SM. Invasive fungal infection in chronic granulomatous disease: insights into pathogenesis and management. Curr Opin Infect Dis. 2012;25:658–669. doi: 10.1097/QCO.0b013e328358b0a4. [DOI] [PubMed] [Google Scholar]
- 28.Grimm MJ, et al. Monocyte- and macrophage-targeted NADPH oxidase mediates antifungal host defense and regulation of acute inflammation in mice. J Immunol. 2013;190:4175–4184. doi: 10.4049/jimmunol.1202800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liese JG, et al. Chronic granulomatous disease in adults. Lancet. 1996;347:220–223. doi: 10.1016/s0140-6736(96)90403-1. [DOI] [PubMed] [Google Scholar]
- 30.Morgenstern DE, Gifford MA, Li LL, Doerschuk CM, Dinauer MC. Absence of respiratory burst in X-linked chronic granulomatous disease mice leads to abnormalities in both host defense and inflammatory response to Aspergillus fumigatus. J Exp Med. 1997;185:207–218. doi: 10.1084/jem.185.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philippe B, et al. Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect Immun. 2003;71:3034–3042. doi: 10.1128/IAI.71.6.3034-3042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ank N, et al. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durbin RK, Kotenko SV, Durbin JE. Interferon induction and function at the mucosal surface. Immunol Rev. 2013;255:25–39. doi: 10.1111/imr.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broggi A, Tan Y, Granucci F, Zanoni I. IFN-lambda suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat Immunol. 2017 doi: 10.1038/ni.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Van Ziffle JA, Lowell CA. Neutrophil-specific deletion of Syk kinase results in reduced host defense to bacterial infection. Blood. 2009;114:4871–4882. doi: 10.1182/blood-2009-05-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hohl TM, et al. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe. 2009;6:470–481. doi: 10.1016/j.chom.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serbina NV, Hohl TM, Cherny M, Pamer EG. Selective expansion of the monocytic lineage directed by bacterial infection. J Immunol. 2009;183:1900–1910. doi: 10.4049/jimmunol.0900612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klover PJ, et al. Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia. 2010;12:899–905. doi: 10.1593/neo.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivera A, Van Epps HL, Hohl TM, Rizzuto G, Pamer EG. Distinct CD4+-T-cell responses to live and heat-inactivated Aspergillus fumigatus conidia. Infect Immun. 2005;73:7170–7179. doi: 10.1128/IAI.73.11.7170-7179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Material and Methods
Fig S1. Lymphoid-deficient and IFN-γ-deficient mice control fungal infection normally.
Fig S2. Global transcriptional response of antifungal neutrophils.
Fig S3. Inflammatory responses in IFNR−/− mice 48 hours post-infection.
Fig S4. Analysis of p47Phox−/− mice susceptibility to Af infection.
Fig S5. Validation of hIFNLR1 antibody.
Fig S6. Analysis of efficient neutrophil-specific gene excision in Mrp8-Cre mice.
Fig S7. Type III IFN transcription in CCR2-depleted mice is rescued by adoptive transfer of CCR2+monocytes.
Excel sheet with differentially expressed gene lists