Abstract
Background
Sexual parasites offer unique insights into the reproduction of unisexual and sexual populations. Because unisexuality is almost exclusively linked to the female sex, most studies addressed host-parasite dynamics in populations where sperm-dependent females dominate. Pelophylax water frogs from Central Europe include hybrids of both sexes, collectively named P. esculentus. They live syntopically with their parental species P. lessonae and/or P. ridibundus. Some hybrid lineages consist of all males providing a chance to understand the origin and perpetuation of a host-parasite (egg-dependent) system compared to sperm-dependent parthenogenesis.
Methods
We focused on P. ridibundus-P. esculentus populations where P. ridibundus of both sexes lives together with only diploid P. esculentus males. Based on 17 microsatellite markers and six allozyme loci, we analyzed (i) the variability of individual genomes, (ii) the reproductive mode(s) of all-male hybrids, and (iii) the genealogical relationships between the hybrid and parental genomes.
Results
Our microsatellite data revealed that P. esculentus males bear Mendelian-inherited ridibundus genomes while the lessonae genome represents a single clone. Our data indicate that this clone did not recently originate from adjacent P. lessonae populations, suggesting an older in situ or ex situ origin.
Conclusions
Our results confirm that also males can perpetuate over many generations as the unisexual lineage and successfully compete with P. ridibundus males for eggs provided by P. ridibundus females. Natural persistence of such sex-specific hybrid populations allows to studying the similarities and differences between male and female reproductive parasitism in many biological settings.
Electronic supplementary material
The online version of this article (10.1186/s13293-018-0172-z) contains supplementary material, which is available to authorized users.
Keywords: Pelophylax, Water frog, Hemiclone, Hybridogenesis, Sexual parasites, Unisexual, All-male lineage
Background
Unisexual animals are model systems for understanding the evolution and maintenance of sex and recombination despite the costs that may arise [1, 2]. Interspecific hybridization between diploid bisexual species is likely the only mechanism by which unisexuality originates in vertebrates [3]. In 46 described cases of squamate reptiles, 51 cases of fish, and 23 cases of amphibians, unisexuality is almost exclusively linked to the female sex [4–6]. Hybrid males are frequently inviable or sterile (e.g., [7]), but interestingly, a few cases of viable and likely fertile male vertebrates have been documented, e.g., in water frogs of the genus Pelophylax [8–10] and fish of the genera Squalius [11], Misgurnus [12, 13], and Hypseleotris [14]. An example from the Squalius alburnoides complex suggests that perpetuation of hybrid males in populations depends on highly complex reproductive mechanisms [11]. Here, hybrid females are sperm-dependent sexual parasites on the males of the host species which reproduces sexually, while hybrid males are rather a by-product of crosses between hybrid females and sexual males. A production of various types of gametes and presence of polyploid inividuals for male maintenance is rather requisite in this mating system. All-male hybridogenetic lineages were found in Pelophylax and Hypseleotris. It is not yet clear what their origin is and what enables the maintenance of male unisexuality in these systems.
Hybridogenesis, combining elements of both clonal and sexual reproduction, seems to be the most promising reproductive mode for the maintenance of male unisexuality as separate, sex-specific lineages [15]. Hybrids with hybridogenetic reproduction usually discard one complete parental genome from their germline prior to meiosis and clonally transmit the remaining one, generally the mother’s one [3, 16], except some modified cases [11, 17, 18]. Hybridity is restored in each generation through fertilization by the sexual species, whose genome has been eliminated in the germline of the hybrid. Hence, inheritance is typically half clonal and half sexual (Mendelian), referred to as hemiclonal [19].
The Pelophylax esculentus complex consists of two sexual species, the pool frog Pelophylax lessonae (genomic composition LL) and the the marsh frog P. ridibundus (RR), and their hybrid taxon, the edible frog P. esculentus (RL). In most parts of Western and Central Europe, P. esculentus lives in sympatry with P. lessonae, in what is known as the “L-E system” [20]. In this system, both male and female hybrids usually exclude the haploid lessonae (L) genome from their germline and transmit the ridibundus (R) genome to their gametes. Hybridity is restored in each generation by heterospecific crosses between P. esculentus and P. lessonae, the donor of the L genome. When a hybrid male mates with a LL or RL female, only female offspring originates. Thus, males originate from matings between hybridogenetic females and heterospecific sexual P. lessonae males producing both hybrid sexes [21].
Conversely, in the R-E system, most P. esculentus exclude the ridibundus genome, transmit their lessonae genome, and mate with P. ridibundus to perpetuate the hybrid lines [21–23]. Special cases are populations which consist of P. ridibundus and only P. esculentus males. Such populations have been reported from the Czech Republic [24], Germany [8, 9, 25], Hungary [26], and Poland [27–29]. In contrast to the L-E system, some P. esculentus males from this population type transmit only the lessonae genome, others only the ridibundus genome, and some males even produce both ridibundus and lessonae gametes [9, 23, 24, 30]. From artificial crossings between P. ridibundus females and P. esculentus males usually two genotypes originate: if the ridibundus (R) eggs are fertilized by sperm with an R genome, RR females originate, while the combination of R eggs an L sperm gives rise to only hybrid (LR) males [9, 10, 30, 31]. This offers a potential explanation for the existence and perpetuation of all-male lineages in natural populations [9]. So far, it is not known whether one or multiple hybridization events have led to the formation of all-male P. esculentus. Consequently, there has been, and still might be, the possibility for de novo formation of P. esculentus males via ongoing primary hybridizations between the two sexual species, thus leaving open the question of their stable persistence in natural populations.
This study was aimed to (1) explore the origin of all male lineages in populations of the R-E system, i.e., whether male unisexuality evolved from one or several hybridization events, and (2) to understand how all-male hybrids are able to persist without hybrid females in natural populations. In pursuing these goals, we sampled 249 water frogs from 16 populations along the upper Oder River. Based on 17 microsatellite markers, we investigated (a) the mode of asexual reproduction, (b) the genetic variability of clonally transmitted genomes, and (c) their genealogical relationships to homotypic genomes from adjacent populations.
Methods
Study species, sampling sites, and taxon assignment
All European water frogs were collected in the upper Oder River drainage (Central Europe; Czech Republic; Fig. 1, Table 1). In total, 249 individuals from all three Pelophylax taxa were caught with a hand net at 16 locations during years 2002 and 2008. Pelophylax kurtmuelleri from Greece, a sister species of European P. ridibundus, was used as an outgroup in a phylogenetic tree construction. Males were distinguished from females by the presence of vocal sacs and nuptial pads. Taxon affiliation was determined on the basis of external morphological characters [22, 32], and identification was later verified genetically with allozyme markers (see the “Protein electrophoresis” section). Tissue samples obtained from the finger tips or muscles were stored at − 20 °C for allozyme analyses and in 96% ethanol for DNA analyses.
Fig. 1.

Map showing the investigated water frog populations in the upper Oder River. Numbers next to the dots correspond with numbers of localities given in Table 1. Dark gray dots refer to the R-E male system, light gray dots to the L-E system. The inset indicates the geographic location of the study area in Europe
Table 1.
Origin and genetic data of 16 water frog populations from the upper Oder River valley. Given are population types, names, and coordinates for sampling sites, genotypes, and numbers of collected females (f)/males (m)/juveniles (-), types and numbers of the detected multilocus genotypes (MLG), and IDs of hemiclones. For more details, see Additional file 2: Table S2
| No.* | Pop. type | Sampling site | Latitude Longitude |
Genotype | No. of f/m/juv | MLG type | No. of MLGs | Hemiclone ID |
|---|---|---|---|---|---|---|---|---|
| 1 | R-E male | Albrechtičky | 49.708056 18.098889 |
RR | 8/9/- | RR | 17 | |
| RL | -/12/- | R | 12 | |||||
| L | 1 | OderL1 | ||||||
| 2 | Bílovec | 49.769722 18.032222 |
RR | 1/-/- | RR | 1 | ||
| RL | -/2/- | R | 2 | |||||
| L | 1 | OderL1 | ||||||
| 3 | Darkovice | 49.757778 18.213056 |
RR | 3/5/- | RR | 8 | ||
| RL | -/4/- | R | 4 | |||||
| L | 1 | OderL1 | ||||||
| 4 | Dolní Benešov | 49.912222 18.120000 |
RR | 4/2/- | RR | 6 | ||
| RL | -/4/- | R | 4 | |||||
| L | 1 | OderL1 | ||||||
| 5 | Ostrava | 49.858889 18.265278 |
RR | 5/4/- | RR | 9 | ||
| RL | -/5/- | R | 5 | |||||
| L | 1 | OderL1 | ||||||
| 6 | L-E | Bravantice | 49.766944 18.084444 |
LL | 1/-/- | LL | 1 | |
| RL | 7/-/- | R | NA | |||||
| L | 7 | |||||||
| 7 | Břidličná | 49.916389 17.359444 |
LL | -/4/- | LL | 4 | ||
| 8 | Český Těšín | 49.764444 18.591667 |
LL | 3/7/- | LL | 10 | ||
| RL | 5/-/- | R | NA | |||||
| L | 5 | |||||||
| 9 | Dobrá | 49.676667 18.392222 |
LL | 7/22/2 | LL | 31 | ||
| 10 | Důl Staříč | 49.667222 18.258611 |
LL | -/2/- | LL | 2 | ||
| RL | 13/-/- | R | NA | |||||
| L | 13 | |||||||
| 11 | Horní Bludovice | 49.744444 18.457500 |
LL | 6/15/- | LL | 21 | ||
| RL | 2/2/- | R | NA | |||||
| L | 4 | |||||||
| 12 | Horní Domaslavice | 49.694722 18.470000 |
LL | 3/4/- | LL | 7 | ||
| RL | 7/1/- | R | NA | |||||
| L | 8 | |||||||
| 13 | Karviná-Doly | 49.825000 18.483056 |
LL | -/2/- | LL | 2 | ||
| RL | 10/3/- | R | NA | |||||
| L | 13 | |||||||
| 14 | Louky | 49.817222 18.576944 |
LL | 1/-/- | LL | 1 | ||
| RL | 10/4/- | R | NA | |||||
| L | 14 | |||||||
| 15 | Prstná | 49.915000 18.560556 |
LL | 3/5/- | LL | 8 | ||
| RL | 3/-/- | R | NA | |||||
| L | 3 | |||||||
| 16 | Trnávka | 49.683333 18.181389 |
LL | 8/10/3 | LL | 21 | ||
| RR | -/-/1 | RR | 1 | |||||
| RL | 4/-/- | R | NA | |||||
| L | 4 | |||||||
| OG | KK | Greece | 39.873166 22.732813 | KK | 1/-/- | KK | 1 |
R-E male P. ridibundus-P. esculentus male populations, L-E P. lessonae-P. esculentus populations, OG used as an outgroup, RR P. ridibundus, RL P. esculentus, LL P. lessonae, KK P. kurtmuelleri, NA not analyzed
*The localities are numbered according to Fig. 1
Protein electrophoresis
For genotype determination, six allozyme loci previously identified to be diagnostic for Pelophylax taxa were used [20, 33]. Approximately 1 g of skeletal muscle (or gonads) was homogenized on crushed ice for 20 s in an equal volume of Tris NaCl extraction buffer (pH 8.5; [34]) using an Ultra-Turrax homogenizator (IKA-WERK). The homogenate was then centrifuged at 13,500g at 4 °C for 20 min. Enzymes obtained from these tissues, namely aspartate aminotransferase (Aat; EC 2.6.1.1), glucose-6-phosphate isomerase (Gpi; EC 5.3.1.9), glycerol-3-phosphate dehydrogenase (G3pdh; EC 1.1.1.8), l-lactate dehydrogenase (Ldh-1; EC 1.1.1.27), phosphoglucomutase (Pgm-2; EC 5.4.2.2), and phosphogluconate dehydrogenase (6-Pgd; EC 1.1.1.44), were analyzed by horizontal potato starch gel electrophoresis [20, 34]. Subsequently, gels were cut into three 2-mm-thick slices and stained with appropriate allozyme chromogenic detection methods [35–37]. Stained gel slices were photographed and the agar layers were transferred to filter paper, then dried and stored as part of the protocol. The visualized allele products were designated from “a” fastest to “e” slowest according to their mobility. Samples which revealed unclear patterns were reprocessed.
DNA extraction, microsatellite genotyping
Genomic DNA was extracted using a NucleoSpin commercial kit (Macherey-Nagel GmbH and Co.) with the epMotion 5075 automated pipetting system (Eppendorf). We amplified 17 microsatellite loci: Ga1a19, Re2caga3, Re1Caga10, RICA1b6 [38], RlCA1b5, RlCA5, RlCA18 [39], RlCA2a34, GA1a23, Rrid169A, Rrid059A, RlCA1a27, Rrid135A [40], Res16, Res20, Res22 [41], and Rrid013A [39, 42], using the redesigned primer sets described in Hermaniuk et al. [43]. We followed Pruvost et al. [31] for species-specific characterization of the aforementioned markers. Polymerase chain reaction (PCR) was performed according to Christiansen and Reyer [40]. Fragment lengths were determined using an ABI 3730 Avant capillary sequencer (Applied Biosystems, Zug, Switzerland) and an internal size standard (GeneScan-500 LIZ); alleles were scored with GeneMapper v. 3.7 (Applied Biosystems, Zug, Switzerland).
Preparing datasets used for all analyses
Due to the presence of null alleles, raw microsatellite genotypes of P. lessonae and P. ridibundus were checked for potential genotyping errors with Micro-Checker version 2.2.3 [44]. This method estimates frequencies of null alleles with the Brookfield 2 null allele estimator, which treats nonamplifications as data and regards them as null homozygotes when calculating null allele frequencies [45]. In each population, Micro-Checker tested every locus for departure from the Hardy-Weinberg (HW) genotypic equilibrium. We found one locus (Res20) in P. lessonae and four loci (Res16, Rrid069A, RICA5, Re1Caga10) in P. ridibundus with a potential presence of null alleles, and therefore applied a correction for null alleles at these loci following Wagner et al. [46]. The after-correction analysis in Micro-Checker did not detect any locus with a presence of null alleles. The software MSA v. 4.05 [47] was used to determine summary statistics like mean microsatellite allele numbers (AN), observed heterozygosities (HOBS), and expected heterozygosities (HEXP) within populations.
Based on the definition of hybridogenetic reproduction, diploid hybrid water frogs from Central Europe transmit one haploid chromosome set clonally to gametes (therefore termed a “hemiclone” [19]), whereas the other set is discarded and regained for the next generation by mating with an individual of the sexual parental species. As our aim was to test the origin of a particular hemiclone in this vertebrate species, we analyzed sexual and clonal genomes of hybrid individuals separately. First, we sorted lessonae and ridibundus genomes according to the allele species specificity known from the literature [31]. Then, the correctness of allele separation was tested visually assuming that one allele per locus was received from a sexual mate, and therefore such allele has to be present (and always was in our study) in the gene pool of a sympatric sexual population. Our approach of separating sexually and clonally inherited alleles in hybrid genomes was 100% successful.
While for hybrids from R-E system populations both ridibundus and lessonae genomes were included in our analyses, we used only lessonae genomes of hybrids from the L-E system populations because in this system we were not interested in the origin of ridibundus genomes. We determined a hemiclone by a multilocus genotype (MLG), defined by the identical combination of alleles found in our microsatellite analyses. A minimum of three samples exhibiting the same allele composition was a clear indication that the genome was inherited clonally (PSEX value mentioned below) and did not originate from a sexual donor [48]. As most statistical programs dealing with microsatellite data—including GenAlEx and Populations used in this study—are not designed to compare haploid and diploid data, we transformed the haploid genotypes to diploid forms adding a second allele as missing data. This was done for lessonae and ridibundus genomes of P. esculentus (MLG’s) separately. To diploidize these haploid genotypes for GeneClone, we simply doubled lessonae and ridibundus genomes. Detailed information about the programs used are given below.
Flow cytometry
In order to determine ploidy levels, all individuals were analyzed by flow cytometry using blood samples following the standardized methodology [49]. A drop of blood was added into 70% ethanol and immediately shaken to prevent clotting. Chicken blood was used as a reference standard for cell size measurement. Relative nuclear DNA content was measured with DAPI fluorochrome using the Cystain two Step High Resolution DNA Staining commercial kit (Partec GmbH, Münster, Germany). Fluorescence intensity of 5000 stained nuclei was measured with a Partec PAII flow cytometer at a speed of 0.5 μl/s. Flow cytometric histograms were evaluated using FloMax 2.52 (Partec GmbH, Münster, Germany).
Statistical analysis of microsatellite data
In order to determine the origin of the all-male P. esculentus lineages, we analyzed genetic relationships between hybrids and their parental species on the basis of allele frequencies of up to 17 microsatellite loci using Structure v. 2.3.1 [50], GenAlEx v. 6.5 [51], Populations v. 1.2.32 [52], and GeneClone v.2.0 [53].
Hybrid origin
The origin of P. esculentus was analyzed with a model-based clustering method as implemented in Structure to infer population structure based on genotypic data consisting of unlinked markers. Structure analyses included datasets of 177 individuals (27 hybrid males from the R-E populations, 108 P. lessonae and 42 P. ridibundus, Additional file 1: Table S1). The analysis was carried out using a burn-in period of 10,000 iterations followed by 100,000 Markov Chain Monte Carlo (MCMC) repeats. The probability of the used admixture model was tested for clusters K = 1 − 10. The most probable number of K populations was estimated using log-likelihood ln P(D) according to Evanno et al. [54], with Structure Harvester [55].
Hemiclonal reproduction
In order to distinguish whether individual genomes are of sexual or clonal origin, we estimated PSEX values using the program GeneClone. When the same genotype is detected more than once, PSEX expresses the probability that this MLG has been derived from distinct reproductive events [56]. We applied the PSEX probability taking into account the FIS estimated of the dataset [57]. FIS was also estimated on the basis of the round-robin method, and further used to estimate a corrected PGEN, calculated as the unique MLG probability [56], which in turn provides a better estimate of the probability of clonal identity PSEX [53]. Considering that the presence of missing data precludes the use of PSEX in the program, we considered 10 loci from which both genomes were amplified (Ga1a19, Re1Caga10, RICA1b6, RlCA1b5, RlCA5, RlCA2a34, GA1a23, Rrid059A, Res16, Rrid013A), i.e., MLGs with missing alleles were eliminated (Additional file 2: Table S2). Therefore, GeneClone analyses were run on 143 MLGs which included 21 hybrid males from the R-E populations, 85 P. lessonae and 22 P. ridibundus individuals (Additional file 1: Table S1).
Hemiclones and their genetic relatedness
To sort out MLGs of 177 individuals (27 hybrid males from the R-E populations, 108 P. lessonae, 42 P. ridibundus), we used multilocus matches analysis as implemented in the program GenAlEx. In order to estimate the genetic relatedness between individual genomes, we compared allele frequencies, heterozygosity, and polymorphism estimates with the program GenAlEx. A centred principal component analysis (PCA) was applied to examine clustering of individuals based on total variation of microsatellite allele frequencies without scaling of alleles. For the PCA, we converted a list of 275 MLG’s from 17 loci (Additional file 2: Table S2) into a genetic distance matrix (covariance matrix with data standardization) and then used standard PCA to visualize the results. The PCA dataset included also the lessonae genome of 71 male and female hybrids from the L-E system (Additional file 1: Table S1).
UPGMA trees (unweighted pair group method with arithmetic mean, [58]) were constructed using Nei’s DA distance [59] calculated on the basis of 205 MLGs (Additional file 1: Table S1) with the program Populations which works with datasets containing rather lesser amount of missing data. Therefore, all loci that amplified only in one parental species (Re2caga3, Res22, Rrid169A, Res20, RICA1a27, RICA18) and a locus Rrid135A which amplified only in some individuals were excluded from the analysis. Support for internal branches was evaluated with 7000 replicates. Distinct genetic groups of individual genomes were identified with PCA on the basis of Nei’s DA.
Results
Taxon determination
In total, we identified 108 individuals of P. lessonae, 42 individuals of P. ridibundus, and 98 individuals of P. esculentus using six diagnostic allozyme loci (Additional file 3: Table S3) and 17 microsatellite loci (Additional file 2: Table S2). In P. lessonae, there were four monomorphic (Aat, Gpi, Pgm, G3pdh) and two polymorphic allozyme loci (Ldh-1, 6-Pgd); in P. ridibundus, two monomorphic (Aat, G3pdh) and four polymorphic loci (Ldh-1, Gpi, 6-Pgd, Pgm) were found. In P. esculentus, all six loci were polymorphic. The allelic products of three loci (G3pdh, Pgm, Gpi) displayed an atypical expression in 12 individuals. Here, four P. ridibundus possessed alleles characteristic of the lessonae genome in G3pdh and Pgm, one P. lessonae possessed alleles characteristic of the ridibundus genome in Pgm, and seven P. esculentus possessed either two lessonae or two ridibundus specific alleles in both Gpi and Pgm (Additional file 3: Table S3). Whether the shared alleles represent introgressions caused by crosses between individuals of the parental species and hybrids or ancestral polymorphisms is not clear.
Genetic diversity
Overall, all 17 microsatellite loci were polymorphic among 249 Pelophylax individuals (Additional file 4: Table S4). The loci Re2caga3, Res22, and Rrid169A amplified only in the P. ridibundus genome while the loci Res20, RICA1a27, and RICA18 amplified only in P. lessonae genome. Two loci (RICA5, Ga1a23) that are assumed to be specific for P. lessonae [39, 40], and one locus (Rrid135A), suggested to be specific for P. ridibundus [40], amplified in both P. lessonae and P. ridibundus samples.
A total of 187 alleles were detected, with 6 - 17 alleles per polymorphic locus (Additional file 2: Table S2, Additional file 4: Table S4 and Additional file 5: Table S5). The allele called 83 in RICA1b6 was marked as non-specific because it was amplified in the genomes of both P. lessonae (12 individuals) and P. ridibundus (four individuals). The presence of one or two alleles per locus (Additional file 2: Table S2) supported the flow cytometric data that all individuals were diploid. In addition, allele-dosage effects as indications of polyploidy were not observed.
Analyses of population structure using microsatellite data
Likelihood values provided by Structure converged during the runs, and results did not notably change between replicates. Evanno et al.’s [54] method implemented in Structure Harvester indicated K = 2 as the most probable number of genetic clusters for the dataset. Three types of diploid individuals were found, i.e., those with a q value ranging between 0.004–0.009 and 0.991–0.996, respectively, representing sexual P. lessonae or P. ridibundus, and those with intermediate q values (0.440–0.560) representing hybrid P. esculentus, which is in accordance with results from the determination based on diagnostic markers (Fig. 2a).
Fig. 2.
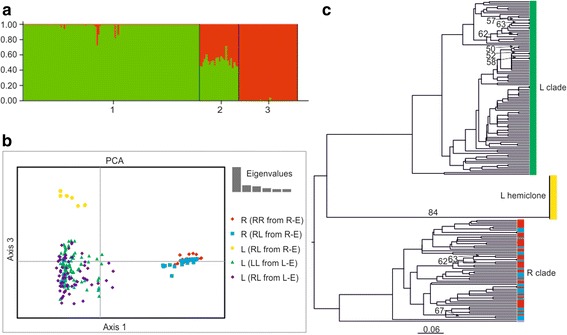
Cluster analyses performed on MLGs of Pelophylax individuals based on microsatellite loci. a Bar plot of 17 microsatellite loci from Bayesian cluster analysis performed in Structure (K = 2). Each vertical line represents one individual, each color represents the species specificity of alleles to one of the parental genomes (green = P. lessonae genome, red = P. ridibundus genome), and each cluster represents a different genotype (cluster 1 = LL, cluster 2 = RL, cluster 3 = RR). b Principal component analysis (PCA) of 17 microsatellite loci performed in GenAlEx. Each point represents an individual MLG, each color and symbol a group of related MLGs (according to allele sharing). Group red diamonds—R (RR from R-E) represents P. ridibundus from R-E system; group blue squares—R (RL from R-E) represents ridibundus genomes of P. esculentus from the R-E system; group yellow circles—L (RL from R-E) represents lessonae genomes of P. esculentus from the R-E system; group green triangles—L (LL from L-E) represents P. lessonae from the L-E system; group violet diamonds—L (RL from L-E) represents lessonae genomes of P. esculentus from the L-E system. Inset screenshot shows the eigenvalues for each axis as principle components of the analysis. c UPGMA tree computed with the program Populations using DA distances which were calculated on the basis of 10 microsatellite loci. Numbers on branches (with or without arrows) indicate bootstrap values > 50%. Each terminal unit represents one individual: green color—L clade represents P. lessonae from the L-E system; yellow color—L hemiclone represents lessonae genomes of P. esculentus from the R-E system; red color—R clade represents P. ridibundus from the R-E system; blue color—R clade represents ridibundus genomes of P. esculentus from the R-E system; violet color—P. kurtmuelleri individual. Detailed information is given in Additional file 6: Figure S1
The PCA identified three groups of microsatellite MLGs, the first two principle components accounted for 62.15% (axis 1) and 11.47% (axis 3) of genetic variation, respectively (Fig. 2b). Cluster 1 (green and dark blue symbols) grouped P. lessonae and lessonae-specific MLGs of P. esculentus sampled in L-E system populations. Cluster 2 (yellow symbols) included lessonae-specific MLGs obtained from P. esculentus in R-E populations while cluster 3 (red and blue symbols) grouped P. ridibundus and ridibundus-specific MLGs of P. esculentus from R-E populations.
Genealogical analyses made with the program Populations revealed two distinct clusters of lessonae-specific MLGs. One is characteristic of lessonae-specific MLGs found in P. esculentus from R-E populations; the second comprises genotypes of P. lessonae individuals (Fig. 2c and Additional file 6: Figure S1). Contrary to the previous pattern, ridibundus-specific MLGs found in P. esculentus from R-E populations did not represent a separate lineage but instead clustered with P. ridibundus.
Identification of MLGs and hemiclones
In the dataset which contained missing data for some loci, GenAlEx estimated a total of 185 MLGs among all Pelophylax individuals investigated. Among 150 P. lessonae and P. ridibundus individuals, the program distinguished 150 MLGs. Considering the 27 P. esculentus from R-E male populations, ridibundus genomes were represented by 27 MLGs, whereas the lessonae genomes exhibited only eight MLGs (Fig. 3, Additional file 7: Table S6). One lessonae-specific MLG was present in 14 lessonae genomes of P. esculentus, while the remaining seven MLGs were discovered in one to three lessonae genomes. In contrast to MLGs obtained from ridibundus genomes, all lessonae-specific MLGs shared the same alleles; their differences are only caused by missing data for some loci (Additional file 7: Table S6). Therefore, we consider the eight lessonae-specific MLGs detected in the hybrid males of the R-E populations as a single MLG. This MLG is further suggested to represent a clone (or a hemiclone from an individual level) designated as ODERL1.
Fig. 3.
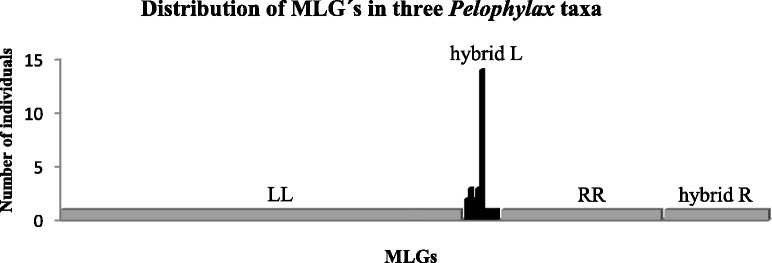
Distribution of 185 microsatellite multilocus genotypes (MLGs) in three Pelophylax taxa generated in GenAlEx (LL, P. lessonae from the L-E system; hybrid L, lessonae genome of P. esculentus from the R-E system; hybrid R, ridibundus genome of P. esculentus from the R-E system; RR, P. ridibundus from the R-E system)
Considering only MLGs with complete allelic data, GenClone generated 122 MLGs among 143 genotypes. Twenty-one lessonae genomes, all originating from P. esculentus of R-E male populations, were represented by only a single MLG. The PSEX value of this MLG ranged from 0.15 to 1.50E−41, indicating clonal inheritance of ODERL1.
Discussion
Detection of all-male P. esculentus lineage
The upper Oder River valley is mainly inhabited by P. ridibundus of both sexes and only males of P. esculentus (R-E system; populations 1-5, Table 1), whereas populations of the L-E system (typically represented by P. lessonae and P. esculentus of both sexes) live outside the valley. Since 2001, no hybrid female was detected among 377 hybrid individuals in localities of the R-E system (Table 1 and unpublished data), which makes this region a newly discovered area for the coexistence of bi-sexual P. ridibundus with diploid all-male P. esculentus hybrids. Together with the occurrence of similar populations downstream the Oder River drainage basin [8, 9, 28, 60], this points to Central Europe as the area of origin of R-E male populations.
P. esculentus males are active in maintaining their own all-male hybrid lineage
Hybrid males are usually infertile [61] and therefore considered as a by-product of hybridizing sexual species or a sexual male species with a unisexual female species [7, 62]. In the L-E system of Western Europe, P. esculentus males usually result only from crosses between P. esculentus females and heterogametic P. lessonae males. Backcrosses between such hybrid males and P. lessonae females result exclusively in P. esculentus females, due to the presence of clonal ridibundus genomes in the sperm that obviously contain female-determining factors [21, 22, 63].
Two lines of evidence support our finding that P. esculentus males from the upper Oder River valley are fertile and maintain the all-male hybrid lineage, by crossing with syntopic P. ridibundus females, i.e., they do not originate from primary hybridizations between their parental sexual species, or between sexual males and hybrid females.
First, our extensive sampling did not reveal the presence of P. lessonae or even a single hybrid female, necessary for the origin of diploid hybrid males [21]. The second line of evidence comes from independent statistical tests of allelic variation of the nuclear loci. The individually specific MLGs and the presence of both sexes indicate that both investigated taxa, P. ridibundus and P. lessonae, came from randomly mating sexual populations. Ridibundus alleles of P. esculentus males were grouped together with alleles of P. ridibundus males and females in two different clustering approaches (Fig. 2a, b). Moreover, 27 ridibundus genomes from P. esculentus males represent 27 MLGs, i.e., again all combinations are unique. It is therefore reasonable to assume that P. esculentus received its haploid ridibundus genomes from sympatric P. ridibundus females. On the other hand, we suggest that the lessonae genomes found in P. esculentus males from the Oder River are transmitted clonally only by these hybrids over the generations because the lessonae genome of P. esculentus males differed not only from the genome of sympatric P. ridibundus individuals but also from the genome of P. lessonae included in this study (Fig. 2a, b). The statistical test for the Psex value of these 27 lessonae genomes (1.50E−41) supports its clonal inheritance.
All-male hybrids represent a single hemiclone
Hemiclonal hybrids are genetically identical for half of the diploid parental genome [64] which they clonally pass on to the next generation. Analyses of ridibundus and lessonae allelic richness in all-male P. esculentus revealed that 13 loci within the ridibundus genome were polymorphic with the observed allelic frequencies (0.036–0.842). Together with the abovementioned observation that single ridibundus MLGs were not shared among individuals at all, it is reasonable to assume that these ridibundus genomes came from recombinant eggs of sexually reproducing P. ridibundus. In the lessonae genome, however, we observed only monomorphic loci with allele frequencies of 1.00. Therefore, it is proven that the lessonae genome is clonally inherited and represents a single clone (Figs. 2 and 3, Additional file 2: Table S2).
The characteristic of the lessonae genome ODERL1 only producing male hybrids is in accordance with the XX/XY type of sex determination hypothesized for the R-E system much earlier by Uzzell et al. [9]. However, our finding that all hybrid males possess an identical lessonae (ODERL1) is new finding. Most animal systems with female unisexuality comprise multiple clonal MLGs, as spined loach fishes of the genera Cobitis [65] and Poeciliopsis [66] and the Phoxinus eos-neogaeus complex [67]. Even where hybrids form a monophyletic group, e.g., Poecilia formosa, populations show a fairly high level of clonal diversity [68]. In Pelophylax populations of the L-E system, where both hybrid males and females coexist, the diversity of transmitted ridibundus clones is high [48, 69].
Origin of the hemiclonal line
Apparently, the existence and maintenance of all-male unisexuality among animals in general, and in Pelophylax water frogs in particular, hinge on the formation of a specific genome from a particular sexual species. In European water frogs, the genome of P. lessonae represented by the MLG ODERL1 seems to play a key role in the formation of all-male P. esculentus lineages. Supporting evidence comes from rare Central- and West-European mixed populations, where all-male P. esculentus are triploid and originate from clonal sperm with a diploid lessonae genome and haploid ridibundus eggs [26].
Despite the evidence for a single origin of the ODERL1 clone and the fact that it has evolved from the P. lessonae genepool, this clone does not cluster with other lessonae-specific MLGs (Fig. 2a, c) indicating clear differences between the ODERL1 clone and other P. lessonae genomes. Due to high distance between the two datasets of lessonae alleles, we hypothesize that present P. lessonae populations inhabiting areas close to the Oder River valley are not the most recent donors of the lessonae genome found in the all-male hybrid lineage of the R-E system populations investigated. It also seems unlikely that the lessonae clone recently originated from local P. lessonae. Because microsatellite markers have high mutation rates [70], an old hemiclone would comprise several MLGs that would merge into one multilocus lineage [65]. In contrast to this, we found a single lessonae-specific MLGs in all hybrids males, indicating a quite recent hybridization event. On the other hand, we cannot exclude a potential selective sweep in the lineage that might be caused by positive natural selection for the ODERL1 hemiclone [65]. Although age estimates of the ODERL1 clone and comparison with more distant P. lessonae populations are yet to be obtained, we hypothesize either an older in situ origin or a possible ex situ origin of the ODERL1 clone, i.e., some distance away from the local P. lessonae genepool.
Conclusions
By studying natural populations of Pelophylax water frogs along the upper Oder River valley, we discovered a clonal lessonae genome represented by a single hemiclone that is present in an all-male P. esculentus lineage living in sympatry with P. ridibundus males and females. This reproductive mode mirrors the one that previously has been identified in some all-female hybrid animals [5, 6, 71]. This hybridogenetic system in which all-male hybrids coexist with sexual males and females offers intriguing opportunities to compare evolutionary forces and genetic factors forming mating systems that may reverse those operating in all-female systems. These include phenomena like egg-dependent instead of sperm-dependent reproduction or male-male rather than female-female competition over the gamete donors. Additionally, this natural system provides a comparative model to a hemiclonal laboratory system developed in Drosophila melanogaster [72] for estimating quantitative genetic parameters in hemiclonal analyses [64]. Elucidating the mechanisms underlying these peculiarities might shed more light on the general processes of evolution of sex or mate-choice theory and contribute to understanding the origin and maintenance of sexual host-parasite dynamics.
Additional files
Table S1. Numbers of individual genomes included in datasets for a given analysis. (PDF 335 kb)
Table S2. DNA microsatellite data file for the 17 loci used in the study. (PDF 996 kb)
Table S3. Allozyme data file for the six loci used in the study. (PDF 514 kb)
Table S4. Species-specificity of microsatellite alleles used in this study. (PDF 259 kb)
Table S5. Summary statistics of microsatellite alleles found in P. lessonae, P. ridibundus, and P. esculentus from the R-E system. (PDF 317 kb)
Figure S1: Phylogenetic tree of DA distance of 10 microsatellite loci reconstructed in Populations (method UPGMA, 7 000 replicates, shown only bootstraps above 50 %, distance scale). One terminal branch represents one individual: Green color – P. lessonae, yellow color - L genome from P. esculentus, red colour – P. ridibundus, blue color – R genome from P. esculentus, violet color – P. kurtmuelleri. (JPEG 3388 kb)
Table S6. Multilocus Genotypes (MLGs) derived from P. lessonae, P. ridibundus and P. esculentus from the R-E system based on 17 microsatellite loci. (PDF 316 kb)
Acknowledgements
We thank Chris Johnson who proofread the manuscript and anonymous reviewers for their helpful comment.
Funding
We are grateful for the financial support from the Czech Science Foundation grant nos. 15-19947Y and 13-12580S, Czech Academy of Sciences of the Czech Republic No. RVO 67985904, Charles University in Prague Grant Agency No. 43-251468, Ministry of Education, Youth and Sports of the Czech Republic EXCELLENCE CZ.02.1.01/0.0/0.0/15_003/0000460 OP RDE. NBMP and H-UR were supported through grant no. 3100A0-120225/1 by the Swiss National Science Foundation.
Availability of data and materials
All the data supporting our results and conclusions along with sufficient details are listed in appendices.
Authors’ contributions
MD participated in the design of the study, collected samples, prepared DNA samples, analyzed microsatellite data, and wrote the initial draft of the manuscript. NBMP performed the amplification of microsatellite loci. HUR, JP, and KJ participated in the data interpretation and helped to draft the manuscript. LC conceived of the study and participated in its design, sampling, and manuscript writing. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All experimental procedures involving water frogs were performed in agreement with directives and under the supervision of the Ethical Committee of the Faculty of Science, Charles University, Prague, according to the directives of the State Veterinary Administration of the Czech Republic, permit number 34711/2010-30 from the Ministry of Agriculture of the Czech Republic. All institutional and national guidelines for the care and use of laboratory animals were followed.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13293-018-0172-z) contains supplementary material, which is available to authorized users.
Contributor Information
Marie Doležálková-Kaštánková, Email: dolezalkova@iapg.cas.cz.
Lukáš Choleva, Email: choleva@iapg.cas.cz.
References
- 1.Smith JM, Maynard-Smith J. The evolution of sex. Cambridge: Cambridge Univ. Press; 1978. [Google Scholar]
- 2.Charney ND. Relating hybrid advantage and genome replacement in unisexual salamanders. Evolution. 2012;66(5):1387–1397. doi: 10.1111/j.1558-5646.2011.01523.x. [DOI] [PubMed] [Google Scholar]
- 3.Dawley RM. An introduction to unisexual vertebrates. In: Dawley RM, Bogard JP, editors. Evolution and ecology of unisexual vertebrates New York State museum. Albany: New York Bull; 1989. pp. 1–18. [Google Scholar]
- 4.Vrijenhoek RC, Dawley RM, Cole CJ, Bogart JP. A list of the known unisexual vertebrates. In: Dawley RM, Bogard JP, editors. Evolution and ecology of unisexual vertebrates New York State museum. Albany: New York Bull; 1989. pp. 19–23. [Google Scholar]
- 5.Kearney M, Fujita MK, Ridenour J. Lost sex: the evolutionary biology of parthenogenesis. 2009. Lost sex in the reptiles: constraints and correlations; pp. 447–474. [Google Scholar]
- 6.Lamatsch DK, Stöck M. Lost sex: the evolutionary biology of parthenogenesis. 2009. Sperm-dependent parthenogenesis and hybridogenesis in teleost fishes; pp. 399–432. [Google Scholar]
- 7.Choleva L, Janko K, De Gelas K, Bohlen J, Šlechtová V, Rábová M, et al. Synthesis of clonality and polyploidy in vertebrate animals by hybridization between two sexual species. Evolution. 2012;66(7):2191–2203. doi: 10.1111/j.1558-5646.2012.01589.x. [DOI] [PubMed] [Google Scholar]
- 8.Günther R. Zum natürlichen Vorkommen und zur Morphologie triploider Teichfrösche,“Rana esculenta”, L., in der DDR (Anura, Ranidae). 1975;(51):145-58.
- 9.Uzzell T, Günther R, Berger L. Rana ridibunda and Rana esculenta: a leaky hybridogenetic system (Amphibia Salientia) Proc Acad Nat Sci Phila. 1977;128:147–171. [Google Scholar]
- 10.Günther R. Zur populationsgenetik der mitteleuropäischen wasserfrösche des Rana esculenta—synkleptons (Anura, Ranidae) Zool Anz. 1983;211(1/2):43–54. [Google Scholar]
- 11.Alves MJ, Coelho MM, Collares-Pereira MJ. Evolution in action through hybridisation and polyploidy in an Iberian freshwater fish: a genetic review. Genetica. 2001;111(1):375–385. doi: 10.1023/A:1013783029921. [DOI] [PubMed] [Google Scholar]
- 12.Morishima K, Oshima K, Horie S, Fujimoto T, Yamaha E, Arai K. Clonal diploid sperm of the diploid-triploid mosaic loach, Misgurnus anguillicaudatus (Teleostei:Cobitidae) J Exp Zool. 2004;301A(6):502–511. doi: 10.1002/jez.a.49. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto T, Yasui GS, Yoshikawa H, Yamaha E, Arai K. Genetic and reproductive potential of spermatozoa of diploid and triploid males obtained from interspecific hybridization of Misgurnus anguillicaudatus female with M. mizolepis male. J Appl Ichthyol. 2008;24(4):430–437. doi: 10.1111/j.1439-0426.2008.01131.x. [DOI] [Google Scholar]
- 14.Schmidt DJ, Bond NR, Adams M, Hughes JM. Cytonuclear evidence for hybridogenetic reproduction in natural populations of the Australian carp gudgeon (Hypseleotris: Eleotridae) Mol Ecol. 2011;20(16):3367–3380. doi: 10.1111/j.1365-294X.2011.05206.x. [DOI] [PubMed] [Google Scholar]
- 15.Lehtonen J, Schmidt DJ, Heubel K, Kokko H. Evolutionary and ecological implications of sexual parasitism. Trends Ecol Evol. 2013;28(5):297–306. doi: 10.1016/j.tree.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Schultz RJ. Hybridization, unisexuality, and polyploidy in the teleost Poeciliopsis (Poeciliidae) and other vertebrates. Am Nat. 1969;103(934):605–619. doi: 10.1086/282629. [DOI] [Google Scholar]
- 17.Bogart JP, Bi K, Fu J, Noble DWA, Niedzwiecki J. Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes. Genome. 2007;136:119–36. [DOI] [PubMed]
- 18.Stöck M, Ustinova J, Betto-Colliard C, Schartl M, Moritz C, Perrin N. Simultaneous Mendelian and clonal genome transmission in a sexually reproducing, all-triploid vertebrate. Proc R Soc B Biol Sci. 2012;279(1732):1293–1299. doi: 10.1098/rspb.2011.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vrijenhoek RC. Factors affecting clonal diversity and coexistence. Integr Comp Biol. 1979;19(3):787–797. [Google Scholar]
- 20.Uzzell T, Berger L. Electrophoretic phenotypes of Rana ridibunda, Rana lessonae, and their hybridogenetic associate, Rana esculenta. Proc Acad Nat Sci Phila. 1975;127(1975):13–24. [Google Scholar]
- 21.Graf JD, Polls PM. Evolutionary genetics of the Rana esculenta complex. In: Dawley RM, Bogard JP, editors. Evolution and ecology of unisexual vertebrates New York State museum. Albany: New York Bull; 1989. pp. 289–302. [Google Scholar]
- 22.Plötner J. Die westpaläarktischen Wasserfrösche. Beih Z Für Feldherpetologie Laurenti-Verl Bielef. 2005. [Google Scholar]
- 23.Günther R, Plötner J. Zur Problematik der klonalen Vererbung bei Rana kl esculenta (Anura). Beitr Zur Biol Bibliogr. 1960-1987 Eur Wasserfrösche. 1988. pp. 23–46. [Google Scholar]
- 24.Doležálková M, Sember A, Marec F, Ráb P, Plötner J, Choleva L. Is premeiotic genome elimination an exclusive mechanism for hemiclonal reproduction in hybrid males of the genus Pelophylax? BMC Genet. 2016;17(1):100. doi: 10.1186/s12863-016-0408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger L, Günther R. Inheritance patterns of water frog males from the environments of nature reserve Steckby, Germany. Zool Pol. 1991;37:87–100. [Google Scholar]
- 26.Tunner H, Heppich-Tunner S. A new population system of water frogs discovered in Hungary. Proc Sixth Ordinary Gen Meet Soc Eur Herpetol. 1992;19-23:453-460.
- 27.Rybacki M. Diploid males of Rana esculenta from natural populations in Poland producing diploid spermatozoa. Zool Pol. 1994;39(3-4).
- 28.Rybacki M. Structure of water frog populations [Rana esculenta complex] of the Wolin Island, Poland. Zool Pol. 1994;39(3-4):345-64.
- 29.Rybacki M, Berger L. Types of water frog populations (Rana esculenta complex) in Poland. Mitt Mus Nat.Kd Berl. Zool R. 2001;77:51–7.
- 30.Vinogradov AE, Borkin LJ, Günther R, Rosanov JM. Two germ cell lineages with genomes of different species in one and the same animal. Hereditas. 1991;114(3):245–251. doi: 10.1111/j.1601-5223.1991.tb00331.x. [DOI] [PubMed] [Google Scholar]
- 31.Pruvost NBM, Hoffmann A, Reyer HU. Gamete production patterns, ploidy, and population genetics reveal evolutionary significant units in hybrid water frogs (Pelophylax esculentus) Ecol Evol. 2013;3(9):2933–2946. doi: 10.1002/ece3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Günther R. Die Wasserfrösche Europas. Neue Brehm-Bücher. 1990;600:1–228. [Google Scholar]
- 33.Beerli P. Genetic isolation and calibration of an average protein clock in western Palearctic water frogs of the Aegean region. Ph.D. dissertation. Switzerland: Universität Zürich; 1994.
- 34.Valenta M, Hyldgaard-Jensen J, Jensen S. Interaction of veronal pyrophosphate citrate and protein with lactate dehydrogenase isoenzyme determination and kinetics. Acta Vet Scand. 1971;12:15. [PubMed] [Google Scholar]
- 35.Harris H, Hopkinson D. Handbook of enzyme electrophoresis in human genetics. 1976. [Google Scholar]
- 36.Buth DG, Murphy RW. Use of nicotinamide adenine dinucleotide (NAD)-dependent glucose-6-phosphate dehydrogenase in enzyme staining procedures. Stain Technol. 1980;55(3):173–176. doi: 10.3109/10520298009067234. [DOI] [PubMed] [Google Scholar]
- 37.Pasteur N, Pasteur G, Bonhomme F. Manuel technique de génétique par électrophorèse des protéines. Paris: Techn and Doc; 1987. [Google Scholar]
- 38.Arioli M, Jakob C, Reyer H-U. Genetic diversity in water frog hybrids (Pelophylax esculentus) varies with population structure and geographic location. Mol Ecol. 2010;19(9):1814–1828. doi: 10.1111/j.1365-294X.2010.04603.x. [DOI] [PubMed] [Google Scholar]
- 39.Garner TWJ, Gautschi B, Röthlisberger S, Reyer HU. A set of CA repeat microsatellite markers derived from the pool frog, Rana lessonae. Mol Ecol. 2000;9(12):2173–2175. doi: 10.1046/j.1365-294X.2000.105311.x. [DOI] [PubMed] [Google Scholar]
- 40.Christiansen DG, Reyer HU. From clonal to sexual hybrids: genetic recombination via triploids in all-hybrid populations of water frogs. Evolution. 2009;63(7):1754–1768. doi: 10.1111/j.1558-5646.2009.00673.x. [DOI] [PubMed] [Google Scholar]
- 41.Zeisset I, Rowe G, Beebee TJC. Polymerase chain reaction primers for microsatellite loci in the north European water frogs Rana ridibunda and R. lessonae. Mol Ecol. 2000;9:1173–1174. doi: 10.1046/j.1365-294x.2000.00954-2.x. [DOI] [PubMed] [Google Scholar]
- 42.Hotz H, Uzzel T, Guex G-T, Alpers D, Semlitsch RD, Beerli P. Microsatellites: a tool for evolutionary genetic studies of western Palearctic water frogs. Mitt Mus Natkd Berl, Zool R. 2001;77(1):43–50. [Google Scholar]
- 43.Hermaniuk A, Pruvost NBM, Kierzkowski P, Ogielska M. Genetic and cytogenetic characteristics of pentaploidy in water frogs. Herpetologica. 2013;69(1):36–45. doi: 10.1655/HERPETOLOGICA-D-12-00037. [DOI] [Google Scholar]
- 44.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4(3):535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- 45.Brookfield JF. A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol. 1996;5(3):453–455. doi: 10.1111/j.1365-294X.1996.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 46.Wagner AP, Creel S, Kalinowski ST. Estimating relatedness and relationships using microsatellite loci with null alleles. Heredity (Edinb) 2006;97(5):336–345. doi: 10.1038/sj.hdy.6800865. [DOI] [PubMed] [Google Scholar]
- 47.Dieringer D, Schlötterer C. MICROSATELLITE ANALYSER (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes. 2003;3(1):167–169. doi: 10.1046/j.1471-8286.2003.00351.x. [DOI] [Google Scholar]
- 48.Pruvost NBM, Mikulíček P, Choleva L, Reyer HU. Contrasting reproductive strategies of triploid hybrid males in vertebrate mating systems. J Evol Biol. 2015;28(1):189–204. doi: 10.1111/jeb.12556. [DOI] [PubMed] [Google Scholar]
- 49.Vinogradov AE, Borkin LJ, Günther R, Rosanov JM. Genome elimination in diploid and triploid Rana esculenta males: cytological evidence from DNA flow cytometry. Genome. 1990;33(5):619–627. doi: 10.1139/g90-092. [DOI] [PubMed] [Google Scholar]
- 50.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peakall R, Smouse PE. GenALEx 6.5: genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28(19):2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langella O. Populations version 1.2.30. Distributed by the author. Gif Sur Yvette: CNRS UPR9034; 1999.
- 53.Arnaud-Haond S, Belkhir K. GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes. 2007;7(1):15–17. doi: 10.1111/j.1471-8286.2006.01522.x. [DOI] [Google Scholar]
- 54.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 55.Earl DA, BM vH. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res. 2001;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 56.Arnaud-Haond S, Alberto F, Teixeira S, Procaccini G, Serrão EA, Duarte CM. Assessing genetic diversity in clonal organisms: low diversity or low resolution? Combining power and cost efficiency in selecting markers. J Hered. 2005;96(4):434–440. doi: 10.1093/jhered/esi043. [DOI] [PubMed] [Google Scholar]
- 57.Young AG, Hill JH, Murray BG, Peakall R. Breeding system, genetic diversity and clonal structure in the sub-alpine forb Rutidosis leiolepis F. Muell. (Asteraceae) Biol Conserv. 2002;106(1):71–78. doi: 10.1016/S0006-3207(01)00230-0. [DOI] [Google Scholar]
- 58.Sokal R, Michener C. A statistical method for evaluating systematic relationships. University of Kansas Science Bull. 1958;38:1409–1438. [Google Scholar]
- 59.Takezaki N, Nei M. Empirical tests of the reliability of phylogenetic trees constructed with microsatellite DNA. Genetics. 2008;178(1):385–392. doi: 10.1534/genetics.107.081505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plötner J, Grunwald C. A mathematical model of the structure and the dynamics of Rana ridibunda/esculenta-♂ ♂-populations (Anura, Ranidae) J Zool Syst Evol Res. 1991;29(3):201–207. doi: 10.1111/j.1439-0469.1991.tb01631.x. [DOI] [Google Scholar]
- 61.Wu CI, Johnson NA, Palopoli MF. Haldane’s rule and its legacy: why are there so many sterile males? Trends Ecol Evol. 1996;11:281–284. doi: 10.1016/0169-5347(96)10033-1. [DOI] [PubMed] [Google Scholar]
- 62.Vasil’ev VP, Akimova NV, Emel’yanova NG, Pavlov DA, Vasil’eva ED. Reproductive capacities in the polyploid males of spined loaches from the unisexual-bisexual complex, occurred in the Moscow river. Folia Biol. 2003;51:67–73. [PubMed] [Google Scholar]
- 63.Christiansen DG. Gamete types, sex determination and stable equilibria of all-hybrid populations of diploid and triploid edible frogs (Pelophylax esculentus) BMC Evol Biol. 2009;9(1):135. doi: 10.1186/1471-2148-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abbott JK, Morrow EH. Obtaining snapshots of genetic variation using hemiclonal analysis. Trends Ecol Evol. 2011;26:359–368. doi: 10.1016/j.tree.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 65.Janko K, Kotusz J, de Gelas K, Šlechtová V, Opoldusová Z, Drozd P, et al. Dynamic formation of asexual diploid and Polyploid lineages: multilocus analysis of Cobitis reveals the mechanisms maintaining the diversity of clones. PLoS One. 2012;7(9):e45384. doi: 10.1371/journal.pone.0045384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Angus RA, Schultz RJ. Clonal diversity in the unisexual fish Poeciliopsis monacha-lucida: a tissue graft analysis. Evolution. 1979;33(1):27–40. doi: 10.2307/2407362. [DOI] [PubMed] [Google Scholar]
- 67.Angers B, Schlosser IJ. The origin of Phoxinus eos-neogaeus unisexual hybrids. Mol Ecol. 2007;16(21):4562–4571. doi: 10.1111/j.1365-294X.2007.03511.x. [DOI] [PubMed] [Google Scholar]
- 68.Stöck M, Lampert KP, Möller D, Schlupp I, Schartl M. Monophyletic origin of multiple clonal lineages in an asexual fish (Poecilia formosa) Mol Ecol. 2010;19(23):5204–5215. doi: 10.1111/j.1365-294X.2010.04869.x. [DOI] [PubMed] [Google Scholar]
- 69.Mikulíček P, Kautman M, Demovič B, Janko K. When a clonal genome finds its way back to a sexual species: evidence from ongoing but rare introgression in the hybridogenetic water frog complex. J Evol Biol. 2014;27(3):628–642. doi: 10.1111/jeb.12332. [DOI] [PubMed] [Google Scholar]
- 70.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 71.Beukeboom LW, Vrijenhoek RC. Evolutionary genetics and ecology of sperm-dependent parthenogenesis. J Evol Biol. 1998;11(6):755–782. doi: 10.1007/s000360050117. [DOI] [Google Scholar]
- 72.Rice WR, Linder JE, Friberg U, Lew TA, Morrow EH, Stewart AD. Inter-locus antagonistic coevolution as an engine of speciation: assessment with hemiclonal analysis. Proc Natl Acad Sci. 2005;102:6527–6534. doi: 10.1073/pnas.0501889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Numbers of individual genomes included in datasets for a given analysis. (PDF 335 kb)
Table S2. DNA microsatellite data file for the 17 loci used in the study. (PDF 996 kb)
Table S3. Allozyme data file for the six loci used in the study. (PDF 514 kb)
Table S4. Species-specificity of microsatellite alleles used in this study. (PDF 259 kb)
Table S5. Summary statistics of microsatellite alleles found in P. lessonae, P. ridibundus, and P. esculentus from the R-E system. (PDF 317 kb)
Figure S1: Phylogenetic tree of DA distance of 10 microsatellite loci reconstructed in Populations (method UPGMA, 7 000 replicates, shown only bootstraps above 50 %, distance scale). One terminal branch represents one individual: Green color – P. lessonae, yellow color - L genome from P. esculentus, red colour – P. ridibundus, blue color – R genome from P. esculentus, violet color – P. kurtmuelleri. (JPEG 3388 kb)
Table S6. Multilocus Genotypes (MLGs) derived from P. lessonae, P. ridibundus and P. esculentus from the R-E system based on 17 microsatellite loci. (PDF 316 kb)
Data Availability Statement
All the data supporting our results and conclusions along with sufficient details are listed in appendices.


