Abstract
Centipedes are one of the oldest venomous animals and use their venoms as weapons to attack prey or protect themselves. Their venoms contain various components with different biomedical and pharmacological properties. However, little attention has been paid to the profiles and diversity of their toxin-like proteins/peptides. In this study, we used a proteotranscriptomic approach to uncover the diversity of centipede toxin-like proteins in Scolopendra subspinipes mutilans. Nine hundred twenty-three and 6,736 peptides, which were separately isolated from venom and torso tissues, respectively, were identified by ESI-MS/MS and deduced from their transcriptomes. Finally, 1369 unique proteins were identified in the proteome, including 100 proteins that exhibited overlapping expression in venom and torso tissues. Of these proteins, at least 40 proteins were identified as venom toxin-like proteins. Meanwhile, transcriptome mining identified ∼10-fold more toxin-like proteins and enabled the characterization of the precursor architecture of mature toxin-like peptides. Importantly, combined with proteomic and transcriptomic analyses, 25 toxin-like proteins/peptides (neurotoxins accounted for 50%) were expressed outside the venom gland and involved in gene recruitment processes. These findings highlight the extensive diversity of centipede toxin-like proteins and provide a new foundation for the medical-pharmaceutical use of centipede toxin-like proteins. Moreover, we are the first group to report the gene recruitment activity of venom toxin-like proteins in centipede, similar to snakes.
Centipedes are one of the oldest and important groups of excellent predatory arthropods, with a fossil record spanning 420 million years (1). They comprise ∼3300–3500 centipede species distributed worldwide and most provinces of China (2). The body length of adult centipedes ranges from 4 to 300 mm, and venom glands are located in the first pair of limbs (3). The venom is used not only to paralyze and kill prey but also for defense against predators (4). Human injuries caused by centipede stings are also frequently reported. Symptoms of the stings include intense local pain, redness, swelling, superficial necrosis, chills, fever, and weakness. Serious secondary infections have even been reported to lead to human death (5). Although a report of a lethal centipede bite has not been reported in humans, their bites kill earthworms, snails, amphibians, reptiles, and even rats (6, 7). Thus, the venoms of centipedes contain diverse components with a variety of functions, providing an efficient way to rapidly paralyze prey. However, the centipede is still regarded as a neglected group of venomous animals and little is known about the protein/peptide toxins in centipedes.
Centipede venoms serve as a rich reservoir containing diverse components with a variety of functions. Centipede venoms are rich source of structurally and pharmacologically diverse peptides (8–10). Centipede venoms contain large amounts of non-protein components, such as biogenic amines (11, 12), polysaccharides and lipids (13). Meanwhile, some enzymes are identified as protein components of centipede venom, such as serine proteases, endopeptidases, carboxypeptidases, esterases and acid or basic phosphatases (14, 15). In recent studies, centipede toxins have attracted increasing attention because of their excellent chemical and pharmacological activities, particularly as ion channel inhibitors and other neurotoxins (16–19). Some specific toxins and several antimicrobial peptides were also identified in centipede venom (17, 20–22). These venom peptides have remarkable chemical, thermal and biological stability, enabling researchers to utilize their desirable functions for therapeutic use.
However, biochemical and pharmacological research of centipedes have lagged well behind studies of other venomous animals, such as snakes, spiders, and scorpions (10). Complete venom peptide or protein sequences and pharmacological data are available for an even smaller number of species (9, 10). Similarly, venom complexity must be further confirmed using new strategies of large-scale sequencing of broad array of centipede venoms, providing new putative proteins and enabling results to be compared among species (2, 16, 23). Therefore, a fully integrated approach that combines transcriptomics and peptidomics/proteomics seems essential for understanding the venom composition, venom maturation and venom production mechanisms. Here, using an in-depth proteotranscriptomic analysis (combined proteomic and transcriptomic analysis) of centipede venoms and torso tissues, we described the protein/peptide composition of the dissected venom gland and torso of Scolopendra subspinipes mutilans. We also present the first complete comparative analysis of the protein content and toxin-like proteins/peptides distribution in the venom gland and torso, based on our RNA-Seq data and MS datasets. We finally provide an overview of toxin production from the torso to the venom gland, initially highlighting the extensive and surprising phenomenon of the toxin-like protein gene recruitment process in centipede. This result provided foundational evidence supporting the gene recruitment hypothesis for venom toxin in which toxin gene recruitment is linked to the functional constraints of the recruited proteins (24–26).
EXPERIMENTAL PROCEDURES
Animals and Ethics
Adult S. mutilans (both sexes) were collected from Hubei province in China. All centipede (S. mutilans) studies were reviewed and approved by the Animal Care and Use Committee of the Kunming Institute of Zoology of the Chinese Academy of Sciences.
Chemicals
Acetonitrile (ACN)1, methanol (MeOH), dichlormethane, trifluoroacetic acid (TFA) were purchased as HPLC or LC-MS grade from Carl Roth (Karlsruhe, Germany). Dithiothreitol (DTT), acetic acid, trypsin (sequencing grade) were purchased from Sigma-Aldrich (Sigma-Aldrich).
Venom Collection
The pool of crude venom was obtained after stimulating the venom glands in the first pair forceps of centipedes with a 3 V alternating current, as described in our previous report (16). Venom secretions were freeze-dried and stored at −20 °C until further use.
RNA Extraction and Sequencing
Venom gland and torso tissues (285 mg, torso tissue from 3–10 body segments) were preserved in liquid nitrogen until RNA extraction. The detailed RNA extraction and cDNA library construction methods were reported in our previous studies (27, 28). cDNAs prepared from venom gland and torso tissues of S. mutilans were sequenced using Illumina HiSeq™ 2000.
Venom and Torso Sample Preparation
Venom were dissolved in 500 μl of 25 mm NH4HCO3 buffer, and then applied to an ultrafiltration tube with a 3 kDa cut-off. The low molecular weight (<3 kDa) fraction was collected after ultrafiltration and desalinated prior to peptidomic analysis. The desalination procedure: the collected peptides were desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density, GE), bed I.D.7 mm, volume 3 ml, Sigma) and concentrated by vacuum centrifugation. Other supernatants with molecular weights greater than 3 kDa were separated on SDS-PAGE gels. Torso tissues were grinded in liquid nitrogen. Then, the homogenate of the tissue was divided into two parts. Extraction buffer (0.25% acetic acid and protease inhibitor mixture) was added to one half and then disrupted using a sonicator (Hielscher Ultrasound Technology, Germany) for 6 cycles on ice. Clear supernatant, which was collected after centrifugation at 20,000 × g for 15 min at 4 °C, was applied to an ultrafiltration tube with a 30 kDa cut-off. Filtrates were desalinated (the same to above desalination procedure) prior to the peptidomic analysis. SDT lysis buffer (4% SDS and 0.1 m DTT in 0.1 m Tris-HCl, pH 7.6) was added to the other sample of torso tissue, incubated for 15 min at 95 °C, ultrafiltrated, and the supernatant was collected after centrifugation, as described above.
Samples were further separated on a 12% gel next to a protein ladder (Thermo, ref. 26614) using SDS-PAGE. Gels were fixed with 50% ethanol/10% acetic acid, stained with Gelcode Blue Stain (Thermo ref. 24592) and destained with MilliQ water (Millipore) (Fig. 1). Six bands were excised from each lane for in-gel digestion with trypsin. After extraction with 100% acetonitrile, samples were desalinated (the same to above desalination procedure), freeze-dried and stored at −80 °C until the subsequent ESI-MS/MS analysis.
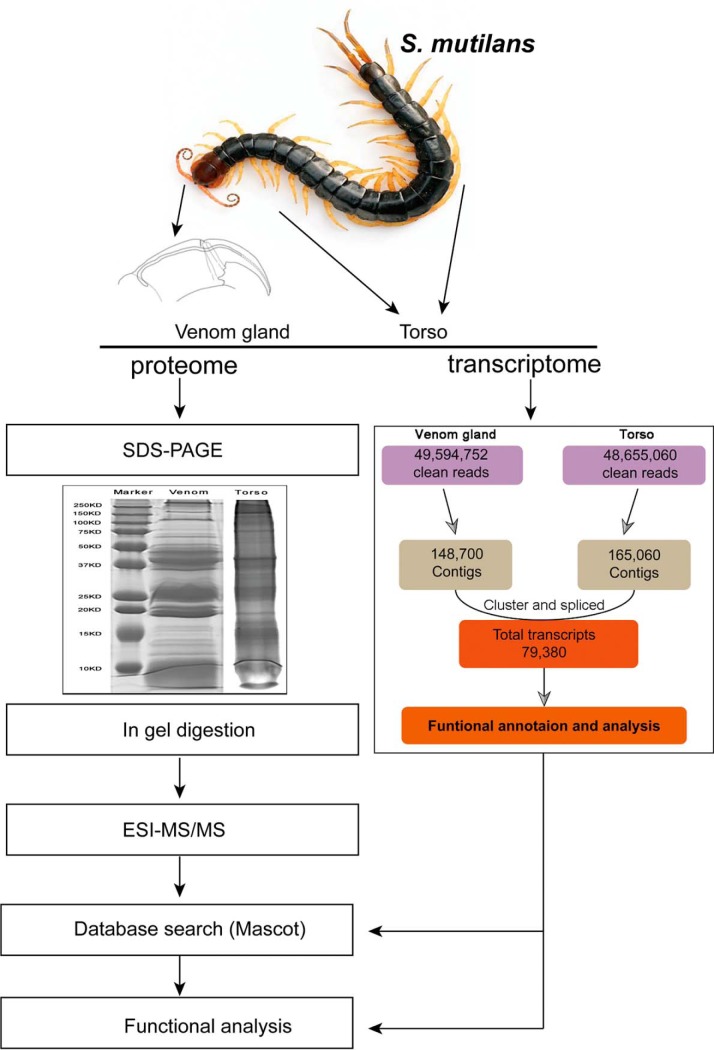
Fig. 1.
Workflow for the isolation and proteomic and transcriptomic analyses of the centipede. The centipede Scolopendra subspinipes mutilans was used for proteomic and transcriptomic analyses. Venoms and torso tissues were processed individually and subjected to SDS-PAGE followed by in-gel digestion. Then, samples were analyzed in a separate ESI-MS/MS assay. For the transcriptomic analysis, venom glands, not venoms, and torso tissues were used for high-throughput sequencing. The functional analysis was combined with proteomic and transcriptomic data.
Transcriptome Analysis
De novo transcriptome assembly was performed with the short-read assembly program SOAP de novo-Trans (http://soap.genomics.org.cn/) using the default parameters. The program combined reads with certain lengths of overlap and connected paired-end reads to form contigs. Further assembly of contigs from each sample was performed by sequence splicing and removing redundant sequences using the sequence clustering software TGICL (29), resulting in the generation of the longest possible non-redundant unigenes. TGICL parameters were identical to the parameters used in our previous study (27).
HPLC Fractionation
Candidate fractionation of samples after in-gel digestion was performed using an EASY-nLC HPLC system (Thermo Fisher Scientific) equipped with a binary rapid separation nano flow pump and ternary loading pump. Mobile phase eluent A consisted of 0.1% TFA (aqueous) and mobile phase eluent B consisted of ACN/ddH2O/TFA 90/10/0.08% (v/v/v). The sample was added to the Thermo Scientific EASY loading column (2 cm × 100 μm, 5 μm -C18) by the auto-sampler and then to the analytical column (75 μm ×100 mm, 3 μm -C18). The flow rate was set to 250 nL/min. Peptide separation on the column was achieved with linear stepwise gradients (0′–5% B, 5′–5% B, 12.5′–20% B, 62.5′–70% B, 63.5′–99% B, 65′–99% B, 66′–5% B and 72′–5% B). Fractions of 1.25 ml (5 min) each, starting at 20% eluent B, were collected and lyophilized.
Mass Spectrometry
The Q Exactive instrument (Thermo Finnigen) was operated in the data-dependent mode to automatically switch between full scan MS and MS/MS acquisition. The survey of full scan MS spectra (m/z 300–1800) was acquired in the Orbitrap with a resolution of 70,000 (m/z 200) after accumulation of ions to a 3 × 106 target value based on predictive automatic gain control (AGC) from the previous full scan. Dynamic exclusion was set to 15 s. The 10 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented by higher-energy collisional dissociation (HCD) with a fixed injection time of 60 ms and a resolution of 17,500 (m/z 200) for the MS2 scanning method. Typical mass spectrometric conditions were: spray voltage, 2 kV; no sheath and auxiliary gas flow; heated capillary temperature, 250 °C; normalized HCD collision energy, 27 eV; and underfill ratio, 0.1%. The MS/MS ion selection threshold was set to 1 × 105 counts.
Data Processing
The RAW data files were processed with Proteome Discoverer (Thermo Scientific, Version 1.4). Generated peak lists were searched against our transcriptome database (79,380 entries) using Mascot software version 2.2. Trypsin was chosen as the enzyme, with 2 missed cleavages allowed. The MS/MS search criteria were: peptides tolerance of 20 ppm for MS, and 0.1 Da for MS/MS mode. Carbamido methylation of cysteine was set as a static modification, and methionine oxidation was set as a dynamic modification. High confidence peptides were used for protein identification, resulting in a false discovery rate (FDR) threshold (FDR ≤ 0.01). Only unique peptides with high confidence were used for protein identification.
Experimental Design and Statistical Rationale
The experimental design and statistical rationale for each of the experiments conducted in this work have been described in each subsection and workflow is shown in Fig. 1. Venoms were collected from three hundred fifty centipedes. Torso tissues were collected from twelve centipedes. Protein extraction was made based on produced biological triplicates. Proteins contained at least one unique peptide with FDR ≤ 0.01 during protein identification (see above). To compare the abundances of phosphopeptides between the control and treatment samples, label-free quantification was performed with a minimum of 1.5-fold changes to determine the differentially-expressed phosphopeptides. In addition, the Student's t test was employed to identify significant changes between the control and treatment sample among the three biological replicates. p values <0.05 were considered to be significant. Quantitative real-time PCR (qPCR) was conducted using five biological and three technological replicates. Comparative gene expression analysis set p values < 0.05 with the level of statistical significance.
Quantitative Real-Time PCR
Total RNA Isolation and cDNA Synthesis
Venom gland and torso RNA was extracted using the Eastep® Super Total RNA Extraction kit (Promega) according to the manufacturer's instructions. RNA sample concentration and quality was determined using the GENEQUANT 100 (Classic, GE). Using primer-blast designed specific primers of toxin-like genes, amplicon lengths varied from 200–300 bp and primer lengths between 18–22 bp (supplementary Data).
Quantitative Real-Time PCR
The quantitative real-time PCR assay was validated using Light Cycler 480 platforms (Roche Diagnostics Corp., Indianapolis, IN), and SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa Biotechnology (Dalian) Co., Ltd., China). The final master mix (10 μl) comprised 5 μl of SYBR enzyme mix, 2.5 μl of PCR-grade water, 1 μl of each primer (at 10 μm working concentration), then 0.5 μl of template (cDNA from venom gland and torso) was added to the master mix in order to give a final reaction volume of 10 μl. The cycling conditions used were pre-incubation 95 °C for 4 min, followed amplification by 45 cycles of 95 °C for 5 s, 60 °C for 30 s, melting curve by 95 °C for 1 s, 65 °C for 15 s and 95 °C for every 5 °C, and a final cooling step of 40 °C for 30 s. Specificity of PCR reactions was verified by melting curve analysis of each sample amplified product. Samples were run in triplicates. For each primer a melting curve of the PCR product was also performed to ensure the absence of artifacts. Expression values were normalized using an endogenous beta-actin.
Data and Bioinformatics Analysis
All unigenes from our centipede database were annotated using BLASTX and searched against known databases, as described in our previous reports (27, 28). Briefly, the analytical method is described below. Unigenes were aligned to a higher priority database and annotated with given descriptions, which were not aligned to lower priority database. The Gene Ontology (GO) annotation was performed using Blast2Go (30) software suite v2.5.0. In these searches, the BLASTX cut-off was set to 1e−6. For the GO mapping process, validated settings were used with a threshold of 1e−6, an annotation cut-off of “45” and a GO weight of “20.” Each annotated sequence was assigned to detailed GO terms.
Comparative Expression Analysis
RNA-Seq data for each tissue (venom gland and torso) were aligned using Bowtie (31) version 0.12.7 and TopHat (32) version 2.0.6 for mapping. Gene expression values were calculated from the expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced (FPKM) (33). We separately calculated the FPKM values for genes from each tissue using Rseq (34). GraphPad Prism version 5.0 (La Jolla, CA) and R version 3.3.2 software were used to plot the graphs and perform statistical analyses.
RESULTS
Isolation of Venom Gland and Workflow of the Analysis Approach
Venom glands and torso tissues were isolated from the centipede S. mutilans using the protocol described in our previous study (Liu et al., 2012). We first carefully selected healthy adult centipedes (n = 350) with no injury to dissect venom glands from their first pair of limbs. We used a 3 V alternating current to stimulate the venom gland and improve the coverage of proteomes by ensuring the inclusion of a greater number of toxin. Torso tissues were dissected from body segments 3–10, which were separate from all gut tissues. Next, the isolated venom gland and torso tissues were further processed (Fig. 1). A portion of each sample was used in the SDS-PAGE analysis to obtain the proteome. The torso tissue had a more complex banding pattern than the venom gland, which contained highly abundant proteins. Protein bands from venom gland and torso tissues were excised for in-gel digestion and subjected to ESI-MS/MS analysis. The other portion of each sample was used to extract RNA and perform an RNA-Seq analysis of the transcriptome.
Identification of Venom Gland and Torso Transcriptome
cDNAs prepared from venom gland and torso tissues of S. mutilans were sequenced using Illumina HiSeq™ 2000. After sequencing and cleaning the low-quality reads, we acquired 49,594,752 clean reads from the venom gland and 48,655,060 clean reads from the torso tissue (Fig. 1). Using the Trinity program for the de novo assembly of clean reads into contigs, 148,700 and 165,060 contigs were separately generated from these two tissues, respectively. Finally, the transcriptome data consisted of 70,380 putative gene objects (all unigenes) ranging from 100 bp to 27,384 bp, with an average length of 290 bp. The size of unigenes larger than 500 bp was 21,647. The largest unigenes contained 27,661 bp, and the N50 of unigenes was 562 bp (Table S1). Notably, 18,566 of the 70,380 (26.4%) unigenes had CDS matches with the direction that possessed significant similarity (cut-off E-value of 1E-6) with best-hit blast results. Moreover, 2429 of 70,380 (3.5%) CDS sequences produced by the ESTScan program were not able to be aligned to known databases. Finally, 20,995 unigene sequences had complete CDS (supplemental Fig. S1).
Identification of Venom Gland and Torso Proteome
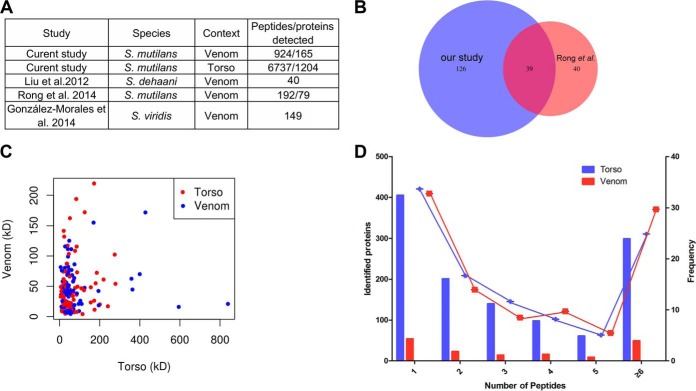
We initially compared our proteome (1369 proteins and 7661 peptides) with previously reported centipede proteomes to assess the comprehensiveness of our centipede proteome (Fig. 2A). Three previous studies profiled the proteomes of venoms from S. dehanii, S. mutilans and S. viridis. 79, 192, and 149 peptides/proteins were identified from the above species, respectively. Of these studies, the study by Rong et al. reported the largest peptidome of 192/79 peptides/proteins identified from the venom of S. mutilans using LC-MS/MS analysis. The comparison of our venom proteome with the largest previous venom proteome of this centipede showed that the 39 previously reported proteins were also detected in our proteome (Fig. 2B). Additionally, our proteome identified 126 proteins that were not detected in the previous venom proteome (2). Based on these data, our proteome represents the most comprehensive proteome of S. mutilans to date.
Fig. 2.
Comprehensive profiles of the centipede proteome. A, Comparison of previously reported centipede proteomes to the proteome described in our study. The numbers of detected proteins/peptides in previous studies are shown. B, Venn diagram comparing identified and shared peptides/proteins from S. mutilans between our proteome and the proteome reported by Rong et al. C, The plot shows the molecular weights of proteins identified in the proteomes from centipede venom and torso tissues. The red or blue dots depict different proteins from venom and torso tissues, separately. D, Histogram displaying the number of peptides matched to proteins. The x axis illustrates the number of identified peptides. The primary y axis indicates the number of identified proteins (bars). The right y axis represents the percent (lines).
Among our venom and torso proteomes, 73% of identified proteins had a molecular weight of less than 50 kDa (Fig. 2C). Thus, centipedes contain much smaller functional molecules as our expected. Based on the peptide detection, 32% of proteins were comprised of one unique peptides. 14% of proteins were comprised of two unique peptides. 8.5% of proteins were comprised of three unique peptides. 8.1% of proteins were comprised of four unique peptides. 5.1% of proteins were comprised five unique peptides. 24.8% of proteins were comprised of at least six unique peptides or more (Fig. 2D). The more enriched peptides were assembled to proteins leading to a more comprehensive proteome.
Identification of Common Proteins in Venom Gland and Torso Tissues
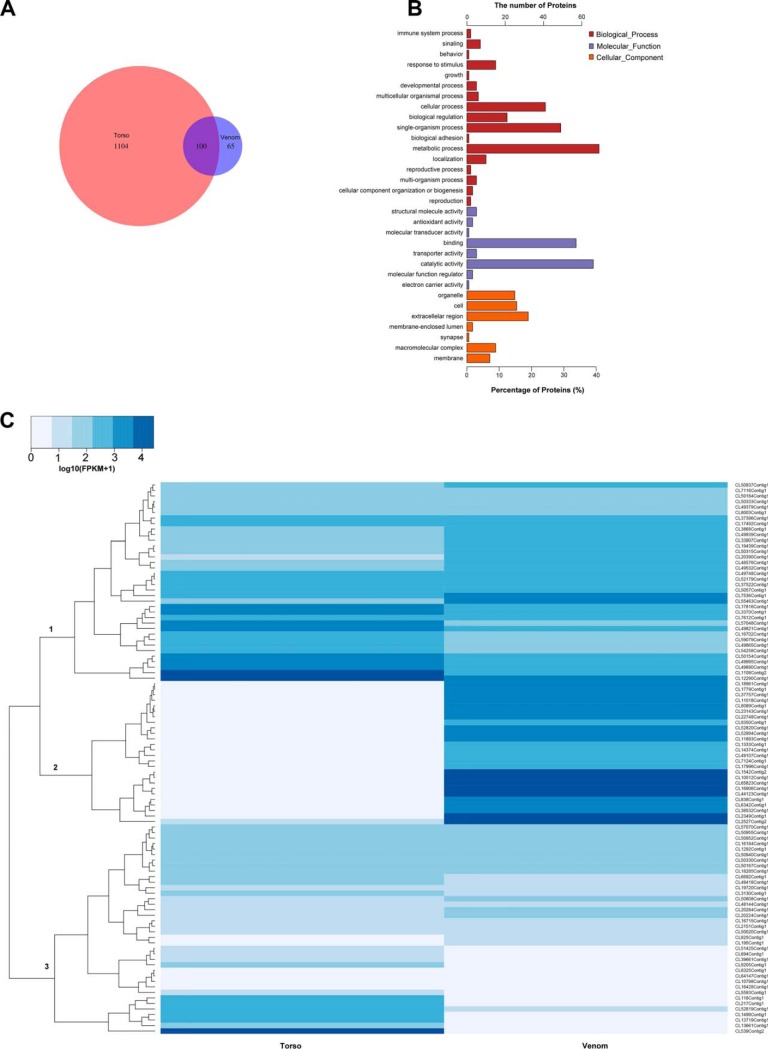
An in depth proteotranscriptomic analysis was performed to determine the proteome of venom and torso tissues. One thousand three hundred sixty-nine unique proteins were identified in the venom and torso tissue proteomes. Among these identified proteins, 1204 unique proteins were identified in torso tissue and 165 unique proteins were identified in the centipede venom. Remarkably, 100 unique proteins were identified as overlapping proteins (proteins determined in each sample) in venom and torso (Fig. 3A).
Fig. 3.
Comparison of the venom and torso proteomes. A, Venn diagram representing the overlapping proteins from the venom and torso tissues. B, Functional annotation of proteins identified in venom. One hundred sixty-five proteins from venom were annotated with GO (Gene Ontology) terms and enriched for biological process, molecular function and cellular component. C, Heat map of 100 overlapping proteins showing differences in expression among venom gland and torso tissues.
We performed functional annotation analyses of these identified proteins to obtain additional insights into these unique proteins. Firstly, the full sequence of proteins was determined by transcriptomic and peptidomic analyses and a database search. Then, all sequences were submitted to the Nr database and annotated with functional descriptions (supplemental Table S2). Secondly, a functional enrichment analysis was performed on each unique protein assigned one or more GO terms, such as “signaling,” “reproduction,” and “organelle” (Figs. 3B and supplemental Fig. S2). The enriched GO terms were divided into “Biological process,” “Molecular function,” and “Cellular component.” As shown in the venom GO enrichment analysis (Fig. 3B and supplemental Table S3), most venom proteins were related to “catalytic activity” (40%) and “binding” (34%) in the “Molecular function” term. Regarding the “Biological process” term, most venom proteins were enriched in “metabolic process” (42%).
Identification of Overlapping Proteins Between Venom and Torso Tissue
Fortunately, using the proteomic analysis, 100 unique proteins were identified as overlapping proteins expressed in both venom and torso tissues (Fig. 3A and supplemental Table S4). These proteins were not only identified by the transcriptomic analysis that provided information about gene expression but peptides were also detected using ESI-MS/MS-based identification. Of these proteins, 66% were low molecular weight proteins (<50 kDa). In addition to cellular/organelle skeleton proteins and enzymes, 40% of proteins were toxin-like proteins. Next, we examined the expression of these proteins among venom gland and torso tissues using the transcriptomic data. Overlapping proteins were differentially expressed in venom gland and torso tissues. Three clusters of differentially expressed proteins were identified (Fig. 3C). The first cluster was comprised constitutive proteins (skeleton proteins or enzymes) with moderate expression in venom gland and torso tissues. Remarkably, proteins belonging to the second cluster were expressed at higher levels in the venom gland than in torso tissues, most of which were venom toxin-like peptides or secreted proteins, such as neurotoxins and venom allergens. The third cluster included constitutive proteins expressed in the centipede torso tissue, such as glycoproteins of the cellular membrane. Among the highly expressed overlapping proteins, in addition to cellular components and catalytic enzymes, 40% of proteins were determined to be toxin-like proteins (Table I). Most toxin-like peptides were neurotoxins and ion channel inhibitors that had been previously identified in S. mutilans and the other species, such as txk1a_scomu, tx6a_scomu, and neurotoxin 3, among others.
Table I. Toxin-like proteins/peptides identified from the proteome of the centipede, S. mutilans.
| Sequence name | Sequence description | Sequence length | Accession Number | E-Value | MW (kDa) | Calc. pI |
|---|---|---|---|---|---|---|
| CL18110Contig1 | alkaline phosphatase | 464 | gi 926637143 | 8.39E-148 | 50.96 | 6.30 |
| CL49379Contig1 | cathepsin b | 330 | gi 31872149 | 2.22E-162 | 36.64 | 6.74 |
| CL50955Contig1 | cathepsin c | 432 | gi 675369694 | 0 | 48.77 | 6.65 |
| CL37522Contig1 | cathepsin d | 382 | gi 336454162 | 6.55E-179 | 41.95 | 7.83 |
| CL50333Contig1 | cathepsin l | 335 | gi 501293796 | 3.07E-159 | 37.29 | 6.54 |
| vg_contig_t204 | γ-glutamyltranspeptidase 1-like | 40 | gi 926653374 | 2.58E-07 | 4.03 | 7.99 |
| CL50454Contig1 | iron zinc purple acid phosphatase-like protein | 424 | gi 193624668 | 0.00E+00 | 49.21 | 6.20 |
| CL3629Contig1 | k+ channel inhibitor 4 | 78 | gi 392295737 | 6.59E-48 | 8.684 | 7.69 |
| CL49825Contig1 | legumain-like | 408 | gi 321469736 | 1.03E-173 | 46.16 | 6.37 |
| CL49890Contig1 | paramyosin | 871 | gi 42559470 | 0 | 102.28 | 5.73 |
| CL14374Contig1 | phospholipase membrane associated | 147 | gi 306755927 | 1.74E-50 | 16.83 | 6.49 |
| CL5057Contig1 | prosaposin isoform x1 | 677 | gi 321471690 | 2.65E-125 | 75.98 | 4.92 |
| CL50315Contig1 | serine protease inhibitor | 369 | gi 344252156 | 3.32E-66 | 41.81 | 6.30 |
| CL13661Contig1 | serpin b3 isoform x1 | 349 | gi 260797582 | 2.34E-66 | 39.37 | 5.20 |
| CL3130Contig1 | serpin b4-like | 374 | gi 532074526 | 9.05E-73 | 42.26 | 5.40 |
| CL8089Contig1 | trypsin | 227 | gi 2853182 | 1.45E-50 | 25.99 | 5.29 |
| vg_contig_t422 | trypsin partial | 38 | gi 403311423 | 1.59E-06 | 4.27 | 11.05 |
| CL65823Contig1 | tx6a_scomu | 68 | gi 657193247 | 2.46E-23 | 7.77 | 7.55 |
| CL7323Contig1 | txk1a_scomu | 74 | gi 586946312 | 1.96E-42 | 8.57 | 7.61 |
| CL18341Contig1 | txk1c_scomu | 70 | gi 586946314 | 9.29E-34 | 8.35 | 5.38 |
| vg_contig_t38 | txk1e_scomu | 65 | gi 586946316 | 2.54E-38 | 7.53 | 7.64 |
| CL10512Contig1 | txk3a_scomu | 84 | gi 586946319 | 7.69E-53 | 9.81 | 5.77 |
| CL38532Contig1 | txk3a_scomu | 83 | gi 586946319 | 4.49E-15 | 9.69 | 8.76 |
| CL44425Contig1 | txo2a_scomu | 76 | gi 586946322 | 3.52E-48 | 8.46 | 4.78 |
| CL12576Contig1 | neurotoxin 3 | 58 | gi 586946326 | 5.60E-05 | 6.68 | 9.66 |
| CL2527Contig2 | neurotoxin 3 | 77 | gi 586946326 | 7.55E-36 | 8.88 | 8.57 |
| CL44123Contig1 | neurotoxin 4 | 64 | gi 586946327 | 2.08E-23 | 6.96 | 8.41 |
| CL61661Contig1 | neurotoxin 5 | 56 | gi 586946328 | 2.11E-32 | 6.07 | 5.76 |
| CL48983Contig1 | Unknown function | 142 | gi 675390069 | 1.97E-05 | 16.50 | 8.37 |
| CL6342Contig1 | Unknown function | 225 | gi 926644759 | 7.44E-11 | 25.27 | 5.80 |
| CL54258Contig1 | Unknown function | 71 | gi 240247657 | 5.89E-06 | 7.50 | 4.98 |
| CL18961Contig1 | Unknown function | 207 | gi 241641704 | 8.63E-08 | 23.42 | 9.48 |
| CL22748Contig1 | Unknown function | 200 | gi 926644759 | 2.93E-14 | 23.20 | 5.02 |
| CL2349Contig1 | Unknown function | 175 | gi 648215761 | 2.12E-05 | 19.98 | 6.49 |
| CL37757Contig1 | Unknown function | 192 | gi 926636983 | 4.65E-05 | 21.78 | 6.40 |
| CL838Contig1 | venom allergen partial | 189 | gi 675367593 | 8.49E-36 | 20.77 | 8.87 |
| CL49773Contig1 | venom carboxylesterase-6-like | 510 | gi 817190507 | 4.01E-80 | 57.95 | 5.20 |
| CL7422Contig2 | venom protease-like | 352 | gi 817060716 | 1.82E-75 | 38.16 | 6.52 |
| CL52994Contig1 | serine protease | 211 | gi 672349346 | 2.49E-136 | 23.55 | 4.78 |
| CL9350Contig1 | zinc metalloproteinase | 312 | gi 321475779 | 8.43E-08 | 34.98 | 8.24 |
Identification of Centipede Toxin-like Proteins in the Proteome
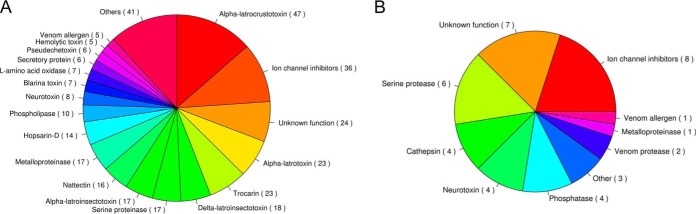
As expected, transcripts for at least 366 toxin-like proteins were identified in our transcriptome database (Fig. 4A and supplemental Table S5). At least 18 kind of toxin-like proteins were identified using the transcriptomic analysis. Among these transcripts, most putative toxins were alpha-latrocrustotoxins (47, 12.8%) and ion channel inhibitors (36, 9.8%). However, only 40 unique proteins representing known toxins were identified in the centipede proteome. Of course, more proteins with unknown function were identified as toxins. Most known toxins were inhibitors of ion channel proteins, such as K+ and Ca2+ channel inhibitors, in addition to toxins with unknown functions (Fig. 4B). These results were consistent with recent studies on the centipede S. mutilans.
Fig. 4.
Identification of venom toxins in centipede proteomes and transcriptomes. A, Pie chart of venom toxin-like proteins/peptides identified in the centipede transcriptome. Three hundred sixty-six venom toxin-like proteins/peptides were identified using the transcriptomic analysis. B, Pie chart of venom toxin-like proteins/peptides identified in the centipede proteome. Forty proteins were venom toxin-like proteins identified by ESI-MS/MS.
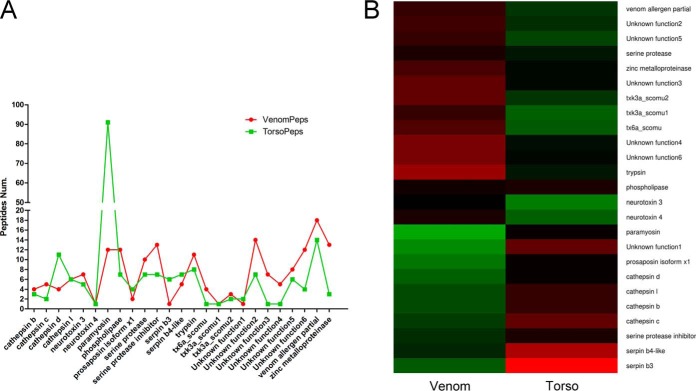
Interestingly, among the 40 identified toxin-like proteins, 25 putative toxins (over-toxins) were overlapping proteins detected in venom and torso tissues by ESI-MS/MS. In the comparison of detected peptide fragments, 2 or more peptides were identified for 90% of overlapping toxins-like peptides in venom and torso, and 10 or more peptides were identified for 10 putative toxins (Fig 5A). Using the gene expression analysis, most overlapping toxin-like peptides (90%) were expressed at higher levels in the venom gland than in torso tissues (Figs 5B, supplemental Figs. S3 and S4). These data from the combined proteomic, transcriptomic, and qRT-PCR gene expression analyses provided the first sound evidence that some centipede toxins were recruited from tissues outside the venom gland.
Fig. 5.
The profiles for overlapping toxin-like proteins/peptides expressed in venom and torso tissues from the centipede. A, Comparison of overlapping putative toxins with one or more detected peptides in venom and torso tissues by ESI-MS/MS. B, Heat map of gene expression profiles for overlapping putative toxins identified in the proteome. Deduced venom toxins were expressed in the torso tissue in addition to the venom gland.
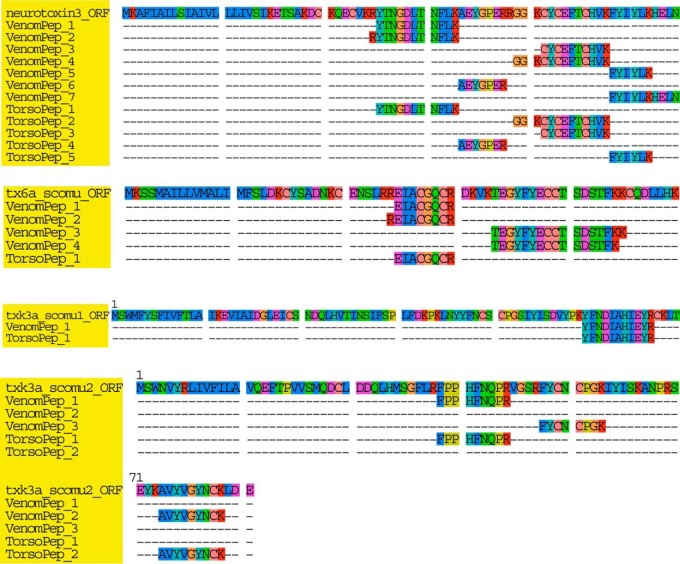
Furthermore, one or more detected peptides were aligned to their full protein sequence from each class of determined toxin-like proteins. Based on the peptide alignment, the detected peptides were 100% similar to their full sequence deduced from transcriptome (Fig. 6). Moreover, when more peptides were detected for a toxin-like protein, then a longer protein sequence was assembled. Importantly, we also identified 15 toxin-like proteins/peptides that were specifically expressed in the venom (Table II). Among these proteins, trypsin, the K+ channel inhibitor and γ-glutamyltranspeptidase-1 were only expressed in venom gland according to the transcriptomic analysis, and short reads were not detected for these genes in the sequencing analysis. In addition, similar to venom proteases, phosphatases, and several ion channel inhibitors, these putative toxins were not overlapping toxins, as peptides were not detected in both tissues using our comprehensive proteomic analysis and sequences were not detected by qRT-PCR.
Fig. 6.
Alignment of detected peptides and transcripts for four toxin-like proteins. Peptides separately detected in venom and torso tissues were aligned to the complete sequence of toxin-like proteins. Different peptides for the toxins were detected in each tissue and were 100% like the precursor identified in the transcriptome. Each toxin-like protein was colored with yellow background.
Table II. Toxin-like proteins/peptides specific-expressed in the venom gland of S. mutilans.
| Sequence name | Sequence description | Peptides | Sequence length | Accession Number | E-Value | MW (kDa) | Calc. pI | FPKM |
|---|---|---|---|---|---|---|---|---|
| CL12576Contig1 | neurotoxin 3 | 4 | 58 | gi 586946326 | 5.60E-05 | 6.68 | 9.66 | 4944.86 |
| CL18110Contig1 | alkaline phosphatase | 7 | 464 | gi 926637143 | 8.39E-148 | 50.96 | 6.30 | 71.03 |
| CL18341Contig1 | K+ channel inhibitor | 1 | 70 | gi 586946314 | 9.29E-34 | 8.35 | 5.38 | 70.95 |
| CL3629Contig1 | K+ channel inhibitor 4 | 1 | 78 | gi 392295737 | 6.59E-48 | 8.68 | 7.69 | 6059.87 |
| CL44425Contig1 | Ca2+ channel inhibitor | 2 | 76 | gi 586946322 | 3.52E-48 | 8.46 | 4.78 | 9079.74 |
| CL48983Contig1 | Unknown function | 7 | 142 | gi 675390069 | 1.97E-05 | 16.50 | 8.37 | 1137.33 |
| CL49773Contig1 | venom carboxylesterase-6-like | 3 | 510 | gi 817190507 | 4.01E-80 | 57.95 | 5.20 | 63.82 |
| CL49825Contig1 | legumain-like | 1 | 408 | gi 321469736 | 1.03E-173 | 46.16 | 6.37 | 109.76 |
| CL50454Contig1 | iron zinc purple acid phosphatase-like protein | 2 | 424 | gi 193624668 | 0.00E+00 | 49.21 | 6.20 | 41.87 |
| CL61661Contig1 | neurotoxin 5 | 1 | 56 | gi 586946328 | 2.11E-32 | 6.069 | 5.76 | 22786.04 |
| CL7323Contig1 | K+ channel inhibitor | 1 | 74 | gi 586946312 | 1.96E-42 | 8.57 | 7.61 | 2111.82 |
| CL7422Contig2 | venom protease-like | 1 | 352 | gi 817060716 | 1.82E-75 | 38.16 | 6.52 | 47.38 |
| vg_contig_t204 | γ-glutamyltranspeptidase 1-like | 1 | 40 | gi 926653374 | 2.58E-07 | 4.03 | 7.99 | 408.92 |
| vg_contig_t38054 | K+ channel inhibitor | 2 | 65 | gi 586946316 | 2.54E-38 | 7.53 | 7.64 | 0.00 |
| vg_contig_t422523 | trypsin partial | 1 | 38 | gi 403311423 | 1.59E-06 | 4.27 | 11.05 | 0.91 |
DISCUSSION
Centipede venom is a rich source of toxins with different pharmacological properties, like other animal venoms. As one of oldest venomous arthropods, centipede venoms have recently been shown to display good biomedical and pharmacological activities for drug development (2, 16, 17, 19, 23). However, no papers or methods have focused on venom diversity and toxin recruitment by combining high throughput proteomic and transcriptomic analyses using ESI-MS/MS and RNA-Seq technology. In this study, we thus searched and determined proteins expressed in or outside the venom gland. We used our established approach to search for proteins by (1) investigating the comprehensive proteomes of venom and torso tissues using ESI-MS/MS; (2) investigating the comprehensive transcriptomes of S. mutilans sequences from venom gland and torso tissues; (3) selecting all overlapping proteins in venom and torso tissues to determine toxin diversity; and (4) performing a toxin recruitment analysis that combined the expression and alignment of overlapping proteins and venom-specific proteins. Using our approach, we identified at least 25 toxin-like proteins/peptides that exhibited overlapping expression in venom gland and torso tissues. The patterns of toxins gene expression provided evidence for venom gene recruitment in centipede.
Previous studies have reported the proteomes of centipede species, particularly S. mutilans (2). However, to our knowledge, no studies have systematically identified numerous toxin-like proteins/peptides and their expression profiles. In particular, a comprehensive transcriptome of centipede assembled de novo by RNA-Seq technology and compared with a cDNA library has not been reported (16, 23). In this study, almost 50% of the proteins detected by Rong et al. were identified in our peptidome/proteome and confirmed to be expressed in our transcriptome. High throughput ESI-MS/MS and RNA-Seq technology provided a powerful platform to investigate the novel proteins and the diversity of venoms, particularly the low abundance peptides/proteins that are not detected using conventional methods (35). Seven thousand six hundred sixty-one unique peptides were identified by ESI-MS/MS. This number is 6-fold higher than the number of proteins that were ultimately identified using our approach. Namely, one detected protein was identified from at least 6 peptides in our proteome data set. Meanwhile, using a transcriptomic analysis, more than 400 toxin-like proteins/peptides were shown to be expressed in this centipede. Compared with the transcriptomic results, only 40 toxin-like proteins/peptides were identified in proteome of this centipede. Thus, most putative toxins may be expressed at low levels in venoms. Overall, centipede venom unexpectedly contains a variety of toxin-like proteins/peptides.
Furthermore, several toxin-like proteins exhibited overlapping expressed in venom gland and torso tissues. Most toxin-like proteins were expressed at higher levels in the venom gland than in the torso tissue. Additionally, some toxin-like proteins were enriched in the torso tissue. Thus, venom gene recruitment was observed in centipede, based on their patterns of transcript expression and peptides detected in the proteome. Researchers have hypothesized that toxin genes display a recruitment process in snake venom (24, 25). Venom gene homologs in snakes were recently shown to be expressed in many different tissues outside of the oral glands (26, 36). Compared with venom gland-specific expression putative toxins, most neurotoxins and ion channel inhibitors from centipede displayed extraordinarily higher levels of expression in venoms than in torso tissues. Thus, our study is the first to report that venom genes were recruited from outside of the centipede venom gland using transcriptomic and proteomic approaches. Additionally, like neurotoxins and ion channel inhibitors, numerous homologs of transcripts or peptides/proteins with different sequences were identified. This result provided evidence that these toxin genes had undergone a gene duplication process in centipede that resulted in the expansion of multilocus venom gene families. This process was consistent with a similar phenomenon observed in snakes in previous reports (36, 37).
Finally, the centipede is a traditional Chinese medicine that has been applied over two thousand years in China (38). In addition to centipede toxin-like proteins/peptides, our results also reveal that additional active molecules are expressed in venom gland and torso tissues. Our results hint that the use of the whole dried centipede body is the proper medication (Pharmacopeia of the Peoples' Republic of China 1992). Importantly, our data provide important clues for methods to improve the application of centipede as a traditional Chinese medicine.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via iProX (http://www.iprox.org/) with the dataset identifier IPX0001117000. Meanwhile, the data was also shared in our Lab website (database.amddlab.org).
Supplementary Material
Acknowledgments
We thank the members of our research groups for providing technical assistance and participating in discussions.
Footnotes
* This work was supported by grants from the Chinese National Natural Science Foundation (81373945, 31560596, U1602225, and 31572268), the Key Research Program of the Chinese Academy of Sciences (KJZD-EW-L03), the “Yunnan Scholar” Program, the Yunnan Applied Basic Research Projects (2016FD076), and Puer University (RCXM003 & CXTD011).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ACN
- acetonitrile
- MeOH
- methanol
- TFA
- trifloroacetic acid
- DTT
- dithiotreitol
- FDR
- false discovery rate.
REFERENCES
- 1. Undheim E. A., and King G. F. (2011) On the venom system of centipedes (Chilopoda), a neglected group of venomous animals. Toxicon 57, 512–524 [DOI] [PubMed] [Google Scholar]
- 2. Rong M., Yang S., Wen B., Mo G., Kang D., Liu J., Lin Z., Jiang W., Li B., Du C., Yang S., Jiang H., Feng Q., Xu X., Wang J., and Lai R. (2015) Peptidomics combined with cDNA library unravel the diversity of centipede venom. J. Proteomics 114, 28–37 [DOI] [PubMed] [Google Scholar]
- 3. Edgecombe G. D., and Giribet G. (2007) Evolutionary biology of centipedes (Myriapoda: Chilopoda). Annu. Rev. Entomol. 52, 151–170 [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y. (2015) Why do we study animal toxins? Zoological Res. 36, 183–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knysak I., Martins R., and Bertim C. R. (1998) Epidemiological aspects of centipede (Scolopendromorphae: Chilopoda) bites registered in greater S. Paulo, SP, Brazil. Rev. Saude Publica 32, 514–518 [DOI] [PubMed] [Google Scholar]
- 6. Uzel A. P., Steinmann G., Bertino R., and Korsaga A. (2009) [Necrotizing fasciitis and cellulitis of the upper limb resulting from centipede bite: two case reports]. Chir. Main 28, 322–325 [DOI] [PubMed] [Google Scholar]
- 7. Mohri S., Sugiyama A., Saito K., and Nakajima H. (1991) Centipede bites in Japan. Cutis 47, 189–190 [PubMed] [Google Scholar]
- 8. Undheim E. A., Fry B. G., and King G. F. (2015) Centipede venom: recent discoveries and current state of knowledge. Toxins 7, 679–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hakim M. A., Yang S., and Lai R. (2015) Centipede venoms and their components: resources for potential therapeutic applications. Toxins 7, 4832–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Undheim E. A., Jenner R. A., and King G. F. (2016) Centipede venoms as a source of drug leads. Expert Opin. Drug Discovery 11, 1139–1149 [DOI] [PubMed] [Google Scholar]
- 11. Mohamed A. H., Zaid E., El-Beih N. M., and Abd El-Aal A. (1980) Effects of an extract from the centipede Scolopendra moristans on intestine, uterus and heart contractions and on blood glucose and liver and muscle glycogen levels. Toxicon 18, 581–589 [DOI] [PubMed] [Google Scholar]
- 12. Welsh J. H., and Batty C. S. (1963) 5-Hydroxytryptamine content of some arthropod venoms and venom-containing parts. Toxicon 1, 165–170 [Google Scholar]
- 13. Nagpal N., and Kanwar U. (1981) The poison gland in the centipede Otostigmus ceylonicus; morphology and cytochemistry. Toxicon 19, 898–902 [DOI] [PubMed] [Google Scholar]
- 14. Freyvogel T. A. (1972) Poisonous and venomous animals in East Africa. Acta Trop. 29, 401–451 [PubMed] [Google Scholar]
- 15. You W. K., Sohn Y. D., Kim K. Y., Park D. H., Jang Y., and Chung K. H. (2004) Purification and molecular cloning of a novel serine protease from the centipede, Scolopendra subspinipes mutilans. Insect Biochem. Mol. Biol. 34, 239–250 [DOI] [PubMed] [Google Scholar]
- 16. Liu Z. C., Zhang R., Zhao F., Chen Z. M., Liu H. W., Wang Y. J., Jiang P., Zhang Y., Wu Y., Ding J. P., Lee W. H., and Zhang Y. (2012) Venomic and transcriptomic analysis of centipede Scolopendra subspinipes dehaani. J. Proteome Res. 11, 6197–6212 [DOI] [PubMed] [Google Scholar]
- 17. Yang S., Liu Z., Xiao Y., Li Y., Rong M., Liang S., Zhang Z., Yu H., King G. F., and Lai R. (2012) Chemical punch packed in venoms makes centipedes excellent predators. Mol. Cell. Proteomics 11, 640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang S., Xiao Y., Kang D., Liu J., Li Y., Undheim E. A., Klint J. K., Rong M., Lai R., and King G. F. (2013) Discovery of a selective NaV1.7 inhibitor from centipede venom with analgesic efficacy exceeding morphine in rodent pain models. Proc. Natl. Acad. Sci. U.S.A. 110, 17534–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang S., Yang F., Wei N., Hong J., Li B., Luo L., Rong M., Yarov-Yarovoy V., Zheng J., Wang K., and Lai R. (2015) A pain-inducing centipede toxin targets the heat activation machinery of nociceptor TRPV1. Nat. Commun. 6, 8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen M., Li J., Zhang F., and Liu Z. (2014) Isolation and characterization of SsmTx-I, a Specific Kv2.1 blocker from the venom of the centipede Scolopendra Subspinipes Mutilans L. Koch. J. Pept. Sci. 20, 159–164 [DOI] [PubMed] [Google Scholar]
- 21. Peng K., Kong Y., Zhai L., Wu X., Jia P., Liu J., and Yu H. (2010) Two novel antimicrobial peptides from centipede venoms. Toxicon 55, 274–279 [DOI] [PubMed] [Google Scholar]
- 22. Hou H., Yan W., Du K., Ye Y., Cao Q., and Ren W. (2013) Construction and expression of an antimicrobial peptide scolopin 1 from the centipede venoms of Scolopendra subspinipes mutilans in Escherichia coli using SUMO fusion partner. Protein Expr. Purif. 92, 230–234 [DOI] [PubMed] [Google Scholar]
- 23. Gonzalez-Morales L., Pedraza-Escalona M., Diego-Garcia E., Restano-Cassulini R., Batista C. V., Gutierrez Mdel C., and Possani L. D. (2014) Proteomic characterization of the venom and transcriptomic analysis of the venomous gland from the Mexican centipede Scolopendra viridis. J. Proteomics 111, 224–237 [DOI] [PubMed] [Google Scholar]
- 24. Alape-Giron A., Persson B., Cederlund E., Flores-Diaz M., Gutierrez J. M., Thelestam M., Bergman T., and Jornvall H. (1999) Elapid venom toxins: multiple recruitments of ancient scaffolds. Eur. J. Biochem. 259, 225–234 [DOI] [PubMed] [Google Scholar]
- 25. Fry B. G., Roelants K., Champagne D. E., Scheib H., Tyndall J. D., King G. F., Nevalainen T. J., Norman J. A., Lewis R. J., Norton R. S., Renjifo C., and de la Vega R. C. (2009) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Ann. Rev. Genomics Human Gen. 10, 483–511 [DOI] [PubMed] [Google Scholar]
- 26. Reyes-Velasco J., Card D. C., Andrew A. L., Shaney K. J., Adams R. H., Schield D. R., Casewell N. R., Mackessy S. P., and Castoe T. A. (2015) Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol. Biol. Evol. 32, 173–183 [DOI] [PubMed] [Google Scholar]
- 27. Zhao F., Yan C., Wang X., Yang Y., Wang G., Lee W., Xiang Y., and Zhang Y. (2014) Comprehensive transcriptome profiling and functional analysis of the frog (Bombina maxima) immune system. DNA Res. 21, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao F., Guo X., Wang Y., Liu J., Lee W. H., and Zhang Y. (2014) Drug target mining and analysis of the Chinese tree shrew for pharmacological testing. PLoS ONE 9:e104191, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pertea G., Huang X., Liang F., Antonescu V., Sultana R., Karamycheva S., Lee Y., White J., Cheung F., Parvizi B., Tsai J., and Quackenbush J. (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652 [DOI] [PubMed] [Google Scholar]
- 30. Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., and Robles M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 31. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trapnell C., Pachter L., and Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang H., and Wong W. H. (2009) Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25, 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Savitski M. M., Nielsen M. L., Kjeldsen F., and Zubarev R. A. (2005) Proteomics-grade de novo sequencing approach. J. Proteome Res. 4, 2348–2354 [DOI] [PubMed] [Google Scholar]
- 36. Vonk F. J., Casewell N. R., Henkel C. V., Heimberg A. M., Jansen H. J., McCleary R. J., Kerkkamp H. M., Vos R. A., Guerreiro I., Calvete J. J., Wuster W., Woods A. E., Logan J. M., Harrison R. A., Castoe T. A., de Koning A. P., Pollock D. D., Yandell M., Calderon D., Renjifo C., Currier R. B., Salgado D., Pla D., Sanz L., Hyder A. S., Ribeiro J. M., Arntzen J. W., van den Thillart G. E., Boetzer M., Pirovano W., Dirks R. P., Spaink H. P., Duboule D., McGlinn E., Kini R. M., and Richardson M. K. (2013) The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. U.S.A. 110, 20651–20656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Casewell N. R., Huttley G. A., and Wuster W. (2012) Dynamic evolution of venom proteins in squamate reptiles. Nat. Commun. 3, 1066. [DOI] [PubMed] [Google Scholar]
- 38. Chen K., and Yu B. (1999) Certain progress of clinical research on Chinese integrative medicine. Chinese Med. J. 112, 934–937 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via iProX (http://www.iprox.org/) with the dataset identifier IPX0001117000. Meanwhile, the data was also shared in our Lab website (database.amddlab.org).