Abstract
Enterococcus faecalis is a bacterial pathogen associated with both endodontic and systemic infections. The biofilm formation ability of E. faecalis plays a key role in its virulence and drug resistance attributes. The formation of E. faecalis biofilms on implanted medical devices often results in treatment failure. In the present study, we report protein markers associated with the biofilm formation ability of E. faecalis using iTRAQ-based quantitative proteomics approach. In order to elucidate the biofilm-associated protein markers, we investigated the proteome of strong and weak biofilm-forming E. faecalis clinical isolates in comparison with standard American Type Culture Collection (ATCC) control strains. Comparison of E. faecalis strong and weak biofilm-forming clinical isolates with ATCC control strains showed that proteins associated with shikimate kinase pathway and sulfate transport were up-regulated in the strong biofilm former, while proteins associated with secondary metabolites, cofactor biosynthesis, and tetrahydrofolate biosynthesis were down-regulated. In the weak biofilm former, proteins associated with nucleoside and nucleotide biosynthesis were up-regulated, whereas proteins associated with sulfate and sugar transport were down-regulated. Further pathway and gene ontology analyses revealed that the major differences in biofilm formation arise from differences in metabolic activity levels of the strong and weak biofilm formers, with higher levels of metabolic activity observed in the weak biofilm former. The differences in metabolic activity could therefore be a major determinant of the biofilm ability of E. faecalis. The new markers identified from this study can be further characterized in order to understand their exact role in E. faecalis biofilm formation ability. This, in turn, can lead to numerous therapeutic benefits in the treatment of this oral and systemic pathogen. The data has been deposited to the ProteomeXchange with identifier PXD006542.
Enterococcus faecalis is a Gram-positive, facultative anaerobic coccus usually considered as a harmless commensal in the gastrointestinal tract of humans (1). However, recent surveillance data indicate that E. faecalis is emerging as a leading pathogen in nosocomial infections accounting for about 90% of all cases of life-threatening enterococcal infections such as bacterial endocarditis, urinary tract infections, wound infections, and bacteremia leading to complications like meningitis, among others (2–4). In addition, E. faecalis is also greatly associated with root canal (endodontic) infections in the field of dentistry (5–7).
The success of E. faecalis as a clinical pathogen lies in its virulence attributes, including biofilm formation. E. faecalis is able to adhere to surfaces and form surface-attached microbial communities embedded in a matrix of extracellular polymeric substances known as biofilms (8–11). The National Institutes of Health estimates that as many as 80% of bacterial infections involve biofilm mode of growth, which contributes significantly to morbidity and mortality (12).
E. faecalis biofilm infections are extremely difficult to treat, especially when biofilms are formed on implanted medical devices. Hence, biofilm formation of E. faecalis is directly associated with therapeutic failure. In the field of dentistry, E. faecalis is known to form biofilms in the root canals of teeth that cause treatment failure, adding heavy economic burden (6, 13–15).
Strategies to combat the biofilm formation of E. faecalis can lead to potential therapeutic benefits. However, there is currently a limited understanding of the factors associated with biofilm formation of E. faecalis. Though, some of the previous genetic screening studies have suggested the role of genes coding for enterococcal surface protein Esp, aggregation substance AS, gelatinase GelE, and quorum sensing system Fsr in E. faecalis biofilm formation, the association has always not been clear due to conflicting literature reports (16–21). Moreover, previous proteomics studies on E. faecalis have been largely focused on the evaluation of the differential protein expression patterns in planktonic E. faecalis cells exposed to antibiotics or to stress conditions (22–26).
In the current study, we have addressed this knowledge gap by employing an iTRAQ-based proteomics analysis of E. faecalis biofilms. The present study took a novel approach to examine the protein expression profiles of E. faecalis biofilms by comparing the proteomics profile of strong and weak biofilm-forming clinical isolates with commonly used ATCC laboratory isolates. This new approach and the comprehensive examination of proteomics profiles helped in obtaining direct evidence on biofilm regulators and the possible mechanisms governing biofilm formation of E. faecalis.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Strong and weak biofilm-forming E. faecalis clinical isolates, namely Ef 63 and Ef 64, derived from patients with failed root canal treatments, were included in the current study (7, 27). Standard laboratory strains E. faecalis ATCC 29212 and E. faecalis ATCC 51299 were used as the control strains. Bacterial strains were cultured overnight in Brain-Heart Infusion broth (Oxoid, Singapore) at 37°C under shaking conditions prior to each experiment.
Biofilm Formation
E. faecalis biofilms were formed as described previously (28). Briefly, overnight cultures of E. faecalis isolates were harvested, washed with PBS, and resuspended in the culture medium to ∼107 cells/ml. The cells were then seeded into the wells of a flat-bottomed, polystyrene microtiter plate and incubated at 37 °C for 48 h to allow for biofilm formation. Biofilm formation for each of the four E. faecalis isolates included in the study was performed in biological replicates.
Protein Extraction from E. faecalis Biofilms
E. faecalis biofilm proteins were extracted by harvesting and pelleting the biofilm cells. The resultant pellets were washed once with PBS and then resuspended in a buffer containing 0.5 m triethylammonium bicarbonate, pH 8.5, and 1% SDS. The resuspended cells were homogenized in a Micro Smash MS-100 beater (Tomy Seiko Co., Ltd., Japan) with 0.5-mm glass beads in screw cap tubes at a pulse of 4,000 rpm for 20 s in 8eight cycles. The homogenized cells were centrifuged to obtain the proteins in the supernatant. Protein estimation was carried out using Bradford assay (Bio-Rad). 100 μg proteins from each sample were used for iTRAQ1 labeling.
iTRAQ Labeling and Sample Cleanup
100 μg of proteins extracted from the E. faecalis isolates 63, 64, 51299, and 29212 were labeled using iTRAQ Reagent 8-plex kit (SCIEX) according to the manufacturer's instructions with slight modifications. The first set of biological replicates for E. faecalis isolates Ef 63, Ef 64, ATCC 51299, and ATCC 29212 was labeled with labeling reagents 113, 114, 115, and 116, respectively. The second set of biological replicates for E. faecalis isolates Ef 63, Ef 64, ATCC 51299, and ATCC 29212 was labeled with labeling reagents 117, 118, 119, and 121, respectively. The iTRAQ labeling workflow is represented in Fig. 1. Briefly, protein samples were reduced with 5 mm TCEP at 65 °C for 60 min, alkylated with 10 mm MMTS for 15 min at room temperature, and then digested with trypsin (Promega) in the ratio of 1:20 at 37 °C for 16 h. The digested protein samples were labeled with iTRAQ reagent labels according to the manufacturer's protocol at room temperature for 2 h. Following this, the eight iTRAQ-labeled peptide samples were combined into a single centrifuge tube and were subjected to strong cation exchange to remove interfering substances such as excess iTRAQ reagent labels, organic solvent, and SDS according to the manufacturer's protocol (SCIEX). The resultant elutes were desalted using Sep-Pak C18 cartridges (Water), and lyophilized before reconstitution in 20 mm ammonium formate in water of pH 10 for two-dimensional liquid chromatography (2D-LC) separation.
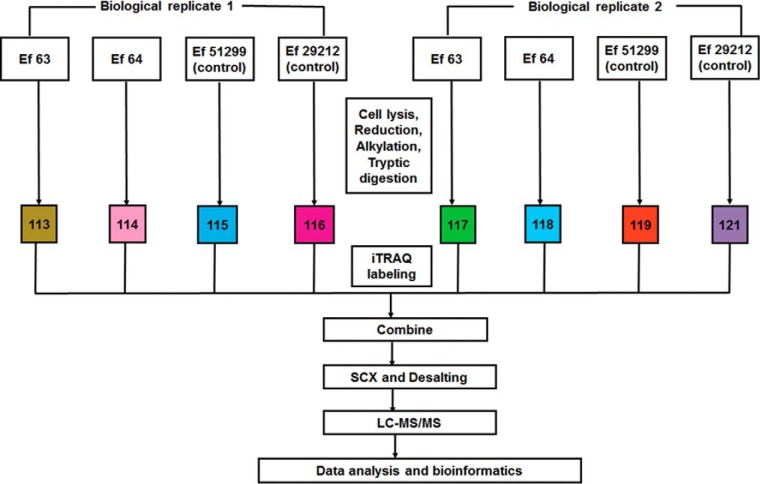
Fig. 1.
Experimental workflow for iTRAQ labeling and analysis: iTRAQ 8-plex labeling was performed for two sets of biological replicates of the strains Ef 63, Ef 64, ATCC 51299, and ATCC 29212. Each strain was labeled as follows for the two biological replicates: Ef 63 (113, 117), Ef 64 (114, 118), ATCC 51299 (115, 119), and ATCC 29212 (116, 121). The labeled fractions were combined and subjected to strong cation exchange chromatography and desalting, followed by separation using liquid chromatography mass spectrometry (LC-MS/MS), and bioinformatics data analysis.
2D LC-MS/MS Analysis
2D LC-MS/MS analysis was carried out as described previously (29, 30). Briefly, the peptides were first separated with the 1290 Infinity LC system (Agilent) linked with a reversed-phase column to achieve the first dimension of the peptide separation. Approximately 100 μg of the labeled peptides mixture reconstituted in 20 mm ammonium formate in water, pH 10, were injected using the micro-pickup loop mode into an Xbridge C18 column (C18, 3.5 μm, 3.0 mm × 150 mm, Waters Corp., Milford, MA). Mobile phase A consisting of 20 mm ammonium formate in water, pH 10, and mobile phase B consisting of 20 mm ammonium formate in 80% acetonitrile, pH 10, were used. The flow rate was set at 0.2 ml/min, and a total of 96 elute fractions were collected on a 96-well v-bottom plate using step gradients of mobile B: 0% for 10 min, 0–80% for 60 min, held at 80% for 5 min, 80–100% for 1 min, held at 100% for 5 min, 100–0% for 1 min, held at 0% for 14 min. The eluted fractions were pooled into ten fractions and desalted with Sep-Pak® tC18 _Elution Plate (186002318, Waters Corp.) using a vacuum manifold (Millipore) before second dimension reversed-phase chromatography. The desalted pooled sample was then lyophilized and reconstituted with 98% water, 2% acetonitrile with 0.1% formic acid. Peptides were separated on the second dimension of peptide separation using nanoLC Ultra and ChiPLC-nanoflex (Eksigent, Dublin, CA) in Trap Elute configuration. The samples were loaded on a 200 μm × 0.5 mm column (ChromXP C18-CL, 3 μm) and eluted on an analytical 75 μm × 15 cm column (ChromXP C18-CL, 3 μm). Mobile phase A containing 2% acetonitrile in H2O and 0.1% formic acid and mobile phase B containing 98% acetonitrile in H2O and 0.1% formic acid were used. The flow rate was set at 0.3 μl/min with the following gradient elution for peptide separation: 5 to 5% of mobile phase B in 1 min, 5 to 12% of mobile phase B in 19 min, 12 to 30% of mobile phase B in 120 min, 30 to 70% of mobile phase B in 10 min, 70 to 90% of mobile phase B in 2 min, 90 to 90% in 7 min, 90 to 5% in 6 min, and held at 5% of mobile phase B for 10 min.
MS analysis was performed in information-dependent mode using a 5600 TripleTOF analyzer (QqTOF; AB SCIEX). Precursor ions were selected across a mass range of 400–1800 m/z using 250 ms accumulation time per spectrum. A maximum of 20 most abundant precursor ions per cycle from each MS spectra were selected for MS/MS analyses with 100 ms accumulation time for each precursor, with a dynamic exclusion of 15 s. MS/MS recording was carried out in high sensitivity mode with rolling collision energy and with the iTRAQ reagent collision energy adjustment turned on.
Peptide and Protein Identification
Peptide identification and quantification was carried out on the ProteinPilot 5.0 software Revision 1656 (AB SCIEX) using the Paragon™ algorithm and the integrated false discovery rate (FDR) analysis function. In addition, further processing was conducting using the Pro Group™ algorithm where isoform-specific quantification was employed to determine potential differences between the expressions of various isoforms. This Pro Group algorithm involves calculating the protein ratios using only ratios generated from the spectra that are unique to each protein or protein isoform. This would eliminate any masking of changes in expression that results from peptides that are shared between proteins. The data were searched against a protein sequence database downloaded from UniProtKB for E. faecalis 29212 on February 2, 2016. To increase the specificity of the search, these protein sequence data are added with the international protein index human protein sequence database from the last release (July 2012). The following user-defined search parameters were used for the obtained MS/MS spectra: sample type: iTRAQ 8-plex (peptide labeled); cysteine alkylation: methyl methane-thiosulfonate (MMTS); digestion: trypsin; instrument: TripleTOF5600; special factors: none; species: none; identification (ID) focus: biological modification; database: uniprot_efaecalis_ipi_human.fasta; search effort: thorough; FDR analysis: yes. For the iTRAQ quantification, the peptide used for quantification was automatically selected based on the Pro Group algorithm to determine the reporter peak area, error factor, and ρ-value. The resultant data were subjected to auto bias-correction to remove any possible variations arising from unequal mixing during the combination of different labeled samples. The ProteinPilot software calculates the median ratio of all identified proteins based on an internal reference channel In order to account for potential iTRAQ ratio compression, background correction was performed to make fold changes closer to their actual values as described by Strbenac et al., and this function of the ProteinPilot software uses one of the best algorithms to serve this purpose (31). The ProteinPilot software counts each iTRAQ-modified peptide as unique to each identified protein. The peak areas and the signal-to-noise ratios were extracted from the database by ProteinPilot to translate the raw data into a quantifiable data for downstream analysis.
The FDR determination of the data was subsequently conducted by searching against the concatenated databases made up of in silico on-the-fly reversal for decoy sequences that were generated automatically by the software based on our combined E. faecalis UniProtKB and human ipi search database. The unused protein score of ≥1.3 was used as the cutoff for correctly identified proteins, corresponding to 0.4% FDR. The FDR was calculated using the number of false positive hits as indicated by the reversed sequences over the total number of proteins after applying the cutoff of ≥1.3 unused protein score. The results were then exported into Microsoft Excel for manual data interpretation. The cutoff threshold for up-regulation and down-regulation of proteins was determined using population statistics applied to the biological replicates as proposed by Gan et al. (32, 33).
After determining the cutoff threshold, the four iTRAQ ratios obtained from each of the biological replicates of Ef 63 or Ef 64 over the biological replicates of the control Ef 51299 or Ef 29212 (e.g. 113/115, 113/116, 117/115, and 117/116) were subjected to the one sample t test analysis to determine significant change in protein abundance. The iTRAQ ratios were transformed by log2 before averaging them. These sets of four iTRAQ ratios were compared with a set of four dummy ratios of zero that represents no changes in protein abundance. A one sample t test was then conducted to evaluate whether the iTRAQ ratio of a protein is truly different from the mean of zero. Only the proteins showing p values < 0.05 for their mean iTRAQ ratios were considered statistically significant and were selected for further analysis.
Gene Ontology Analysis
The identified proteins were subjected to gene ontology analysis using Cytoscape (v2.8.3) (34) with Biological Network Gene Ontology Tool (BINGO) plugin (v2.44) (35). The resultant network figures were exported and visualized for overrepresentation of pathways as described previously (36).
RNA Extraction and Real-time PCR
RNA was extracted from 48 h E. faecalis biofilms using Qiagen RNeasy Protect Bacteria Mini Kit, followed by reverse transcription and real-time PCR analysis to test for the gene expression levels as previously described (27). Primer sequences for the genes of interest were generated using National Center for Biotechnology Information (NCBI) primer-Basic Local Alignment Search Tool (BLAST) search. The list of primers used in this study is provided in Supplemental Table 1.
Experimental Design and Statistical Rationale
48 h grown E. faecalis biofilms were collected for iTRAQ-based mass spectrometry analysis. Biofilm proteins were extracted from two independent sets of each of the four strains Ef 63, Ef 64, Ef 51299, and Ef 29212 and were included as biological replicates in the iTRAQ study (Fig. 1). Protein expression profiles of the two control strains Ef 51299 and Ef 29212 were taken as the basis for determining the up-regulation/down-regulation patterns in the strains Ef 63 and Ef 64. Proteins meeting the required cutoff threshold ratios (Figs. 2 and 3) and showing significant differential expression (One sample t test p value < 0.05) were considered as up-regulated or down-regulated based on their expression patterns. Only the proteins that are up-regulated/down-regulated wrt both Ef 51299 and 29212 were included for further gene ontology and pathway enrichment analysis. For real-time PCR analysis, RNA was extracted from four different biological replicates of the strains Ef 63, Ef 64, Ef 51299, and Ef 29212. Real-time PCR analysis was performed using four biological replicates and two technical replicates each. Significance of the results was analyzed following analysis of variance. Differences were considered significant when p values are less than 0.05.
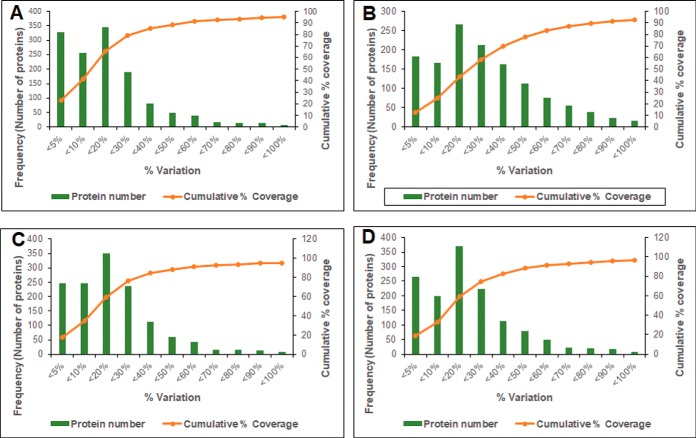
Fig. 2.
Determination of experimental variation for the identified proteins: (A) experimental variation of Ef 63 compared with ATCC 29212; (B) experimental variation of Ef 64 compared with ATCC 29212; (C) experimental variation of Ef 63 compared with ATCC 51299; and (D) experimental variation of Ef 64 compared with ATCC 51299. The horizontal axis represents % variation of iTRAQ ratios. The primary vertical axis represents the corresponding number of proteins (bars) having different % variation. The secondary vertical axis represents the cumulative % of the counted proteins. Variation against 88% coverage of population was considered as the cutoff value.
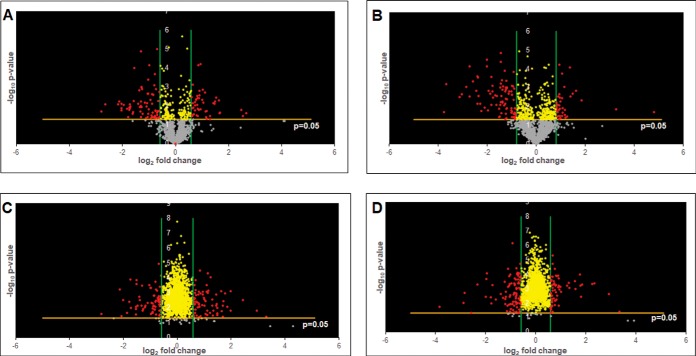
Fig. 3.
Volcano plots of differentially regulated proteins reveal significance patterns: (A) volcano plots of differentially regulated proteins of Ef 63 compared with ATCC 29212; (B) volcano plots of differentially regulated proteins of Ef 64 compared with ATCC 29212; (C) volcano plots of differentially regulated proteins of Ef 63 compared with ATCC 51299; and (D) volcano plots of differentially regulated proteins of Ef 64 compared with ATCC 51299. Threshold cutoffs determined for log2 fold change ratios are represented by green lines. p value cutoff of 0.05 is represented by a yellow line. Gray dots represent proteins that do not show significant differences in expression, yellow dots represent proteins that show significant differences in expression but within the cutoff log2 fold change ratios, and red dots represent proteins that show significant differences in expression outside the cut-off log2 fold change ratios.
RESULTS
iTRAQ-based Quantitative Analysis of the Proteome of E. faecalis Isolates Ef 63 and Ef 64
In order to identify the biomarkers associated with the biofilm formation ability of E. faecalis, an iTRAQ-based mass spectrometry quantitative analysis was performed. Previously, we found that E. faecalis clinical isolates Ef 63 and Ef 64 had significantly stronger and weaker biofilm formation ability, respectively, when compared with that of the two ATCC strains of E. faecalis (27). E. faecalis ATCC strains 51299 and 29212 showed comparable levels of biofilm formation, and both were included for iTRAQ analysis in order to increase the robustness of comparison with the strong and the weak biofilm forming E. faecalis isolates (Fig. 1). A qualification criterion of unused protein score ≥1.3 (was used to obtain a total of 1412 proteins from the analysis. The corresponding global FDR for the unused score of ≥1.3 was 0.4%. The differential expression analysis of the proteome was performed by comparing the strong and the weak biofilm-forming E. faecalis isolates Ef 63 and Ef 64 against each of the ATCC strains individually. Analysis of fold changes of Ef 63 versus ATCC 51299 (113:115, 113:119, 117:115, 117:119), Ef 63 versus ATCC 29212 (113:116, 113: 121, 117:116, 117:121), Ef 64 versus ATCC 51299 (114:115, 114:119, 118:115, 118:119), and Ef 64 versus ATCC 29212 (114:116, 114:121, 118:116, 118:121) was carried out to get the differential expression profiles. The obtained fold changes were then filtered using population statistics to get the significant threshold cutoff of up-regulated and down-regulated proteins for each of the comparisons performed in the study. Percentage variations corresponding to 88% coverage were taken as the threshold cutoff and any values outside 88% were considered as significantly altered. Based on these population statistics, the following cutoff fold change values were obtained for Ef 63 versus ATCC 51299 set: >1.5-fold and <0.67 (1/1.5)-fold for up-regulated and down-regulated proteins corresponding to 50% variation, Ef 64 versus ATCC 51299: >1.5-fold and <0.67 (1/1.5)-fold for up-regulated and down-regulated proteins corresponding to 50% variation, Ef 63 versus ATCC 29212: >1.5-fold and <0.67 (1/1.5)-fold for up-regulated and down-regulated proteins corresponding to 50% variation, and Ef 64 versus ATCC 29212: >1.75-fold and <0.57 (1/1.75)-fold for up-regulated and down-regulated proteins corresponding to 75% variation (Fig. 2).
The profile of differentially expressed proteins in each comparison were represented using volcano plots by applying the fold change and p value cutoff values (Fig. 3). The proteins with fold changes beyond the cutoff values obtained from Fig. 2 and with p values < 0.05 were considered as the most significantly affected proteins in each comparison.
Identification of Differentially Regulated Proteins in the Strong and Weak Biofilm-forming E. faecalis Isolates Ef 63 and Ef 64
In order to identify the proteins that could be associated with the biofilm formation ability of E. faecalis, the list of differentially regulated proteins obtained from the previous analysis were further scrutinized and combined for a more robust comparison. Following a comparison of the strong and the weak biofilm-forming strains with both the ATCC control strains, only the list of proteins that were showing significant up-regulation or down-regulation with respect to both 51299 and 29212 were selected as significantly up-regulated and down-regulated in the strong and the weak biofilm formers.
(a) Up-regulated Proteins in the Strong Biofilm Former Ef 63
The proteins that are significantly up-regulated in the strong biofilm former Ef63 in comparison with both the ATCC strains are listed in Table I. Of these proteins, shikimate kinase encoded by aroK, and 5-enolpyruvylshikimate-3-phosphate synthase encoded by aroA are enzymes involved in the intermediate and penultimate steps of the shikimate pathway, which is essential for the synthesis of aromatic amino acids in bacteria (37, 38). Trehalose-6-phosphate encoded by trePP is a part of trehalose metabolic pathway (39). Other up-regulated proteins include prepentate dehydratase encoded by pheA, which is a part of the phenylalanine biosynthesis pathway and a sulfate permease protein encoded by ychM, belonging to sulfate transporter/secondary anion transporters superfamily (40, 41). Members of the phage family protein, a putative oxidoreductase DR75_1429, and hypothetical proteins DR75_2587, DR75_1646, and DR75_1626 were also up-regulated. Hypothetical protein DR75_2587 belongs to a putative lipoprotein family based on NCBI BLAST results.
Table I. Functional classification of the significantly up-regulated proteins of Ef 63 in comparison with both ATCC 29212 and 51299 strains meeting both p value cutoff and log2 fold-change cutoff criteria.
| Proteins upregulated in 63 | ||||||
|---|---|---|---|---|---|---|
| Accession # | Protein name | Gene name | Unused protein score | Peptides (95%) | Fold change wrt 29212 | Fold change wrt 51299 |
| UPI00032E9DD8 | 3-phosphoshikimate 1-carboxyvinyl transferase; 5-enolpyruvylshikimate-3-phosphate synthase | aroA; DR75_569 | 26.4 | 26 | 3.110713011 | 3.409029024 |
| UPI000291C4DE | Trehalose 6-phosphate phosphorylase | trePP | 18.68 | 13 | 3.023335133 | 1.983570371 |
| UPI00019F352C | Shikimate kinase (SK) (EC 2.7.1.71); shikimate kinase family protein | aroK; DR75_570 | 10.04 | 5 | 2.480368467 | 3.124256418 |
| UPI000005C662 | Putative oxidoreductase YcjS; oxidoreductase, NAD-binding Rossmann fold family protein | DR75_1429 | 10 | 5 | 1.923075843 | 2.021658161 |
| UPI0001E1A33D | Prephenate dehydratase | pheA; DR75_571 | 8 | 5 | 2.891981312 | 3.264071801 |
| UPI0002910B7F | Phage major tail, phi13 family protein | DR75_1505 | 3.8 | 2 | 3.200903531 | 2.780474197 |
| UPI00019F3544 | Hypothetical protein | DR75_2587 | 2.18 | 2 | 2.823765229 | 1.523319053 |
| UPI000291BF2D | Hypothetical protein | DR75_1646 | 2 | 2 | 2.783602486 | 2.472863241 |
| UPI0001B1D951 | Sulfate permease; sulfate permease family protein | ychm; DR75_1732 | 2 | 2 | 1.752083299 | 1.823848359 |
| UPI0001B2BBD4 | Hypothetical protein DR75_1626 | DR75_1626 | 2 | 1 | 17.41533347 | 9.888407625 |
(b) Up-regulated Proteins in the Weak Biofilm Former Ef 64
The proteins that are up-regulated in the weak biofilm former Ef 64 are listed in Table II. Of these, orotidine-5-phosphate decarboxylase encoded by pyrF, dihydroorotase encoded by pyrA, orotate-phosphoribosyltransferase encoded by pyrE, and carbamoyl synthase encoded by carA are enzymes belonging to de novo pyrimidine nucleobase and nucleotide biosynthetic process. Bifunctional protein encoded by pyrR is a transcriptional regulator of the pyrimidine nucleotide operon. Other up-regulated proteins include ribosome recycling factor encoded by frr, which helps in the dissociation of ribosomes from the mRNA upon completion of translation and is essential for cell growth, and hypothetical proteins DR75_2329 and DR75_910 (42).
Table II. Functional classification of the significantly up-regulated proteins of Ef 64 in comparison with both ATCC 29212 and 51299 strains meeting both p value cutoff and log2 fold-change cutoff criteria.
| Proteins upregulated in 64 | ||||||
|---|---|---|---|---|---|---|
| Accession # | Protein names | Gene names | Unused protein score | Peptides (95%) | Fold change wrt 29212 | Fold change wrt 51299 |
| UPI000005C524 | Ribosome-recycling factor (RRF) (ribosome-releasing factor) | frr;DR75_1144 | 36.78 | 64 | 1.906634 | 1.764784 |
| UPI00019F32AD | Orotidine 5′-phosphate decarboxylase (EC 4.1.1.23) (OMP decarboxylase) | pyrF; DR75_712 | 20.48 | 12 | 2.080761 | 1.604881 |
| UPI000005C2BD | Bifunctional protein PyrR | pyrR; DR75_720 | 18.04 | 14 | 2.086637 | 1.805877 |
| UPI0001A5BFBD | Carbamoyl-phosphate synthase, small subunit | carA; DR75_716 | 18 | 11 | 1.980158 | 1.743035 |
| UPI000175BF0F | Dihydroorotase(DHOase) (EC3.5.2.3) | pyrC; DR75_717 | 10 | 5 | 2.627464 | 2.391439 |
| UPI000005C2B4 | Orotate phosphoribosyl transferase (OPRT) (OPRTase) (EC 2.4.2.10) | pyrE; DR75_711 | 2.66 | 2 | 2.216797 | 1.530228 |
| UPI0001B6DEC1 | Hypothetical protein DR75_2329 | DR75_2329 | 2.04 | 1 | 1.812651 | 7.603089 |
(c) Down-regulated Proteins in the Strong Biofilm Former Ef 63
The proteins that are down-regulated in the strong biofilm former Ef 63 are listed in Table III. Of these, DNA protection during starvation protein encoded by dps confers DNA protection during stationary phase, and galactose mutarotase encoded by galM is part of galactose metabolism (43, 44). Aspartate-1-decarboxylase encoded by panD, dihydroneopterin aldolase encoded by folB, and serine O-acetyltransferase encoded by cysE are enzymes involved in metabolic pathways such as biosynthesis of secondary metabolites, cofactor biosynthesis, and tetrahydrofolate biosynthesis. DR75_2966 and DR75_2967 are replication-associated proteins belonging to plasmids or mobile genetic elements of Gram-positive bacteria (45, 46). DR75_634 represents an ethanolamine two-component response regulator, which belongs to a phosphor-relay system. DR75_1696 and DR75_1697 are proteins belonging to the phosphoenol-pyruvate-dependent sugar transport system (PTS) responsible for the transport of ascorbate and lactose respectively.
Table III. Functional classification of the significantly down-regulated proteins of Ef 63 in comparison with both ATCC 29212 and 51299 strains meeting both p value cutoff and log2 fold-change cutoff criteria.
| Proteins downregulated in 63 | ||||||
|---|---|---|---|---|---|---|
| Accession # | Protein names | Gene names | Unused protein score | Peptides (95%) | Fold change wrt 29212 | Fold change wrt 51299 |
| UPI000005C830 | Dps family protein; DNA protection during starvation protein | dps; DR75_1942 | 25.85 | 74 | 0.158975959 | 0.141553384 |
| UPI0001B1DD9C | Galactose mutarotase -aldose epimerase conserved domain | galM | 19.17 | 13 | 0.649019509 | 0.549455743 |
| UPI0000059961 | Replication-associated protein RepB; cobQ/CobB/MinD/ParA nucleotide binding domain protein (plasmid) | DR75_2966 | 10.19 | 7 | 0.505973488 | 0.505026395 |
| UPI00019C6B36 | PTS system ascorbate-specific IIB component; PTS system, lactose/cellobiose specific IIB subunit | DR75_1696 | 8 | 8 | 0.242902381 | 0.518277773 |
| UPI000005C334 | Aspartate 1-decarboxylase (EC4.1.1.11) (aspartate alpha-decarboxylase) | panD; DR75_827 | 6.65 | 9 | 0.405554528 | 0.274218075 |
| UPI000005C853 | 7,8-dihydroneopterin aldolase (EC4.1.2.25); dihydroneopterin aldolase | folB; DR75_1970 | 2.47 | 2 | 0.631259615 | 0.438989396 |
| UPI0000059960 | Replication-associated protein B; putative replication-associated protein (plasmid) | DR75_2967 | 2.08 | 1 | 0.388184377 | 0.292597088 |
| UPI000005C269 | Ethanolamine two-component response regulator; ANTAR domain protein | DR75_634 | 2 | 1 | 0.546653837 | 0.60537555 |
| UPI0001B2588A | Glycerophosphoryl diester phosphodiesterase family protein | DR75_1000 | 2 | 1 | 0.57470862 | 0.297523575 |
| UPI000005BCA4 | Serine O-acetyltransferase | cysE; DR75_2093 | 2 | 1 | 0.399293872 | 0.34637114 |
(d) Down-regulated Proteins in the Weak Biofilm Former Ef 64
The proteins that are down-regulated in the weak biofilm former Ef 64 are listed in Table IV. Of these, UDP-N-acetylenolpyruvoylglucosamine reductase encoded by murB is involved in peptidoglycan biosynthesis pathway, guanosine monophosphate reductase encoded by guaC is involved in purine ribonucleotide interconversion, and aspartate semialdehyde dehydrogenase encoded by asd is involved in amino acid biosynthesis. DR75_2512 represents a PTS sugar transport system responsible for sorbose transport and DR75_2078 represents an ROK family protein responsible for repressing xylose utilization. Sulfate permease and trehalose-6 phosphate phosphorylase proteins encoded by ychM and trePP are down-regulated in the weak biofilm former Ef 64 in contrast to their up-regulation in the strong biofilm former Ef 63. Aspartate 1-decarboxylase encoded by panD is down-regulated in both Ef 64 and Ef 63. Other down-regulated proteins include a putative lipoprotein (DR75_1236), hydrolase (DR75_684), hypothetical proteins DR75_2043, and DR75_1811, which is a probable ABC transporter permease belonging to yitT family based on NCBI BLAST results.
Table IV. Functional classification of the significantly down-regulated proteins of Ef 64 in comparison with both ATCC 29212 and 51299 strains meeting both p value cutoff and log2-fold change cutoff criteria.
| Proteins downregulated in 64 | ||||||
|---|---|---|---|---|---|---|
| Accession # | Protein name | Gene name | Unused protein score | Peptides (95%) | Fold change wrt 29212 | Fold change wrt 51299 |
| UPI000005C57D | UDP-N-acetylenolpyruvoylglucosamine reductase | murB; DR75_1237 | 26.08 | 16 | 0.49488141 | 0.525412 |
| UPI000291C4DE | Trehalose 6-phosphate phosphorylase | trePP | 18.68 | 13 | 0.50414138 | 0.688992 |
| UPI000175BDE7 | Guanosine monophosphate reductase | guaC; DR75_1175 | 12.15 | 7 | 0.43528692 | 0.420406 |
| UPI00019C74B7 | PTS system IIB component; PTS system sorbose subIIB component family protein | DR75_2512 | 8.88 | 7 | 0.42363621 | 0.450195 |
| UPI000005C334 | Aspartate 1-decarboxylase (EC4.1.1.11) (aspartate alpha-decarboxylase) | panD; DR75_827 | 6.65 | 9 | 0.20464538 | 0.135274 |
| UPI000005C0BA | Aspartate-semialdehyde dehydrogenase | asd | 6 | 4 | 1.70472867 | |
| UPI0001E19F8C | Hypothetical protein DR75_1765-possible peptidase | DR75_1765 | 5.77 | 3 | 0.3620089 | 0.300302 |
| UPI000005BE6D | Xylose repressor; ROK family protein | DR75_2508 | 4.04 | 2 | 0.41415556 | 0.526703 |
| UPI000005BC96 | TdcF protein; reactive intermediate/imine deaminase family protein | DR75_2078 | 4.03 | 2 | 0.41691202 | 0.480218 |
| UPI000290D069 | Phage regulatory, Rha family protein | DR75_1539 | 4 | 3 | 0.83902121 | 0.87443 |
| UPI0001B1D951 | Sulfate permease | ychm; DR75_1732 | 2 | 2 | 0.522333 | 0.553958 |
| UPI0001E1A077 | Lipoprotein, putative | DR75_1236 | 2 | 1 | 0.3670319 | 0.382167 |
| UPI000005C88A | Hypothetical protein | DR75_2043 | 2 | 1 | 0.15033957 | 0.139521 |
Pathway Enrichment Analysis of the Up-regulated and the Down-regulated Proteins of Ef 63 and Ef 64
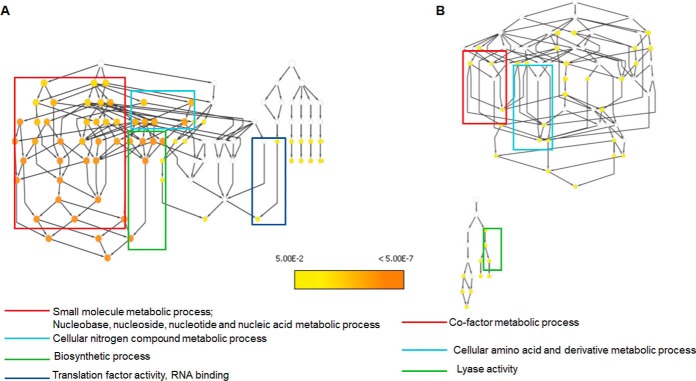
In order to test for pathway overrepresentation/enrichment, gene ontology analysis for the up-regulated and down-regulated proteins of the strong and weak biofilm formers was performed using BINGO Cytoscape plugin. The pathway enrichment analysis for each comparison was done under conditions GO_full and GO_slim categories. The GO_full analysis represents the full annotation of all the genes involved in various biological processes and molecular functions, whereas the GO_slim analysis represented only the generic pathways affected in each comparison set, corresponding to the bigger picture of pathways affected. The pathway enrichment analysis of up-regulated proteins in Ef 63 and down-regulated proteins in Ef 64 with respect to the two ATCC control strains did not result in any pathway mapping in the GO_slim analysis, whereas a network of biological processes and molecular functions, which were significantly affected was obtained in the GO_full analysis (Figs. S1 and S4). The analysis of the up-regulated proteins in Ef 64 with respect to the two control strains in GO_slim mode showed the pathways relating to small molecule metabolic process and nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, cellular nitrogen compound metabolic process, biosynthetic process, and RNA-binding activity to be significantly up-regulated (Fig 4A). The full list of up-regulated biological processes and molecular functions is provided in Fig. S2. The analysis of the down-regulated proteins in Ef 63 with respect to the two control strains in GO_slim mode showed the pathways relating to cofactor metabolic process, cellular amino acid and metabolic derivative process, and lyase activity to be significantly down-regulated (Fig 4B). The full list of down-regulated biological processes and molecular functions is provided in Fig. S3. Based on this analysis, there are a higher number of metabolic processes up-regulated in Ef 64 and down-regulated in Ef 63, indicating higher metabolic activity in the weak biofilm former.
Fig. 4.
Gene ontology analysis of differentially regulated proteins reveals significantly affected pathways in Ef 63 and Ef 64: (A) biological processes, molecular functions, and cellular components that are significantly up-regulated in Ef 64 in comparison with ATCC 29212 and 51299 strains and (B) biological processes, molecular functions, and cellular components that are significantly down-regulated in Ef 63 in comparison with ATCC 29212 and 51299 strains.
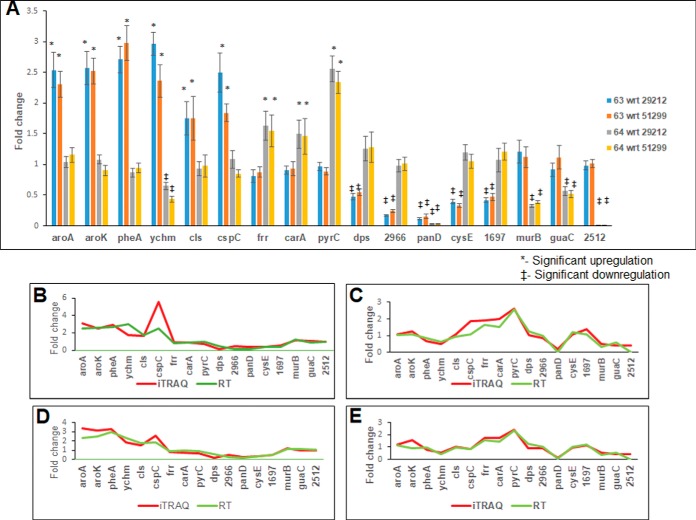
Correlation of iTRAQ Data with Quantitative Real-time PCR (qPCR) Results
In order to verify the reliability of the iTRAQ analysis, gene expression profiles of selected genes were obtained using qPCR. Representative genes for qPCR analysis were selected based on the most important pathways affected in each comparison and some of the unique transporter class proteins that are not represented in the pathway enrichment list. The significantly up-regulated and down-regulated gene expression profiles were obtained from qPCR (Fig 5A). The results obtained from qPCR analysis were then checked for their correlation with iTRAQ data by Pearson's correlation analysis. A significant positive correlation was obtained between the iTRAQ data and the qPCR results, which indicates that the gene expression profile and the protein expression profile follow a similar trend ensuring the reliability of iTRAQ analysis (Figs. 5B–5E).
Fig. 5.
Quantitative real-time PCR analysis and correlation with iTRAQ data: (A) quantitative real-time PCR analysis of selected significantly regulated genes obtained from iTRAQ analysis; (B) correlation of real-time gene expression data from A to iTRAQ protein expression data of 63 wrt 29212; (C) correlation of real-time gene expression data from A to iTRAQ protein expression data of 64 wrt 29212; (D) correlation of real-time gene expression data from A to iTRAQ protein expression data of 63 wrt 51299; and (E) correlation of real-time gene expression data from A to iTRAQ protein expression data of 64 wrt 51299.
DISCUSSION
Bacterial-biofilm-associated clinical problems have been described in a wide spectrum of Gram-positive as well as Gram-negative organisms, including Staphylococcus aureus, Streptococcus mutans, E. faecalis, Pseudomonas aeruginosa, and Escherichia coli (47, 48). However, the molecular markers and mechanism involved in E. faecalis biofilm formation is still not clearly understood. Previously, we found significant changes in the transcriptomic expression profile of E. faecalis isolates pertaining to several cellular and metabolic processes and transporters of the E. faecalis biofilm versus planktonic modes (27). Taking a step ahead, in this iTRAQ-based proteomics study, we specifically focused on the protein expression profiles of the strong and weak biofilm formers of E. faecalis in comparison to the ATCC laboratory strains.
Analysis of the protein profiles signatures across different strains elucidated interesting observations on metabolic processes, biosynthetic processes, and transport systems such as sugar transporters and sulfate permease involved in E. faecalis biofilm formation. For instance, the up-regulation of shikimate pathway proteins encoded by aroA and aroK in the strong biofilm former indicates an increased synthesis of aromatic amino acids in the strong biofilm former Ef 63. Certain studies have shown that the production of 5-enolpyruvylshikimate-3-phosphate synthase by aroA can lead to glyphosphate resistance in pathogenic bacteria such as S. aureus and the virulence, growth, and UV tolerance of the plant pathogen Burkholderia glumae (38, 49). This suggests that the strong biofilm former Ef 63 can display increased levels of glyphosphate resistance and pathogenesis mechanisms, possibly resulting in higher biofilm formation. Similarly, up-regulation of sulfate permease encoded by ychM in the strong biofilm former Ef 63, and down-regulation of sulfate permease in the weak biofilm former Ef 64, suggests that sulfur uptake/secondary anions transport have a role in E. faecalis biofilm formation ability. Sulfur uptake has been shown to be important for the virulence of certain pathogenic bacteria such as Mycobacterium tuberculosis (41). In addition, links between sulfur uptake, c-di-GMP signaling and biofilm formation have been found in Shewanella oneidensis, where biofilm formation is reduced due to decrease in sulfur uptake (48). Therefore, the increased and decreased levels of sulfate permease found in the biofilms of Ef 63 and Ef 64 could be associated with the strong biofilm formation capability of Ef 63, and the weak biofilm formation capability of Ef 64, respectively, indicating the possible role of ychM as a regulator of biofilm formation in E. faecalis.
Sugar transporter systems such as PTS system, lactose/cellobiose-specific IIB subunit and PTS system sorbose subIIB component family protein are down-regulated in Ef 63 and Ef 64, respectively. This suggests that the active transport of lactose and sorbose are down-regulated in the strong and the weak biofilm formers, respectively. Though the exact correlation between the expression of sugar transporters and biofilm formation is still not clear, previous studies of E. faecalis proteome in response to bile stress have shown up-regulation of the IIAB component of a mannose-specific PTS system (26). Similarly, comparative proteomics of E. faecalis clinical isolates and food isolates showed IIA, IIBC, and IIATP-binding cassette (ABC) components of the PTS system to be up-regulated in the food isolates of E. faecalis (25). Taken together, this indicates that sugar transport systems are differentially regulated according to the stress response and the isolate nature and may be possibly linked to the biofilm formation ability of E. faecalis, depending on the nature of the sugar transport system affected.
The pathway enrichment analysis of the differentially regulated proteins using Cytoscape revealed interesting insights into the metabolic profiles of the strong and the weak biofilm formers Ef 63 and Ef 64. In the isolate Ef 64, small molecule, nucleotide, and nitrogen compound metabolic processes and biosynthetic pathways are significantly up-regulated, indicating a higher metabolic activity profile of the weak biofilm former. In the isolate Ef 63, cofactor metabolic process, cellular amino acid and derivative metabolic process, and lyase activity are significantly down-regulated, indicating a lower metabolic activity profile of the strong biofilm former. Previous studies comparing biofilm and planktonic states of Streptococcus pneumoniae have suggested that biofilm cells exhibit low metabolic activity (50–52). In addition, certain studies on Burkholderia cenocepacia and P. aeruginosa have shown that persister cells, which represent a subpopulation of biofilm cells, show a lowered metabolic activity state capable of conferring protection against antibiotics and harmful environmental conditions (10, 53–55). It is possible that the metabolically active biofilm cells promote dispersal instead of adhesion, resulting in biofilms with less mass. Hence, the differences in the metabolism may have contributed to the strong and weak biofilm-forming ability of Ef 63 and Ef 64, respectively. Metabolic regulation is also observed in some multispecies oral biofilm models. In a mixed-species biofilm model of S. mutans, differences in fatty acid biosynthesis and branched chain amino acids metabolism were observed in the mixed-species biofilms in comparison with single-species biofilms (56). Similarly, in a mixed-species biofilm study of Anaeroglobulus geminatus, differentially regulated proteins were found to be involved in carbon metabolism, iron transport, and proteolysis. These studies show that constant changes in metabolic responses occur in bacterial biofilm systems in order to cope with a dynamically changing environment (57).
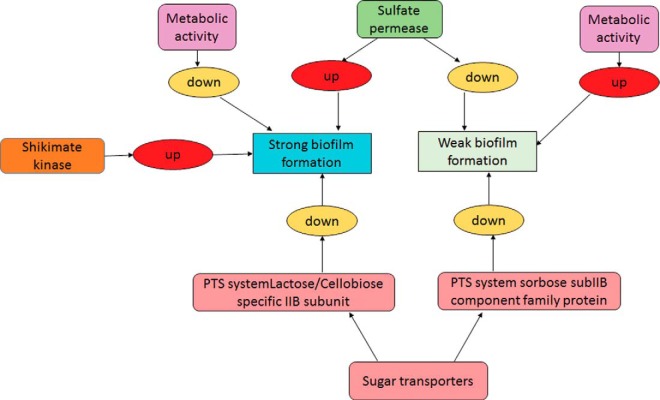
Correlation of the obtained iTRAQ data with RT-PCR results confirms the reliability of the analysis, serving as a validation for the novel regulators of biofilm formation identified from this study. In conclusion, this pioneering proteomics study has shed novel insights into the biofilm-forming ability of E. faecalis. It was found that the shikimate pathway, sugar and sulfate permease transporter systems, and metabolic profiles of E. faecalis may have a significant role in the biofilm formation of E. faecalis and its pathogenic traits (Fig. 6). Further studies using genetic knock-out mutants are warranted to elucidate the roles of the identified biomarkers associated with E. faecalis biofilm, which may possibly provide a new tool set to design novel treatment strategies for this recalcitrant bacterial pathogen.
Fig. 6.
Novel regulators of E. faecalis biofilm formation identified from iTRAQ analysis.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD006542.
Footnotes
* This study was supported by NUS-Start-up (R-221-000-064-133) and NMRC (R-221-000-086-511) to C.J.S.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- iTRAQ
- isobaric tags for relative and absolute quantitation
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- TOF
- time of flight
- 2D-LC
- two-dimensional liquid chromatography
- ABC
- ATP-binding cassette
- ATCC
- American Type Culture Collection
- BINGO
- Biological Network Gene Ontology tool
- BLAST
- Basic Local Alignment Search Tool
- ID
- identification
- iTRAQ
- isobaric tags for relative and absolute quantitation
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MMTS
- Methyl methane-thiosulfonate
- NCBI
- National Center for Biotechnology Information
- ROK
- repressor, open reading frame, kinase
- TOF
- time of flight
- UDP
- uridine diphosphate.
REFERENCES
- 1. Fisher K., and Phillips C. (2009) The ecology, epidemiology and virulence of Enterococcus. Microbiology. 155, 1749–1757 [DOI] [PubMed] [Google Scholar]
- 2. Hidron A. I., Edwards J. R., Patel J. R., Horan T. C., Sievert D. M., Pollock D. A., and Fridkin S. K. (2008) NHSN annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 29, 996–1011 [DOI] [PubMed] [Google Scholar]
- 3. Hollenbeck B. L., and Rice L. B. (2012) Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 3, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anders D., and Bruun N. E. (2013) Enterococcus faecalis infective endocarditis: Focus on clinical aspects. Expert Rev. Cardiovasc. Ther. 11, 1247–1257 [DOI] [PubMed] [Google Scholar]
- 5. Stuart C. H., Schwartz S. A., Beeson T. J., and Owatz C. B. (2006) Enterococcus faecalis: Its role in root canal treatment failure and current concepts in retreatment. J. Endod. 32, 93–98 [DOI] [PubMed] [Google Scholar]
- 6. Ricucci D., and Siqueira J. F. Jr. (2010) Biofilms and apical periodontitis: Study of prevalence and association with clinical and histopathologic findings. J. Endod. 36, 1277–1288 [DOI] [PubMed] [Google Scholar]
- 7. Zhu X., Wang Q., Zhang C., Cheung G. S., and Shen Y. (2010) Prevalence, phenotype, and genotype of Enterococcus faecalis isolated from saliva and root canals in patients with persistent apical periodontitis. J. Endod. 36, 1950–1955 [DOI] [PubMed] [Google Scholar]
- 8. Mohamed J. A., and Huang D. B. (2007) Biofilm formation by enterococci. J. Med. Microbiol. 56, 1581–1588 [DOI] [PubMed] [Google Scholar]
- 9. Garsin D. A., Willems R. J. (2010) Insights into the biofilm lifestyle of enterococci. Virulence 1, 219–221 [DOI] [PubMed] [Google Scholar]
- 10. Donlan R. M., and Costerton J. W. (2002) Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Høiby N., Bjarnsholt T., Givskov M., Molin S., and Ciofu O. (2010) Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332 [DOI] [PubMed] [Google Scholar]
- 12. Cook L. C., and Dunny G. M. (2013) Effects of biofilm growth on plasmid copy number and expression of antibiotic resistance genes in Enterococcus faecalis. Antimicrob. Agents Chemother. 57, 1850–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chávez de Paz L. (2007) Redefining the persistent infection in root canals: Possible role of biofilm communities. J. Endod. 33, 652–662 [DOI] [PubMed] [Google Scholar]
- 14. Preethee T., Kandaswamy D., and Hannah R. (2012) Molecular identification of an Enterococcus faecalis endocarditis antigen efaA in root canals of therapy-resistant endodontic infections. J. Conserv. Dent. 15, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Al-Ahmad A., Ameen H., Pelz K., Karygianni L., Wittmer A., Anderson A. C., Spitzmüller B., and Hellwig E. (2014) Antibiotic resistance and capacity for biofilm formation of different bacteria isolated from endodontic infections associated with root-filled teeth. J. Endod. 40, 223–230 [DOI] [PubMed] [Google Scholar]
- 16. Paganelli F. L., Willems R. J., and Leavis H. L. (2012) Optimizing future treatment of enterococcal infections: Attacking the biofilm? Trends Microbiol. 20, 40–49 [DOI] [PubMed] [Google Scholar]
- 17. Shankar N., Lockatell C. V., Baghdayan A. S., Drachenberg C., Gilmore M. S., and Johnson D. E. (2001) Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69, 4366–4372, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tendolkar P. M., Baghdayan A. S., Gilmore M. S., and Shankar N. (2004) Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 72, 6032–6039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toledo-Arana A., Valle J., Solano C., Arrizubieta M. J., Cucarella C., Lamata M., Amorena B., Leiva J., Penadés J. R., and Lasa I. (2001) The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67, 4538–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duggan J. M., and Sedgley C. M. (2007) Biofilm formation of oral and endodontic Enterococcus faecalis. J. Endod. 33, 815–818 [DOI] [PubMed] [Google Scholar]
- 21. Sandoe J. A., Witherden I. R., Cove J. H., Heritage J., and Wilcox M. H. (2003) Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J. Med. Microbiol. 52, 547–550 [DOI] [PubMed] [Google Scholar]
- 22. Arntzen MØ Karlsk I. L., Skaugen M., Eijsink V. G., and Mathiesen G. (2015) Proteomic investigation of the response of Enterococcus faecalis V583 when cultivated in urine. PLoS One 10, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Giard J. C., Laplace J. M., Rincé A., Pichereau V., Benachour A., Leboeuf C., Flahaut S., Auffray Y., and Hartke A. (2001) The stress proteome of Enterococcus faecalis. Electrophoresis. 22, 2947–2954 [DOI] [PubMed] [Google Scholar]
- 24. Wang X., He X., Jiang Z., Wang J., Chen X., Liu D., Wang F., Guo Y., Zhao J., Liu F., Huang L., and Yuan J. (2010) Proteomic analysis of the Enterococcus faecalis V583 strain and clinical isolate V309 under vancomycin treatment. J. Proteome Res. 9, 1772–1785 [DOI] [PubMed] [Google Scholar]
- 25. Pessione A., Lamberti C., Cocolin L., Campolongo S., Grunau A., Giubergia S., Eberl L., Riedel K., and Pessione E. (2012) Different protein expression profiles in cheese and clinical isolates of Enterococcus faecalis revealed by proteomic analysis. Proteomics. 12, 431–447 [DOI] [PubMed] [Google Scholar]
- 26. Bøhle L. A., Færgestad E. M., Veiseth-Kent E., Steinmoen H., Nes I. F., Eijsink V. G., and Mathiesen G. (2010) Identification of proteins related to the stress response in Enterococcus faecalis V583 caused by bovine bile. Proteome Sci. 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seneviratne C. J., Suriyanarayanan T., Swarup S., Chia K. H. B., Nagarajan N., and Zhang C. (2017) Transcriptomics analysis reveals putative genes involved in biofilm formation and biofilm-associated drug resistance of Enterococcus faecalis. J. Endod. 43, 949–955 [DOI] [PubMed] [Google Scholar]
- 28. Seneviratne C. J., Yip J. W., Chang J. W., Zhang C. F., and Samaranayake L. P. (2013) Effect of culture media and nutrients on biofilm growth kinetics of laboratory and clinical strains of Enterococcus faecalis. Arch. Oral Biol. 58, 1327–1334 [DOI] [PubMed] [Google Scholar]
- 29. Ghosh D., Li Z., Tan X. F., Lim T. K., Mao Y., and Lin Q. (2013) iTRAQ based quantitative proteomics approach validated the role of calcyclin binding protein (CacyBP) in promoting colorectal cancer metastasis. Mol. Cell. Proteomics 12, 1865–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao Y., Lim T. K., Lin Q., and Li S. F. Y. (2016) Identification of cypermethrin induced protein changes in green algae by iTRAQ quantitative proteomics. J. Proteomics 139, 67–76 [DOI] [PubMed] [Google Scholar]
- 31. Strbenac D., Zhong L., Raftery M. J., Wang P., Wilson S. R., Armstrong N. J., Wang P., Wilson S. R., Armstrong N. J., and Yang J. Y. H. (2017) Quantitative performance evaluator for proteomics (QPEP): Web-based application for reproducible evaluation of proteomics preprocessing methods. J. Proteome Res. 16, 2359–2369 [DOI] [PubMed] [Google Scholar]
- 32. Ghosh D., Yu H., Tan X. F., Lim T. K., Zubaidah R. M., Tan H. T., Chung M. C., and Lin Q. (2011) Identification of key players for colorectal cancer metastasis by iTRAQ quantitative proteomics profiling of isogenic SW480 and SW620 cell lines. J. Proteome Res. 10, 4373–4387 [DOI] [PubMed] [Google Scholar]
- 33. Gan C. S., Chong P. K., Pham T. K., and Wright P. C. (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 6, 821–827 [DOI] [PubMed] [Google Scholar]
- 34. Christmas R., Avila-Campillo I., Bolouri H., Schwikowski B., Anderson M., Kelley R., Landys N., Workman C., Ideker T., Cerami E., Sheridan R., Bader G. D., and Sander C. (2005) Cytoscape: A software environment for integrated models of biomolecular interaction networks. AACR Educ. Book 2005, 12–16 [Google Scholar]
- 35. Maere S., Heymans K., and Kuiper M. (2005) BiNGO: A Cytoscape plugin to assess overrepresentation of bene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 36. Li P., Seneviratne C. J., Alpi E., Vizcaino J. A., and Jin L. (2015) Delicate metabolic control and coordinated stress response critically determine antifungal tolerance of Candida albicans biofilm persisters. Antimicrob. Agents Chemother. 59, 6101–6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Griffin H. G., and Gasson M. J. (1995) The gene (aroK) encoding shikimate kinase I from Escherichia coli. DNA Seq. 5, 195–197 [DOI] [PubMed] [Google Scholar]
- 38. Priestman M. A., Funke T., Singh I. M., Crupper S. S., and Schönbrunn E. (2005) 5-enolpyruvylshikimate-3-phosphate synthase from Staphylococcus aureus is insensitive to glyphosate. FEBS Lett. 579, 728–732 [DOI] [PubMed] [Google Scholar]
- 39. Andersson U., Levander F., and Rådström P. (2001) Trehalose-6-phosphate phosphorylase is part of a novel metabolic pathway for trehalose utilization in Lactococcus lactis. J. Biol. Chem. 276, 42707–42713 [DOI] [PubMed] [Google Scholar]
- 40. Karinou E., Compton E. L., Morel M., and Javelle A. (2013) The Escherichia coli SLC26 homologue YchM (DauA) is a C4-dicarboxylic acid transporter. Mol. Microbiol. 87, 623–640 [DOI] [PubMed] [Google Scholar]
- 41. Zolotarev A. S., Unnikrishnan M., Shmukler B. E., Clark J. S., Vandorpe H., Grigorieff N., Rubin E. J., and Alper S. L. (2009) Increased sulfate uptake by E. coli overexpressing the SLC26-related SulP protein Rv1739c from Mycobacterium tuberculosis. Comp. Biochem. Physiol. 149, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Janosi L., Shimizu I., and Kaji A. (1994) Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl. Acad. Sci. U.S.A. 91, 4249–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Almiron M., Link A. J., Furlong D., and Kolter R. (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6, 2646–2654, [DOI] [PubMed] [Google Scholar]
- 44. Vaillancourt K., Moineau S., Frenette M., Lessard C., and Vadeboncoeur C. (2002) Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: Organization, sequence, transcription, and activity of the gal gene produc. J. Bacteriol. 184, 785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mikalsen T., Pedersen T., Willems R., Coque T. M., Werner G., Sadowy E., van Schaik W., Jensen L. B., Sundsfjord A., and Hegstad K. (2015) Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genomics. 16, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hegstad K., Mikalsen T., Coque T. M., Werner G., and Sundsfjord A. (2010) Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 16, 541–554. [DOI] [PubMed] [Google Scholar]
- 47. Bryers J. D. (2009) Medical biofilms. Biotechnol. Bioeng. 100, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alexander J. W. (1973) Nosocomial infections. Curr. Probl. Surg. 10, 1–54 [DOI] [PubMed] [Google Scholar]
- 49. Karki H. S., and Ham J. H. (2014) The roles of the shikimate pathway genes, aroA and aroB, in virulence, growth and UV tolerance of Burkholderia glumae strain 411gr-6. Mol. Plant Pathol. 15, 940–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Orihuela C. J., Gao G., Francis K. P., Yu J., and Tuomanen E. I. (2004) Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 190, 1661–1669 [DOI] [PubMed] [Google Scholar]
- 51. Ogunniyi A. D., Mahdi L. K., Trappetti C., Verhoeven N., Mermans D., Van der Hoek M. B., Plumptre C. D., and Paton J. C. (2012) Identification of genes that contribute to the pathogenesis of invasive pneumococcal disease by in vivo transcriptomic analysis. Infect. Immun. 80, 3268–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iyer R., and Camilli A. (2007) Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol. Microbiol. 66, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mah T. F., and O'Toole G. A. (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39 [DOI] [PubMed] [Google Scholar]
- 54. Van Acker H., Sass A., Bazzini S., De Roy K., Udine C., Messiaen T., Riccardi G., Boon N., Nelis H. J., Mahenthiralingam E., and Coenye T. (2013) Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS ONE 8, e58943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang L., Chiang W. C., Gao Q., Givskov M., Tolker-Nielsen T., Yang L., and Zhang G. (2012) The catabolite repression control protein Crc plays a role in the development of antimicrobial-tolerant subpopulations in Pseudomonas aeruginosa biofilms. Microbiology 158, 3014–3019 [DOI] [PubMed] [Google Scholar]
- 56. Klein M. I., Xiao J., Lu B., Delahunty C. M., Yates J. R. 3rd, and Koo H. (2012) Streptococcus mutans protein synthesis during mixed-species biofilm development by high-throughput quantitative proteomics. PLoS ONE 7, e45795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bao K., Bostanci N., Thurnheer T., and Belibasakis G. N. (2017) Proteomic shifts in multi-species oral biofilms caused by Anaeroglobus geminatus. Sci. Rep. 7, 4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD006542.