Abstract
Proteases are in the center of many diseases, and consequently, proteases and their substrates are important drug targets as represented by an estimated 5–10% of all drugs under development. Mass spectrometry has been an indispensable tool for the discovery of novel protease substrates, particularly through the proteome-scale enrichment of so-called N-terminal peptides representing endogenous protein N termini. Methods such as combined fractional diagonal chromatography (COFRADIC)1 and, later, terminal amine isotopic labeling of substrates (TAILS) have revealed numerous insights into protease substrates and consensus motifs. We present an alternative and simple protocol for N-terminal peptide enrichment, based on charge-based fractional diagonal chromatography (ChaFRADIC) and requiring only well-established protein chemistry and a pipette tip. Using iTRAQ-8-plex, we quantified on average 2,073 ± 52 unique N-terminal peptides from only 4.3 μg per sample/channel, allowing the identification of proteolytic targets and consensus motifs. This high sensitivity may even allow working with clinical samples such as needle biopsies in the future. We applied our method to study the dynamics of staurosporine-induced apoptosis. Our data demonstrate an orchestrated regulation of specific pathways after 1.5 h, 3 h, and 6 h of treatment, with many important players of homeostasis targeted already after 1.5 h. We additionally observed an early multilevel modulation of the splicing machinery both by proteolysis and phosphorylation. This may reflect the known role of alternative splicing variants for a variety of apoptotic genes, which seems to be a driving force of staurosporine-induced apoptosis.
Proteolysis plays a crucial role in maintaining cellular homeostasis by modulating protein function and activity and its dysregulation underlies many diseases such as cancer and Alzheimer (1–3). Through proteolytic cleavage, novel protein N termini are generated. The identification of these so-called neo N termini is an important step toward understanding which proteins are substrates of a specific protease and revealing regulatory proteolytic networks in health and disease. Moreover, it also allows identifying protease cleavage motifs, which is important for developing protease inhibitors or chemical proteomics tools.
As protein N termini and, more importantly, neo N termini are significantly underrepresented in the proteome, specific methods have been developed for the enrichment of N-terminal peptides followed by mass spectrometry (N-terminomics) to enable the system-wide identification of protease substrates and cleavage patterns (4). Several methods, namely combined fractional diagonal chromatography (COFRADIC) (5), subtiligase N-terminal labeling and enrichment (6) and, later, terminal amine isotopic labeling of substrates (TAILS) (7) pioneered the field of N-terminomics. COFRADIC and TAILS utilize the specific labeling of protein N termini (and Lys residues) as an initial step of the enrichment procedure. Upon proteolytic cleavage as part of the common bottom-up proteomics strategy, this labeling allows quantifying N-terminal peptides but also distinguishing them from internal peptides with free N termini generated during in vitro digestion. Both methods have been used in numerous studies providing novel insights into proteases and their substrates (1, 8–10). Nevertheless, likely due to challenges in technical and (particularly in the past) data analysis aspects, the number of labs worldwide applying N-terminomics methods to screen for protease substrates is still limited.
We recently introduced an alternative HPLC-based strategy for N-terminal peptide enrichment, charge-based fractional diagonal chromatography (ChaFRADIC) (11). The method depends on the separation of peptides into distinct charge-state fractions at pH 2.7, where a peptide's net charge in solution is mainly defined by the number of positively (Arg, Lys, His residues and free N termini) and negatively (e.g. phosphorylation) charged groups (Fig. 1 A) (12). Though providing a high sensitivity for N-terminomics studies, the protocol requires a dedicated, highly reproducible HPLC system with automatic fractionation, which is associated with great costs and maintenance expenses. We therefore advanced the method into charge-based fractional diagonal chromatography in a pipette tip (ChaFRAtip). ChaFRAtip only requires well-established protein chemistry and minimal equipment: A pipette tip with a cellulose frit, strong cation exchange chromatography (SCX) beads and common buffers. We demonstrate that the tip-based approach is efficient and reproducible and that, in conjunction with iTRAQ-8-plex labeling, it allows quantifying more than 2,000 N-terminal peptides across eight samples using only as little as 4.3 μg of protein per condition/channel.
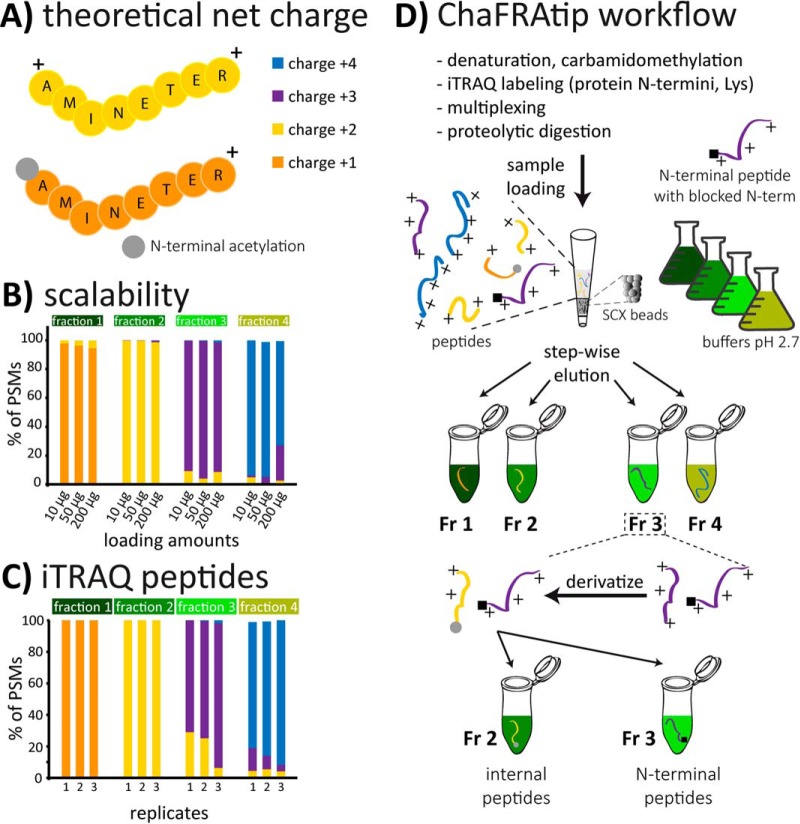
Fig. 1.
(A) The theoretical net charge (TNC) of a fully tryptic peptide at pH 2.7 is typically +2. Presence of His residues or missed cleavages leads to increased TNC, N-terminal acetylation to reduced TNC. (B) Tip-based fractionation of peptides according to their TNC can be scaled to the amount of cell lysate without compromising separation power and (C) also works for iTRAQ-labeled peptides. (D) For ChaFRAtip, free protein N termini and lysines are labeled with iTRAQ/TMT on the protein level; samples are pooled and digested with trypsin. Peptides are fractionated on-tip according to their TNC. Each collected fraction is acetylated, thus reducing the TNC of internal peptides by 1. In a second tip-based fractionation, internal peptides with reduced TNC are removed and N-terminal peptides are collected for subsequent LC-MS analysis.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
The experiments to evaluate the performance and scalability of charge-based separation of peptides in a tip (Fig. 1) were done in technical triplicates using aliquots of the same HeLa digest (see Supplemental Methods for details). Data are represented as means of the three respective replicates (Fig. 1B, scalability unlabeled peptides) or as individual replicates (Fig. 1C, iTRAQ-labeled peptides) and all relative standard deviations (peptide spectrum matches (PSMs) charge-state distribution per fraction) were below 8.5% for the unlabeled peptides and below 12.5% for the iTRAQ-labeled peptides.
For the comparison between HPLC-based ChaFRADIC and ChaFRAtip, three biological replicates of SH-SY5Y cells, each, were either treated with staurosporine in DMSO to induce apoptosis or with DMSO as controls. After labeling and pooling, the samples were split in six aliquots, three of which were used for HPLC-based ChaFRADIC and three for ChaFRAtip. Identified N-terminal peptides were compared based on their concatenated sequences and Uniprot accessions. Only unique N termini that were found in at least two out of three replicates per approach (HPLC versus tip) were used for the comparison (Fig. 2). To generate a consensus motif and a network, only unique N termini that were at least threefold regulated in at least three out of the six datasets replicates were considered.
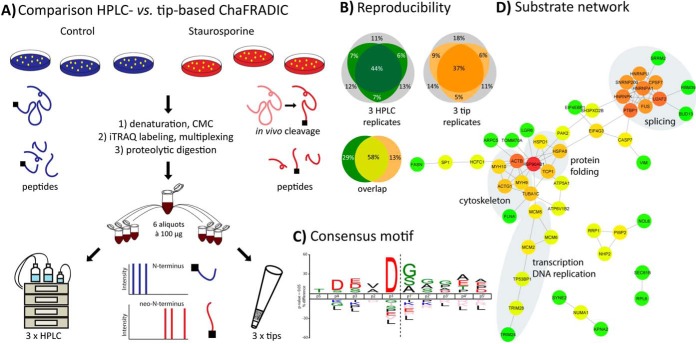
Fig. 2.
(A) Workflow comparing HPLC- and tip-based ChaFRADIC using SH-SY5Y cells that were either treated with vehicle or with staurosporine to induce an apoptotic proteolytic cascade. (B) Both strategies show similar reproducibility across technical replicates and a certain degree of complementarity for all quantified N-terminal peptides. Those quantified in at least two out of three replicates are color-coded. (C) Neo-N-terminal peptides (>2-fold up-regulation, p < 0.05) clearly point toward caspase activity (45). (D) STRING network of identified proteolytic substrates. Colors represent the degree of connectivity (red high, green low).
For the study of the temporal dynamics of staurosporine-induced apoptosis, SH-SY5Y cells were treated with staurosporine for 1.5 h, 3 h, and 6 h or served as controls, in biological duplicate per time point (iTRAQ 8-plex labeling). Considering only N termini that were quantified in at least three replicates per time point, the median log2 ratio and a p value based on a two-sided t test were determined per biological replicate (i.e. 1.5 h A and B, 3 h A and B, 6 h A and B). Only (i) unique N-terminal peptides that were (ii) at least twofold regulated compared with control and (iii) had a p value <0.05 were considered as regulated for the respective time point.
Tip-Based Separation of Peptides (Unlabeled and iTRAQ Labeled)
Details are given in the Supplemental Methods.
Comparison of HPLC and Tip-Based ChaFRADIC
Cell Culture and Sample Preparation
Six biological replicates of SH-SY5Y cells were grown to more than 90% confluency in Dulbecco's Modified Eagle's Medium (PAN-Biotech). The medium was replaced by fresh medium containing either 1 μm staurosporine in DMSO (Sigma Aldrich) or DMSO (both 10 μl) to three replicates, each, followed by incubation for 6 h at 37 °C. The supernatant of each flask containing floating cells was discarded and cells were washed with ice-cold PBS, scraped, and collected by centrifugation with 300 g for 5 min at RT. Cell pellets were washed with 1 ml PBS and collected by another centrifugation step. The final cell pellets were lysed, carbamidomethylated and subjected to BCA assay for protein concentration (see Supplemental Methods). Afterward, 100 μg (according to BCA) of protein were taken from each sample and subjected to ethanol precipitation. Protein pellets were resolubilized with 10 μl of 10% SDS and 40 μl of iTRAQ labeling buffer (0.5 m triethylammonium bicarbonate (TEAB) in 20% (v/v) of isopropanol) were added in order to have a final concentration of 2% SDS. Next, 80 μl of isopropanol was added to six iTRAQ 8-plex (113, 114, 115, 116, 119, 121) reagents, each label was added to a different sample, and incubated at room temperature (RT; 25 °C) for 2 h. Reactions were quenched with 60 mm glycine for 10 min at RT, followed by addition of 130 mm hydroxylamine for 10 min at RT. Afterward, samples were pooled and directly subjected to another ethanol precipitation step. Protein pellets were resolubilized with 150 μl of 6 m guanidinium chloride (GuHCl), which was diluted to 0.2 m with 50 mm TEAB, pH 7.8. Next, acetonitrile (ACN) and CaCl2 were added to a final concentration of 5% and 2 mm, respectively. Finally, trypsin was added in 1:20 (w/w, enzyme to substrate) ratio and incubated at 37 °C for 14 h. Proteolytic digests were controlled using monolithic reversed phase chromatography, as described previously (13). Peptides were desalted by solid-phase extraction (Oasis HLB Extraction Cartridge, 10 mg). After desalting, 1 μg aliquot was analyzed by nano-LC-MS/MS for a global analysis in order to (i) determine systematic errors from sample preparation based on iTRAQ protein ratios and to (ii) determine labeling efficiency. The rest of the sample was dried, resolubilized in 310 μl SCX buffer A, and split into six aliquots à 51 μl corresponding to ∼100 μg of peptide, each.
Enrichment of N-Terminal Peptides Using HPLC ChaFRADIC
Three technical aliquots (100 μg each, BCA results) were fractionated using HPLC-based ChaFRADIC and the other three (100 μg each, BCA) using ChaFRAtip. HPLC separations were performed using a U3000 HPLC system (Thermo Scientific) and a 150 × 1 mm POLYSULFOETHYL A column (PolyLC, Columbia, U.S., 5 μm particle size, 200 Å pore size), in combination with a tertiary buffer system consisting of SCX buffer A (10 mm KH2PO4, 20% ACN, pH 2.7), SCX buffer B (10 mm KH2PO4, 250 mm KCl, 20% ACN, pH 2.7), and SCX buffer C (10 mm KH2PO4, 600 mm NaCl, 20% ACN, pH 2.7). 51 μl of the resolubilized peptides were separated at a flow rate of 80 μl/min. Peptides were separated with an optimized gradient to efficiently separate different charge states, and fractions were automatically collected using the U3000 fractionation option. The gradient was as follows: 100% A for 10 min followed by a linear increase from 0% to 15% B in 9.3 min. Afterward, B was kept at 15% for 8.7 min and then the gradient linearly increased from 15% to 30% B for 8 min. Then B was kept constant at 30% for 11 min and linearly increased to 100% in 5 min. After 5 min at 100% B, C was increased to 100% in 1 min and kept constant for 5 min. Finally, A was increased to 100% in 1 min, and the column was re-equilibrated at 100% A for 20 min. Per replicate, four fractions corresponding to charge states +1, +2, +3, and +4 were collected, as indicated in Supplemental Fig. 1. Collected fractions were completely dried under vacuum and brought to 300 μl with 200 mm Na2HPO4, pH 8.0, to a final pH of ∼7.0. Free N termini of internal peptides were derivatized with N-Hydroxysuccinimide (NHS)-trideutero acetate in two steps, modified from Staes et al. (14) Initially NHS-trideutero acetate was added to a final concentration of 20 mm, and samples were incubated at 37 °C for 1 h, followed by addition of another 10 mm NHS-trideutero acetate under the same conditions. After 2 h of total incubation time, the reaction was quenched using 60 mm glycine for 10 min at RT, followed by incubation for 5 min at 95 °C. Fractions were acidified with 10% triflouroacetic acid (TFA) and desalted using Poros Oligo R3 reversed-phase material. Bound peptides were washed twice with 0.1% of formic acid (FA) and afterward eluted with 100 μl of 70% (v/v) ACN, 0.1% FA. After vacuum drying peptides were resolubilized in 51 μl of SCX buffer A. Each first-dimension fraction was separated and fractionated in a second SCX dimension under exactly the same conditions as mentioned above. Whereas N-terminal peptides retained their retention time window (i.e. charge-state window), internal peptides shifted to earlier retention times based on the reduction in theoretical net charge induced by the N-terminal acetylation. After collection, fractions were desalted, dried, and resolubilized in 22 μl TFA 0.1%. From each fraction, 2/3 were taken for nano-LC-MS/MS.
Enrichment of N-terminal Peptides Using ChaFRAtip
Column preparation (25 μl of SCX material), column conditioning, sample loading (resolubilized in 50 μl SCX buffer A), and first-dimension fractionation were done as described in the Supplemental Methods and Table S1 in the Supplemental Methods. Collected fractions were dried and brought to 300 μl with 200 mm Na2HPO4, pH 8.0. Next, free N termini of internal peptides were derivatized as described above, followed by desalting. After vacuum drying peptides were resolubilized in 50 μl of SCX buffer A. Second-dimension tip-based fractionation was conducted as described in Table S2 in the Supplemental Methods Fractions were desalted, dried, and resolubilized in 22 μl TFA 0.1%. From each fraction 2/3 were taken for nano-LC-MS/MS.
LC-MS Analysis
Fractions were analyzed by nano-LC-MS/MS using an Orbitrap Fusion Lumos (Thermo Scientific), online-coupled to a U3000 HPLC system using a charge-state-dependent decision tree (15), as detailed in the Supplemental Methods. Notably, per peptide, two fragment ion spectra were acquired, one high-resolution MS/MS in the Orbitrap that was used for quantification and one low-resolution MS/MS in the ion trap that was used for identification (see Supplemental Methods).
Data Analysis
Raw data were searched against an Uniprot human database (July 2015, 20,207 target sequences) using Mascot 2.4 (Matrix Science) and the Proteome Discoverer software v1.4 including the reporter ion quantifier and percolator nodes, employing a two-step strategy and semi-ArgC enzyme specificity with a maximum of two missed cleavage sites. To enable the quantification of both classes of N-terminal peptides, those with N-terminal iTRAQ label and those with endogenous N-acetylation, we performed a two-step search strategy: First, data were searched with iTRAQ 8-plex (+304.2053 Da) at peptide N termini and iTRAQ 8-plex at Lys as fixed modifications. Second, N-terminal acetylation (+42.0105 Da) and iTRAQ 8-plex at Lys were set as fixed modifications. In both cases, carbamidomethylation of Cys (+57.0214 Da) was selected as fixed and oxidation (+15.9949 Da) of methionine as variable modifications. As filters, high confidence corresponding to a false discovery rate < 1% and search engine rank 1 were used. Mass tolerances were set to 10 ppm for MS and 0.5 Da for MS/MS. PSMs representing the same (i) amino acid sequence and (ii) protein accession were grouped and their corresponding reporter ion areas summed (Supplemental Table 1). Summed areas were normalized using normalization factors derived from the global analysis in order to compensate for systematic differences in sample amount across channels. Normalized areas were used to calculate the ratios of staurosporine-treated to control cells (average reporter ion area of three staurosporine replicates/average reporter ion area of three control replicates) and to determine p values based on a two-sided t test. Data analysis was first conducted for each set individually, and datasets were merged based on unique N-terminal peptides (i.e. concatenated sequence, Uniprot accession, modification set). Based on the corresponding unique entries, the overlap between the three HPLC and the three tip replicates was assessed (Fig. 2B). To compare HPLC and tip-based enrichment, only N-terminal peptides quantified at least twice with either of the two methods were considered (Fig. 2B). Next, (i) only unique N-terminal peptides (i.e. representing only a single Uniprot entry) that were (ii) at least threefold regulated in (iii) at least three out of the six datasets were used to determine a consensus motif considering the amino acids p5-p5′ according to Schechter and Berger (16) (Fig. 2C). Corresponding protein entries were used to generate a high confidence STRING protein network using Cytoscape (17) (http://www.cytoscape.org/).
Studying the Dynamics of Staurosporine-Induced Apoptosis Using ChaFRAtip
Cell Culture, Sample Preparation, ChaFRAtip, nano-LC-MS/MS, Data Analysis
Neuroblastoma SH-SY5Y cells were grown to more than 70% confluency (107 cells per T75-flask) in Dulbecco's Modified Eagle's Medium (PAN-Biotech). The medium was replaced by fresh medium containing either staurosporine 1 μm in DMSO (Sigma Aldrich) or DMSO (both 10 μl) and incubated for: 15 min, 1.5 h, 3 h, and 6 h in order to check for progression of apoptosis and activation of caspase proteases. At each time point, cells were examined under a microscope for changes in cell shape and detachment as indication for apoptosis (Supplemental Fig. 2). The medium supernatant of each flask, which contained floating cells, was collected, and cells were washed with ice-cold PBS. Cells were then treated with trypsin (0.025% in EDTA PBS) for 2–3 min at 37 °C. The medium supernatant and the trypsinized cells were pooled and cells isolated by centrifugation at 270 g for 5 min. Cell pellets were lysed in PBS containing 0.1% Triton X-100 and incubated with compound SV149 (an activity-based probe for labeling active caspase proteases) at RT for 45 min. Labeling reactions were quenched by adding Laemmli sample buffer, and half of the sample was resolved by SDS-PAGE. Active caspases were visualized by scanning of the carboxytetramethylrhodamine (TAMRA) fluorescence using a Typhoon Trio+ (Supplemental Fig. 2). For the MS samples, a similar procedure was applied. The cell pellets collected after centrifugation at 270 g were flash-frozen in liquid nitrogen and stored at −80 °C for subsequent MS preparations. Only the last three time points (1.5, 3, and 6 h) were harvested, including two biological replicates per time point. Two additional T75 flasks treated with DMSO for 6 h were used as controls. Lysis, carbamidomethylation, and iTRAQ labeling were conducted as above, however, including eight samples (4 conditions × 2 biological replicates) and iTRAQ labels. Protein concentrations were determined by amino acid analysis, and the total amount per sample was 210 μg. After pooling, the sample was divided into six aliquots à 35 μg. Then, in order to assess the robustness of the method three aliquots, each was enriched for N-terminal peptides with ChaFRAtip by two different individuals. nano-LC-MS/MS measurements were done as described in the Supplemental Methods. Per technical replicate, four fractions were measured by LC-MS (90 min total MS time per fraction; total length of HPLC program: 155 min, including equilibration, separation, extensive washing, and re-equilibration on a 50 cm column). For data analysis, endogenously acetylated peptides were excluded, even if quantified based on iTRAQ-labeled Lys residues. Each replicate was individually analyzed (Supplemental Table 2). PSMs quantifying the same unique entry (i.e. concatenated Uniprot accession and sequence) were grouped, and the corresponding reporter ion intensities summed. Normalization was done as described above. For each biological replicate (A and B), the four corresponding reporter ion sums were used to calculate a median intensity (A and B). These median intensities were used to scale the corresponding reporter ion intensities. Based on the unique entries, the six datasets were merged. For each replicate and time point, ratios compared with the corresponding control were determined using the reporter ion intensities and log2-transformed. Considering only N termini that were quantified in at least three replicates per time point, the median log2 ratio and a p value based on a two-sided t test were determined per biological replicate (i.e. 1.5 h A and B, 3 h A and B, 6 h A and B). Only (i) unique N-terminal peptides that were (ii) at least twofold regulated compared with control and (iii) had a p value <0.05 were considered as regulated for the respective time point. These regulated N-terminal peptides were considered to generate a heatmap using Perseus (18) and a STRING protein network using Cytoscape (Fig. 3).
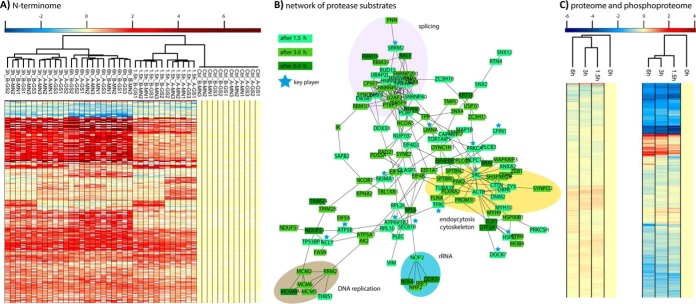
Fig. 3.
(A) Temporal profiles of N-terminal peptides regulated upon staurosporine treatment for 0, 1. 5, 3, and 6 h. Log2-fold changes (fc) compared with the 0 h controls are shown across a total of two biological (A, B) and six technical replicates (GS1/2/3, MN1/2/3) per time point. (B) Protein interaction network analysis of regulated protease substrates found at least twofold regulated with a p value <0.05 in both biological replicates. Several key players of cellular homeostasis are cleaved already after 1.5 h. In particular, the splicing machinery, the cytoskeleton, and endocytosis are early targets, whereas DNA replication proteins are cleaved at later stages. (C) Whereas the >5,500 proteins quantified with at least two unique peptides do not show major changes in abundance, most of the ∼5,900 high-confidence phosphopeptides show a significant down-regulation. Interestingly, a group of up-regulated phosphopeptides comprises many proteins of the splicing machinery. Heatmaps were generated using Perseus (18).
Quantitative Proteome and Phosphoproteome: Sample Preparation and Nano-LC-MS/MS
Sample preparation and digestion were performed using the filter-aided sample preparation (FASP) (19, 20) protocol with minor changes. Briefly, cell lysates corresponding to 50 μg (amino acid analysis quantified) of protein were diluted to 450 μl with freshly prepared 8 m urea/100 mm Tris-HCl, pH 8.5 (21), and placed on a Nanosep centrifugal device (30 kDa, Pall). The devices were centrifuged at 13,900 g at RT for 20 min. All the following centrifugation steps were performed at 13,900 g, RT, 15 min. Residual SDS was removed by three washing steps with 100 μl of 8 m urea/100 mm Tris-HCl, pH 8.5, followed by three steps with 100 μl of 50 mm TEAB, pH 7.8. Next, 100 μl of digestion buffer comprising 1:20 (w/w) sequencing grade trypsin, 0.2 m GuHCl, 2 mm CaCl2 in 50 mm TEAB, pH 7.8, were added and incubated at 37 °C for 14 h. The generated tryptic peptides were recovered by centrifugation followed by two consequent washing steps with 50 μl of 50 mm TEAB, pH 7.8, and 50 μl of ultrapure water. Peptides were acidified with 10% TFA (pH < 3.0), and digests were quality controlled as described previously (13). All samples were completely dried and resolubilized in 0.5 m TEAB, pH 8.5. The peptide concentration was determined using a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Scientific, Germany). Before labeling with iTRAQ 8-plex reagents (22), each sample (∼1 μg) was analyzed by nano-LC-MS/MS to adjust minor differences in sample amounts due to pipetting errors or incorrect protein/peptide concentration estimation based on the alignment of total ion chromatograms. Each sample was labeled with iTRAQ reagents according to the manufacturer's instructions (AB SCIEX). The multiplexed sample (∼400 μg) was divided into two parts, i.e. 50 μg for global proteome analysis and 350 μg for phosphopeptide enrichment, and both parts were dried completely in a SpeedVac. Phosphopeptide enrichment was done using TiO2 (23, 24) beads followed by hydrophilic interaction chromatography (25). In total three hydrophilic interaction chromatography fractions were collected, dried and stored at −80 °C until further use. For the global proteome analysis, dried peptides were resolubilized in 200 μl of 0.5% TFA (pH < 3.0) and desalted using C18 SPEC tips (4 mg, Varian). Eluted peptides were dried and resolubilized in 10 mm ammonium formate, pH 8.0. 25 μg of multiplexed sample was fractionated by reversed-phase chromatography at pH 8.0 on a Biobasic C18, 0.5 × 150 mm, 5 μm particle size column using an UltiMate 3000 LC system (both Thermo Scientific, Germany) with buffers A: 10 mm ammonium formate, pH 8.0, and B: 84% ACN in 10 mm ammonium formate, pH 8.0. Peptides were loaded onto the column with buffer A at a flow rate of 12.5 μl/min and separated using the following gradient: 3% B for 10 min, 3–45% B in 40 min, 45–60% B in 5 min, 60–95% B in 5 min, 95% B hold for 5 min, 95–3% B in 5 min, re-equilibration of the column with 3% B for 20 min. 16 fractions were collected at 1 min intervals from min 5 to 80 in a concatenation mode and all fractions were completely dried and stored at −80 °C until further use. Dried hydrophilic interaction chromatography fractions were resolubilized in 15 μl of 0.1% TFA and analyzed by nano-LC-MS/MS using an Ultimate 3000 nano RSLC system coupled to a Q Exactive HF (both Thermo Scientific), as detailed in the Supplemental Methods. The Q Exactive HF was operated in data-dependent acquisition mode, and MS survey scans were acquired from m/z 300 to 1,500 at a resolution of 60,000 using the polysiloxane ion at m/z 371.1012 as lock mass (26). The 15 most intense ions were isolated with a 0.4 m/z window and fragmented by higher energy collisional dissociation (HCD) with a normalized collision energy of 33%, taking into account a dynamic exclusion of 20 s. All 16 pH 8.0 fractions were resolubilized in 45 μl of 0.1% TFA, and 1/3 of each sample was analyzed by nano-LC-MS/MS as above, with minor changes (see Supplemental Methods).
Quantitative Proteome and Phosphoproteome: Search Parameters
All iTRAQ raw data were processed simultaneously using the MudPIT option with Proteome Discoverer 1.4 and searched against the same database as used above. To maximize the number of PSMs, we included three different search algorithms (Mascot (27), SEQUEST (28), and MS Amanda (29)) using the same set of parameters. Mass tolerances were set to 10 ppm and 0.02 Da for MS and MS/MS, respectively; trypsin as enzyme with a maximum of two missed cleavages; carbamidomethylation of Cys (+57.0214 Da) and iTRAQ-8-plex on N terminus and Lys (+304.2053 Da) as fixed modifications; oxidation of Met (+15.9949 Da); and for the phosphoproteome data phosphorylation of Ser/Thr/Tyr (+79.9663 Da) as variable modifications. All data were exported with the following filter criteria: peptide PSMs with false discovery rate < 1% (high confidence setting), search engine rank 1. For the phosphoproteome data, we additionally used the phosphoRS node (version 3.1)(30) for calculating site localization probabilities.
Quantitative Proteome and Phosphoproteome: iTRAQ Data Normalization and Statistical Evaluation
For the global proteome, instead of working with seven iTRAQ ratios against one reference sample, as provided by the Proteome Discoverer software, here ratios were transformed into normalized abundance values (NAVs). Thus, for statistical comparison a hypothetical ratio (i.e. 113/113) was generated in order to have eight data points in an experiment. The individual channel ratios of each protein were log2 transformed, and for each channel a median over all proteins was (MD1) calculated. Next, a second median (MD2) was generated by taking the medians across all channels. MD2 was then subtracted from individual MD1s to deduce the normalization values (NVs) for each channel. These NVs were subsequently used to normalize the respective log2 transformed ratios for each protein/channel (normalized ratios, NR). Next, a third median (MD3, scaling factor) was calculated by taking the NR for each protein across all channels. This MD3 was then subtracted from the NR of each protein to get NAVs for each protein. Finally, ratios were calculated by dividing the average NAVs or each time point, i.e. 1.5 h staurosporine, by the average NAVs of the controls. Only proteins quantified by at least two unique peptides were considered (Supplemental Table 3). A similar strategy was employed for the phosphoproteome data analysis. However, reporter ion areas of unique phosphorylated PSMs were considered instead of ratios. Next, for unique phosphopeptides PSMs representing the same protein, peptide sequence, and phosphorylation site, areas were summed per channel, and ratios were calculated as aforementioned. Only unique phosphopeptides with phosphoRS probabilities >90% were considered (Supplemental Table 3). The ratios of the 5,877 and 5,547 quantified high-confidence phosphopeptides and proteins, respectively, were used to generate heatmaps using Perseus (Fig. 3).
Deposition of Data
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (31) partner repository with the dataset identifiers PXD005954, PXD006594, PXD006595.
RESULTS
After systematic optimization of the SCX tip conditions, namely (i) preparation of the tips including frit type/design and (ii) SCX beads to peptide ratio, as well as for the individual (elution and washing) steps of the protocol: (iii) buffer compositions and combinations, (iv) volumes, and (v) centrifugation conditions, the final protocol allowed us to reproducibly separate unlabeled (Fig. 1B) and iTRAQ-labeled (Fig. 1C) tryptic peptides. The obtained fractions were on average 85–90% enriched in distinct charge states +1, +2, +3, and +4. The fractionation is scalable (Fig. 1B), can be applied to complex cell lysates and purified proteins, and allowed us to completely transfer the originally HPLC-based N-terminal ChaFRADIC (11) enrichment to the simplified ChaFRAtip setup (Fig. 1D). This comes along with a substantial reduction of time and costs, as dedicated and reproducible HPLC instrumentation is no longer required and samples can be processed in parallel.
We wondered whether ChaFRAtip can provide results comparable to the original HPLC-based setup but at a fraction of the costs and effort. Therefore, we cultivated SH-SY5Y cells in biological triplicates and treated each replicate either with vehicle or staurosporine to induce apoptosis. The resulting six samples (100 μg of protein each, based on BCA) were individually labeled with iTRAQ-8-plex reagents on the protein level, multiplexed, enzymatically digested with trypsin, and separated into six aliquots of 100 μg peptide each. Three aliquots each were used for HPLC-based ChaFRADIC and ChaFRAtip, followed by nano-LC-MS/MS of 2/3 per enriched fraction (Fig. 2A). Indeed, we achieved a similar level of technical reproducibility for both methods (Fig. 2B) and quantified 975 ± 24 (HPLC) and 783 ± 71 (tips) unique N-terminal peptides at 1% false discovery rate. iTRAQ-based quantification was highly reproducible across technical and biological replicates as well as both methods (Supplemental Table 1). Significantly differential neo N-terminal peptides clearly point to the expected caspase activity (Fig. 2C), and the identified substrates are involved e.g. in splicing, protein folding, translation, and transcription (Fig. 2D). Thus, ChaFRAtip enables highly sensitive N-terminomics with similar performance as the much more costly HPLC-based workflow.
We decided to illustrate the power of the ChaFRAtip method by studying the dynamics of staurosporine-induced apoptosis in SH-SY5Y cells. Based on initial temporal profiles of staurosporine-induced caspase activation (Supplemental Fig. 2), we selected four time points: 0.0 h (control), 1.5 h, 3.0 h, and 6.0 h of staurosporine treatment, each in biological duplicates. Following iTRAQ labeling of 26 μg (based on amino acid analysis) of peptide per sample (8 × 26 = 208 μg in total), multiplexing, and trypsin digestion, we split the resulting sample in six aliquots à 35 μg, corresponding to 4.3 μg of protein per time point and replicate. To assess the potential for a method transfer to other labs, we had two different individuals conduct the entire ChaFRAtip enrichment with three samples independently, resulting in a total of six technical replicates. On average, we quantified 2,073 ± 52 unique N-terminal peptides representing only unique protein entries with a strong correlation across biological and technical replicates (Supplemental Tables 2 and 3, Fig. 3A). This unprecedented sensitivity, corresponding to the quantification of 482 unique N-terminal peptides per μg of protein lysate, will allow future quantitative N-terminomics from limited sample amounts, such as clinical biopsies. A total of 288 N termini (220 proteins) showed significant regulation after applying stringent cutoffs: Only unique N-terminal peptides were considered regulated if (i) they were quantified in at least three technical replicates of both biological replicates and showed (ii) at least a twofold median regulation compared with the controls (iii) with a p value < 0.05 (iv) in both biological replicates. We furthermore determined the inter (between all six technical replicates) and intra (between averages of biological replicates) relative standard deviations from our quantitative data presented in Supplemental Table 2 (sheet “i - summary raw”) and obtained average relative standard deviations of 5% (inter) and 9% (intra).
After 1.5 h of staurosporine treatment, 81 protease substrates were identified, followed by 109 and 30 additional substrates after 3 h and 6 h, respectively (Supplemental Tables 2 and 3). These substrates clearly confirm the expected caspase activity and furthermore an orchestrated regulation of specific pathways (Fig. 3B). The 1.5 h substrates comprise several members of the splicing and translation machinery but also central components of intracellular protein translocation (SEC61B, TOMM70, HSPD1, RTN4), endocytosis/vesicle transport (CLTA, DBNL, CTTN, SEC22B, SNX12, SNX2, RAB12), cytoskeletal organization (SEPT2, STMN1, CLASP1), and proteins central to the lipid metabolism (LPIN1, ACLY). Additionally, after 3 h of staurosporine treatment DNA replication (MCM2, 5 and 6, RRM2) and the nuclear envelope (LMNA, TMPO, TPR) were increasingly targeted by proteolysis, whereas substrates cleaved after 6 h confirmed an amplification of the preceding effects.
We complemented our N-terminomics study with quantitative proteomics and phosphoproteomics data, quantifying 5,547 proteins with at least two unique peptides as well as 5,877 high-confidence phosphopeptides (≥90% site localization probability; Fig. 3C; Supplemental Table 4). Notably, only few significant changes in protein abundance were observed. In contrast, as an unspecific kinase inhibitor staurosporine led to a massive reduction in protein phosphorylation levels, with almost 2,000 phosphopeptides showing at least a 2twofold reduction after 1.5 h (Fig. 3C). Interestingly, 31 phosphopeptides from 29 proteins showed a more than threefold up-regulation after 1.5 h. These proteins comprised a set of spliceosome components (XAB2, DDX23, PRPF4B, DDX23, SRRM1, SRRM2, SRSF7, SON). This early multilevel modulation of the splicing machinery by proteolysis and phosphorylation may reflect the known role of alternative splicing variants for a variety of apoptotic genes (32, 33), which seems to be a driving force of staurosporine-induced apoptosis.
DISCUSSION
Our novel ChaFRAtip approach allows the ultrasensitive identification of proteolytic targets and consensus motifs on a proteome-wide scale. The protocol is straightforward, low cost, and requires only minimal equipment that is standard in proteomics laboratories. Therefore, the protocol should be easily transferrable to other laboratories around the globe and may facilitate the generation of novel and expansion of existing knowledge on proteases, their targets, and dynamics. As tip-based procedure, ChaFRAtip further has potential for automation using liquid-handling systems for the purpose of parallelized high throughput label-free quantitative N-terminomics. As demonstrated, the protocol can be adapted to different amounts of starting material and also works with further chemical labeling strategies such as dimethyl or tandem mass tag (TMT) labeling (data not shown). Moreover, it can be further adapted for straightforward and sensitive protease specificity profiling using peptide libraries (34).
The sensitivity demonstrated here is extremely high for N-terminomics and may allow studying samples with severely limited amounts such as biopsies but also combining different analysis strategies as demonstrated. Thus, quantifying the N-terminome (2,073 N-terminal peptides), the proteome (5,547 proteins with at least two unique peptides), and the phosphoproteome (5,900 high-confidence phosphopeptides) with considerable depth required only 4.3 μg, 6.25 μg, and 43.75 μg of protein per time point and biological replicate (protein amounts based on amino acid analysis). Notably, these N-terminomics results were achieved after labeling 26 μg of protein per condition (total of 208 μg), which were later split into six aliquots (each ∼35 μg in total) corresponding to 4.3 μg of protein per condition/channel. We quantified the average of 2,073 N-terminal peptides by measuring four fractions each for 90 min by LC-MS (6 h of total MS time, corresponding to 345.5 quantified unique N-terminal peptides per hour) and anticipate that this can be further improved by gradient optimization, for instance, using dedicated software tools (35). Nevertheless, given state-of-the-art LC-MS equipment and sampling protocols, the presented workflow might even work with as little as 5 μg of starting material, although lower numbers of quantified N-terminal peptides and higher technical variations may be expected partially due to inevitable losses during lysis and the following sample preparation steps. Nevertheless, combined with optimal sampling strategies and standardization, the sensitivity presented here may even allow N-terminomics of needle biopsies. We also analyzed our data for potential physicochemical biases that may arise from the enrichment as this is not uncommon for PTM-enrichment procedures (36). We therefore compared the 3,086 unique identified N-terminal peptides identified in this study to N-terminal peptides in the Peptide Atlas (October 2017, 1,222,862 peptide entries in total), as described in the Supplemental Methods. We could only find slight differences in peptide length and net charge-state distributions that, however, can be attributed to the use of iTRAQ labeling (37) rather than our ChaFRAtip enrichment (see Supplemental Figs. 3–6).
The combined analysis of proteome, phosphoproteome, and N-terminome is particularly interesting as the complex interplay between multiple posttranslational modifications (38–40) but also between different biomolecular species (41) becomes increasingly evident. It has been shown that proteases are regulated by phosphorylation (42), whereas kinases can be regulated by proteolytic cleavage (43). So-called phosphodegrons directly regulate degradation of protease substrates (44). Highly sensitive approaches allowing the multilevel study of patient samples will be an important step toward precision medicine, as it may allow identifying novel disease mechanisms (25), drug targets, and biomarkers for therapy control.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (31) partner repository with the dataset identifiers PXD005954, PXD006594, PXD006595.
Supplementary Material
Acknowledgments
We thank Dr. Julia Burkhart for critical reading of the manuscript.
Footnotes
* This study was supported by the Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen, the Regierende Bürgermeister von Berlin-inkl. Wissenschaft und Forschung, the Bundesministerium für Bildung und Forschung, and KU Leuven (C12/16/020 to SV).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- COFRADIC
- combined fractional diagonal chromatography
- ChaFRAtip
- charge-based fractional diagonal chromatography in tip format
- SCX
- strong-cation-exchange chromatography
- ChaFRADIC
- charge-based fractional diagonal chromatography
- TAILS
- terminal amine isotopic labeling of substrates
- TEAB
- triethylammonium bicarbonate
- BCA
- bicinchoninic acid assay
- TNC
- theoretical net charge.
REFERENCES
- 1. Vögtle F. N., Wortelkamp S., Zahedi R. P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., and Meisinger C. (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 [DOI] [PubMed] [Google Scholar]
- 2. Quirós P. M., Langer T., and López-Otín C. (2015) New roles for mitochondrial proteases in health, ageing and disease. Nature Rev. Cell Biol. 16, 345–359 [DOI] [PubMed] [Google Scholar]
- 3. López-Otín C., and Hunter T. (2010) The regulatory crosstalk between kinases and proteases in cancer. Nature Rev. Cancer 10, 278–292 [DOI] [PubMed] [Google Scholar]
- 4. Agard N. J., and Wells J. A. (2009) Methods for the proteomic identification of protease substrates. Curr. Opin. Chem. Biol. 13, 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., and Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nature Biotech. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 6. Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., and Wells J. A. (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., and Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nature Biotech. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 8. Prudova A., Serrano K., Eckhard U., Fortelny N., Devine D. V., and Overall C. M. (2014) TAILS N-terminomics of human platelets reveals pervasive metalloproteinase-dependent proteolytic processing in storage. Blood 124, e49–e60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prudova A., Gocheva V., Auf dem Keller U., Eckhard U., Olson O. C., Akkari L., Butler G. S., Fortelny N., Lange P. F., Mark J. C., Joyce J. A., and Overall C. M. (2016) TAILS N-terminomics and proteomics show protein degradation dominates over proteolytic processing by cathepsins in pancreatic tumors. Cell Rep. 16, 1762–1773 [DOI] [PubMed] [Google Scholar]
- 10. Gawron D., Ndah E., Gevaert K., and Van Damme P. (2016) Positional proteomics reveals differences in N-terminal proteoform stability. Mol. Syst. Biol. 12, 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Venne A. S., Vögtle F. N., Meisinger C., Sickmann A., and Zahedi R. P. (2013) Novel highly sensitive, specific, and straightforward strategy for comprehensive N-terminal proteomics reveals unknown substrates of the mitochondrial peptidase Icp55. J. Proteome Res. 12, 3823–3830 [DOI] [PubMed] [Google Scholar]
- 12. Ballif B. A., Villen J., Beausoleil S. A., Schwartz D., and Gygi S. P. (2004) Phosphoproteomic analysis of the developing mouse brain. Mol. Cell Proteomics 3, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 13. Burkhart J. M., Schumbrutzki C., Wortelkamp S., Sickmann A., and Zahedi R. P. (2012) Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J. Proteomics 75, 1454–1462 [DOI] [PubMed] [Google Scholar]
- 14. Staes A., Impens F., Van Damme P., Ruttens B., Goethals M., Demol H., Timmerman E., Vandekerckhove J., and Gevaert K. (2011) Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protocols 6, 1130–1141 [DOI] [PubMed] [Google Scholar]
- 15. Mischerikow N., van Nierop P., Li K. W., Bernstein H. G., Smit A. B., Heck A. J., and Altelaar A. F. (2010) Gaining efficiency by parallel quantification and identification of iTRAQ-labeled peptides using HCD and decision tree guided CID/ETD on an LTQ Orbitrap. Analyst 135, 2643–2652 [DOI] [PubMed] [Google Scholar]
- 16. Schechter I., and Berger A. (1968) On the active site of proteases. 3. Mapping the active site of papain; specific peptide inhibitors of papain. Biochem. Biophys. Res. Comm. 32, 898–902 [DOI] [PubMed] [Google Scholar]
- 17. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 [DOI] [PubMed] [Google Scholar]
- 19. Manza L. L., Stamer S. L., Ham A.-J., Codreanu S. G., and Liebler D. C. (2005) Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 5, 1742–1745 [DOI] [PubMed] [Google Scholar]
- 20. Wiśniewski J. R., Zougman A., Nagaraj N., and Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 21. Kollipara L., and Zahedi R. P. (2013) Protein carbamylation: In vivo modification or in vitro artefact? Proteomics 13, 941–944 [DOI] [PubMed] [Google Scholar]
- 22. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., and Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 23. Palmisano G., Parker B. L., Engholm-Keller K., Lendal S. E., Kulej K., Schulz M., Schwämmle V., Graham M. E., Saxtorph H., Cordwell S. J., and Larsen M. R. (2012) A novel method for the simultaneous enrichment, identification, and quantification of phosphopeptides and sialylated glycopeptides applied to a temporal profile of mouse brain development. Mol. Cell. Proteomics 11, 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dickhut C., Radau S., and Zahedi R. (2014) Fast, efficient, and quality-controlled phosphopeptide enrichment from minute sample amounts using titanium dioxide. In Shotgun Proteomics, Martins-de-Souza D., ed. pp. 417–430, Springer; New York. [DOI] [PubMed] [Google Scholar]
- 25. Solari F. A., Kollipara L., Sickmann A., and Zahedi R. P. (2016) Two birds with one stone: Parallel quantification of proteome and phosphoproteome using iTRAQ. Meth. Mol. Biol. 1394, 25–41 [DOI] [PubMed] [Google Scholar]
- 26. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., and Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 27. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 28. Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 29. Dorfer V., Pichler P., Stranzl T., Stadlmann J., Taus T., Winkler S., and Mechtler K. (2014) MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J. Proteome Res. 13, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taus T., Kocher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 31. Vizcaíno J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paronetto M. P., Passacantilli I., and Sette C. (2016) Alternative splicing and cell survival: From tissue homeostasis to disease. Cell Death Different. 23, 1919–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., and Gevaert K. (2005) Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods 2, 771–777 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen M. T. N., Shema G., Zahedi R. P., and Verhelst S. H. L.. Protease specificity profiling in a pipette tip using “charge-synchronized” proteome-derived peptide libraries. Submitted. [DOI] [PubMed] [Google Scholar]
- 35. Moruz L., and Käll L. (2014) GradientOptimizer: An open-source graphical environment for calculating optimized gradients in reversed-phase liquid chromatography. Proteomics 14, 1464–1466 [DOI] [PubMed] [Google Scholar]
- 36. Solari F. A., Dell'Aica M., Sickmann A., and Zahedi R. P. (2015) Why phosphoproteomics is still a challenge. Mol. BioSystems 11, 1487–1493 [DOI] [PubMed] [Google Scholar]
- 37. Thingholm T. E., Palmisano G., Kjeldsen F., and Larsen M. R. (2010) Undesirable charge-enhancement of isobaric tagged phosphopeptides leads to reduced identification efficiency. J. Proteome Res. 9, 4045–4052 [DOI] [PubMed] [Google Scholar]
- 38. Peng M., Scholten A., Heck A. J., and van Breukelen B. (2014) Identification of enriched PTM crosstalk motifs from large-scale experimental data sets. J. Proteome Res. 13, 249–259 [DOI] [PubMed] [Google Scholar]
- 39. Venne A. S., Kollipara L., and Zahedi R. P. (2014) The next level of complexity: Crosstalk of posttranslational modifications. Proteomics 14, 513–524 [DOI] [PubMed] [Google Scholar]
- 40. Zahedi R. P. (2016) Joining forces: studying multiple post-translational modifications to understand dynamic disease mechanisms. Expert Rev. Proteomics 13, 1055–1057 [DOI] [PubMed] [Google Scholar]
- 41. Coman C., Solari F. A., Hentschel A., Sickmann A., Zahedi R. P., and Ahrends R. (2016) Simultaneous metabolite, protein, lipid extraction (SIMPLEX): A combinatorial multimolecular omics approach for systems biology. Mol. Cell Proteomics 15, 1453–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cardone M. H., Roy N., Stennicke H. R., Salvesen G. S., Franke T. F., Stanbridge E., Frisch S., and Reed J. C. (1998) Regulation of cell death protease caspase-9 by phosphorylation. Science 282, 1318–1321 [DOI] [PubMed] [Google Scholar]
- 43. Chan K. T., Bennin D. A., and Huttenlocher A. (2010) Regulation of adhesion dynamics by calpain-mediated proteolysis of focal adhesion kinase (FAK). J. Biol. Chem. 285, 11418–11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ye X., Nalepa G., Welcker M., Kessler B. M., Spooner E., Qin J., Elledge S. J., Clurman B. E., and Harper J. W. (2004) Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 279, 50110–50119 [DOI] [PubMed] [Google Scholar]
- 45. Colaert N., Helsens K., Martens L., Vandekerckhove J., and Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (31) partner repository with the dataset identifiers PXD005954, PXD006594, PXD006595.