Abstract
The copper-containing superoxide dismutases (SODs) represent a large family of enzymes that participate in the metabolism of reactive oxygen species by disproportionating superoxide anion radical to oxygen and hydrogen peroxide. Catalysis is driven by the redox-active copper ion, and in most cases, SODs also harbor a zinc at the active site that enhances copper catalysis and stabilizes the protein. Such bimetallic Cu,Zn-SODs are widespread, from the periplasm of bacteria to virtually every organelle in the human cell. However, a new class of copper-containing SODs has recently emerged that function without zinc. These copper-only enzymes serve as extracellular SODs in specific bacteria (i.e. Mycobacteria), throughout the fungal kingdom, and in the fungus-like oomycetes. The eukaryotic copper-only SODs are particularly unique in that they lack an electrostatic loop for substrate guidance and have an unusual open-access copper site, yet they can still react with superoxide at rates limited only by diffusion. Copper-only SOD sequences similar to those seen in fungi and oomycetes are also found in the animal kingdom, but rather than single-domain enzymes, they appear as tandem repeats in large polypeptides we refer to as CSRPs (copper-only SOD-repeat proteins). Here, we compare and contrast the Cu,Zn versus copper-only SODs and discuss the evolution of copper-only SOD protein domains in animals and fungi.
Keywords: copper, superoxide dismutase (SOD), metalloprotein, superoxide ion, redox signaling
Introduction
Superoxide dismutase (SOD)2 was discovered in 1969 by McCord and Fridovich (1) as a copper metalloprotein present in bovine erythrocytes that can disproportionate superoxide anion radicals with incredible catalytic efficiency (two-step reaction below).
Not long afterward, zinc was also detected in this cuproprotein, establishing mammalian SOD1 as a copper and zinc bimetalloenzyme (2). Since the discovery of Cu,Zn-SODs, distinct classes of SODs that use iron, manganese, or nickel as co-factors have been identified. These SODs are unrelated to Cu,Zn-SODs in primary sequence and structure but share in common the use of a redox-active metal co-factor to disproportionate superoxide (3). SODs protect cells from oxidative stress, particularly in the removal of superoxide produced during metabolism (4, 5), and also have key roles in cell signaling through the local production of H2O2 (6–8). In addition, many SODs are virulence factors for pathogens, allowing them to survive the oxidative burst of macrophages and neutrophils at the host–pathogen interface (9, 10).
Cu,Zn-SODs are the only SODs known to function as bimetalloenzymes, requiring copper for catalysis and zinc to enhance catalytic efficiency and stabilize the protein (11–14). For many decades, all members of this SOD family from bacteria to humans were believed to require both copper and zinc. This dogma was challenged first in 2004 with the discovery of a mycobacterial copper-only SOD (15), and then in 2014 with the identification of a large family of copper-only SODs in fungi that not only lacked zinc but contained an unusually open-access copper site (16). Very recent bioinformatics analyses have revealed that copper-only SOD-like protein sequences also occur as repeated protein domains in large molecules we call CSRP (copper-SOD-repeat protein). In this review, we shall compare and contrast the copper-only versus Cu,Zn-SOD proteins and discuss the utility of the copper-only SOD protein domain in biology.
Ubiquitous Cu,Zn-SODs
The bimetallic copper- and zinc-containing SODs are widely dispersed in biology from bacteria to mammals and are found in both intracellular and extracellular locations. Virtually all eukaryotes express an abundant intracellular Cu,Zn-SOD typically known as SOD1 (1–3, 17–19). This ubiquitous enzyme is found in various intracellular compartments, primarily in the cytosol (19) but also in the mitochondrial inter-membrane space (20–23), the secretory pathway (24, 25), and even the nucleus (26, 27). SOD1 protects against oxidative damage from metabolic sources of superoxide, including that from the mitochondrial respiratory chain, and also functions in cell signaling involving its H2O2 product and peroxide-sensitive kinases and phosphatases (6, 7). In the nucleus, SOD1 can participate in controlling gene expression as has been shown in baker's yeast with gene responses to DNA damage and copper starvation stress (26, 27). SOD1 is a highly abundant protein, and in humans, mutant versions of SOD1 have been linked to an inherited form of amyotrophic lateral sclerosis (ALS). SOD1 misfolding has been implicated in ALS disease, and many excellent reviews have been written on this topic (28–32).
Cu,Zn-SODs can also be found in extracellular locations, and because superoxide does not generally cross biological membranes, the substrate for the extracellular SOD must originate outside the cell. Certain bacteria express Cu,Zn-SODs in their periplasmic space (33–35), and in the case of pathogenic bacteria, these SODs protect against the oxidative burst of the host immune system (9, 10). Many eukaryotes also express a Cu,Zn-SOD distinct from SOD1 that is extracellular. This so-called ecSOD was first discovered by Marklund in 1982 (36) and is a secreted tetrameric protein in extracellular fluid or anchored to the extracellular matrix (3, 36–41). The superoxide substrate for ecSOD is derived from NADPH oxidase (NOX) enzymes (42) that are flavin- and heme-dependent transmembrane enzymes that reduce oxygen to superoxide (42–44). Together, ecSOD and NOX can function in signaling involving reactive oxygen and reactive nitrogen species (8).
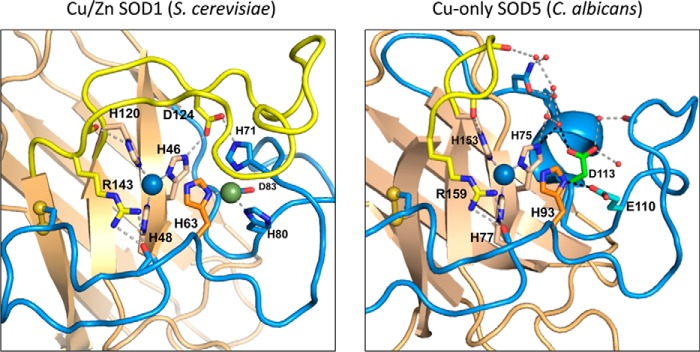
Cu,Zn-SODs can be homodimeric (SOD1), tetrameric (ecSOD), or in rare cases monomeric (Escherichia coli SodC) (3, 45). Each monomer has several landmark features as follows: a Greek key β-barrel fold; highly conserved copper- and zinc-binding residues; a conserved disulfide; active-site arginine; and an extended loop VII, also known in eukaryotic Cu,Zn-SODs as the electrostatic loop (ESL) (46). The catalytic copper in the oxidized Cu(II) state is coordinated in a distorted square planar geometry to an axial water molecule and four histidines, one of which (His-63 in the case of yeast and human SOD1) also coordinates zinc (Fig. 1). For the purpose of this review, we shall refer to this bridging His-63 as the “dynamic” histidine based on its on-and-off coordination to copper during catalysis. As superoxide is oxidized in the first step of catalysis, the dynamic bridge between His-63 and copper is broken as Cu(II) is reduced to Cu(I) and detached from His-63, resulting in a trigonal planar geometry for Cu(I). The zinc co-factor remains bound to the dynamic His-63 during catalysis and is additionally coordinated to two other histidines and an aspartate (Fig. 1) (46–48). Although zinc does not directly interact with the superoxide substrate during catalysis, its coordination with the dynamic His-63 assists in the re-oxidation of Cu(I) to Cu(II) in the second step of catalysis and accounts for the large pH independence of SOD activity (11, 12, 14, 49). The zinc co-factor is also important for stabilizing protein structure (13, 14, 50). Additional invariant features of Cu,Zn-SODs include an intermolecular disulfide (48) and an active-site arginine (Arg-143 in SOD1) positioned at the end of the ESL, which attracts and stabilizes the anionic superoxide substrate over the copper-metal center (Fig. 1) (51). With many charged amino acids residing in the ESL, it is believed to create an electrostatic network to funnel the highly solvated superoxide into the active site and accounts for the remarkably rapid rates of superoxide disproportionation (37, 51–54). The ESL is also believed to play a role in stabilizing copper and zinc binding through a network (or series) of hydrogen bonds (11, 50).
Figure 1.
Active site of Cu,Zn versus copper-only SODs. Left and right, comparison of Saccharomyces cerevisiae Cu,Zn-SOD1 (left) and C. albicans copper-only SOD5 (right) active site with key features highlighted as follows: Greek key β-barrel core (tan), ESL (yellow) with SOD1 Asp-124, active-site arginine (SOD1 Arg-143 and SOD5 Arg-159), disulfide loop (blue) with cysteines as yellow spheres, copper ion (blue), and zinc ion (green) with coordinating residues labeled by number. The dynamic histidine (SOD1 His-63 and SOD5 His-93) is orange, and SOD5 Glu-110 and Asp-113 are cyan and green. Dotted lines represent hydrogen bond networks and small red spheres represent water molecules.
Bacterial copper-SOD functions without zinc
For 35 years following the discovery of Cu,Zn-SODs, all copper-containing SOD enzymes were thought to require zinc. However, in 2004 the copper-containing SodC from Mycobacterium tuberculosis (MtSodC) was reported to lack zinc and to function with only a single copper atom (15). This copper-only SOD retains the Greek key β-barrel backbone of Cu,Zn-SODs as well as the same copper-coordination site, disulfide, active-site arginine, and extended loop VII covering the active site (equivalent to the ESL in eukaryotic Cu,Zn-SODs). However, MtSodC is missing the two non-dynamic histidines needed to bind zinc, with one substituted with an alanine and one missing due to a seven-amino acid deletion in the zinc loop (15). Structural analyses indicate zinc is missing from the active site of MtSodC (15).
As mentioned above, the zinc co-factor in Cu,Zn-SODs promotes pH independence in the enzyme and assists in SOD folding/stability. As would be expected for a zinc-less SOD, MtSodC demonstrates diffusion-limited catalysis from pH 6.0 to 8.0, but catalytic efficiency rapidly decreases above pH 8.0 (15). Because M. tuberculosis can exist in environments of low pH, e.g. the macrophage phagolysosome (55), this sensitivity to alkaline pH is likely a non-issue. To circumvent the requirement for zinc in protein stability, MtSodC shows an altered dimer interface with a long and rigid loop that is thought to stabilize the protein (15). Copper-only SodC is the only periplasmic SOD in M. tuberculosis and has been shown to protect the pathogen from the superoxide bursts of NOX enzymes in activated macrophages (56).
Of note, many SodC sequences in the Mycobacterium genus appear to lack the same zinc-binding histidines, indicating that diverse species of Mycobacterium SodCs are copper-only (15). Even so, within the eubacterial kingdom, copper-only SOD enzymes appear unique to the Mycobacterium genus, as all other periplasmic SODs characterized to date have both copper and zinc (9, 34, 45, 57).
Copper-only SOD in a fungal pathogen
Fungal pathogens, like bacteria, utilize their extracellular SODs as a first line of defense against superoxide generated by macrophage and neutrophil NOX enzymes (9). One of the best studied cases is the extracellular SOD5 from the opportunistic fungal pathogen, Candida albicans (58). C. albicans expresses three extracellular SODs (SOD4–6) that are members of the Cu,Zn-SOD family and are linked to the cell wall through GPI anchors (59). Of these, SOD5 is the most abundantly expressed in numerous models of candidiasis (60–65) and contributes to virulence in a mouse model of disseminated candidiasis (58). The enzyme is induced during the transition to the hyphal filamentous form required for host invasion (58, 66) and protects C. albicans from the superoxide burst of macrophages and neutrophils (63, 67–69). For many years, SOD5 was described as a bimetallic Cu,Zn-SOD (58, 66).
In 2014, three-dimensional structure and biochemical analyses of C. albicans SOD5 revealed that it is not a canonical Cu,Zn-SOD (16). Although SOD5 shares the overall Greek key β-barrel fold of Cu,Zn-SODs and exhibits a similar copper-binding geometry, it is missing the same two zinc-binding histidines as M. tuberculosis SodC, although in this case both due to amino acid substitutions, not deletions. Furthermore, SOD5 is missing an extensive portion of the ESL/loop VII (Fig. 1) (16). The absence of ESL sequences creates a uniquely open active site where copper is much more accessible to solvent. Attempts to load SOD5 with zinc were unsuccessful, demonstrating that SOD5 functions with a single atom of copper and no other metals (16). Despite such striking deviations from canonical Cu,Zn-SODs, copper-only SOD5 is an extremely efficient SOD enzyme capable of disproportionating superoxide at maximum rates approaching diffusion limits, 1.8 × 109 m−1 s−1 at pH 6.0 (16, 70).
How does copper-only SOD5 function in the absence of zinc and the ESL? As mentioned above, zinc promotes pH-independent catalysis of Cu,Zn-SODs through interactions with the dynamic histidine that also binds Cu(II) (11, 12, 14, 49). In lieu of zinc, the dynamic His-93 of copper-only SOD5 interacts with a conserved glutamate (Glu-110) in the active site (Fig. 1). Disruption of this interaction through Glu-110 mutations alters the orientation of the dynamic His-93 and dramatically decreases the pH range of activity (70). Thus, SOD5 Glu-110 appears to act analogous to zinc, interacting with and correctly orienting the dynamic His-93 to promote rapid catalysis up to pH 8 (70).
The absence of the ESL is perhaps the most striking feature of fungal copper-only SODs, as this highly charged loop was previously reported to be critical for substrate guidance and efficient catalysis in Cu,Zn-SODs (51–54). Despite no ESL, SOD5 shows a strong catalytic dependence on ionic strength, indicative of an alternative form of electrostatic substrate guidance (16). In Cu,Zn-SOD1, the ESL also helps stabilize the copper site through interactions involving ESL Asp-124 and copper coordinating His-46 (Fig. 1, left) (11, 50). In copper-only SOD5, the equivalent copper coordinating His-75 interacts with SOD5 Asp-113 through a hydrogen bond network (Fig. 1, right). Evidence indicates that Asp-113 in the SOD5 active site helps circumvent the need for the ESL in stabilizing copper binding (70). Interestingly, Asp-113 is invariant among all copper-containing SODs reported to date; in Cu,Zn-SODs, this aspartate is a zinc ligand and in copper-only SODs the aspartate functions through interactions with the copper site. This re-purposing of Asp-113 together with Glu-110 represents novel adaptations in the active site of eukaryotic copper-only SODs.
Of note, the features of Glu-110 and Asp-113 described above for fungal SOD5 may not extend to prokaryotic copper-only SODs. Mycobacterium SodC retains loop VII/ESL sequences and shows similar loop VII–copper site interactions as seen with Cu,Zn-SOD1 (15). There is no equivalent to SOD5 Glu-110 in mycobacterial SodC; instead, the invariant aspartate (equivalent to SOD5 Asp-113) interacts with the dynamic histidine (15, 70). MtSodC appears to be a hybrid of Cu,Zn and fungal copper-only SODs.
What is the advantage of expressing a copper-only SOD versus a bimetallic Cu,Zn-SOD? Clues may be obtained from examining the metallation process. In mammals, the extracellular Cu,Zn-SOD acquires copper and zinc in the secretory pathway and arrives at the cell surface in an enzymatically active form (71, 72). By contrast, characterization of C. albicans SOD5 indicated that the apoprotein is secreted without copper and is only activated upon scavenging copper from the extracellular environment (16). The open active site may help promote such capture of copper outside the cell. With no zinc site, mis-metallation events, such as copper migrating to the zinc site as is seen with Cu,Zn-SODs (49), are obviated. Additionally, the copper site of C. albicans SOD5 appears refractory to mis-metallation by non-native metals such as zinc (16). As such, enzyme activity remains intact regardless of fluctuations in environmental zinc. The copper-only design may indeed be advantageous to controlling enzyme maturation outside the cell.
Role of copper-only SODs in fungal pathogenesis and signaling
Copper-only SODs are widely distributed throughout the fungal kingdom and, in all cases examined thus far, are predicted to be extracellular and attached to the cell wall through GPI anchors (70). Like C. albicans SOD5, all fungal extracellular SODs lack zinc binding and ESL sequences and retain the equivalents to Glu/Gln-110 and Asp-113 at the active site. Copper-only SODs are found in many fungal pathogens where they combat the oxidative burst of the host and promote virulence, as has been seen with C. albicans, the pulmonary pathogen Histoplasma capsulatum (73), and systemic mycosis pathogen Paracoccidioides brasillienssis (74). Because of their extracellular location, fungal copper-only SODs may be uniquely positioned with their open active site to acquire copper from the host (16). Curiously, copper-only SODs are also found in non-pathogenic fungi that are not subject to host oxidative attacks, such as the Tuber melanosporum truffles fungus (70). With these non-pathogens, the SODs may react with superoxide derived from the fungus itself, similar to how copper- and zinc-containing ecSODs partner with NOX enzymes in mammals as part of signaling through reactive oxygen and reactive nitrogen species (see above). Multicellular fungi are indeed known to use NOX enzymes to signal differentiation (75–77).
Unlike multicellular organisms, unicellular microbes are not generally thought to use NOX enzymes and extracellular SODs for signaling. However, we recently found that copper-only SOD5 from unicellular C. albicans can act in signaling involving ROS and a fungal NOX enzyme known as FRE8 (78). C. albicans FRE8 and SOD5 together generate H2O2 that can help drive morphogenesis of the fungus into an invasive filamentous state (78). Therefore, SOD5 can react with superoxide generated from either the host or the fungal pathogen itself. During infection, one can envision a “superoxide superstorm,” with ROS coming from both the host and the fungal sides of the infection battleground and SOD5 operating at the interface (Fig. 2).
Figure 2.
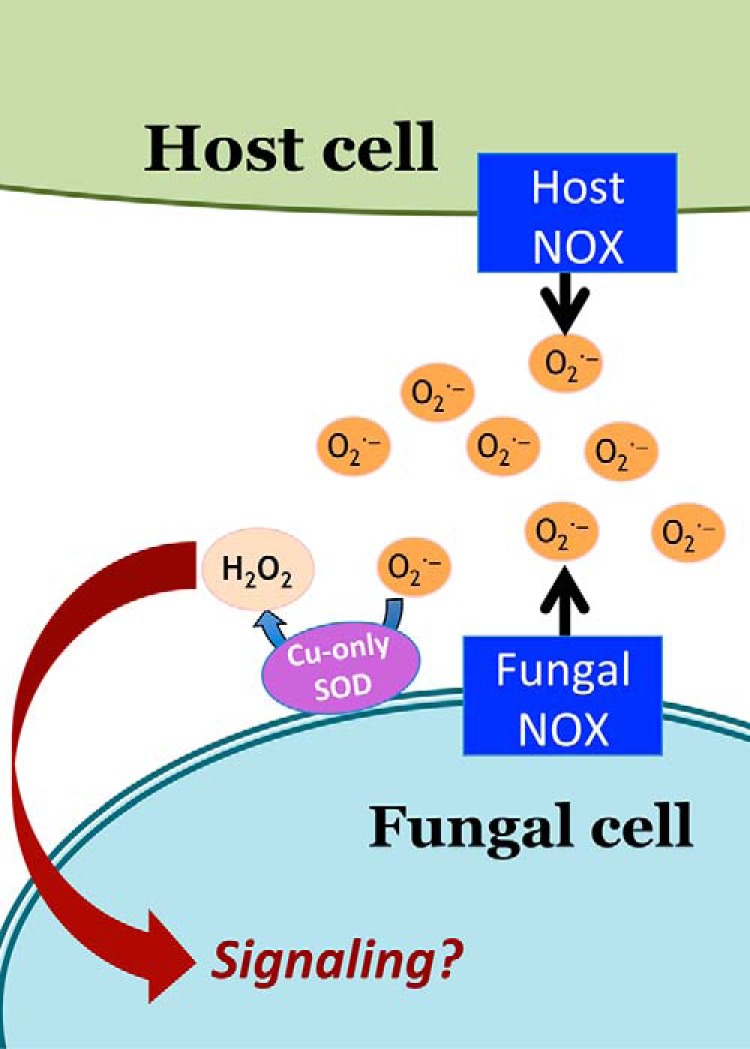
Copper-only SODs can react with superoxide from both the host and fungal pathogen. Shown is a model depicting a fungal copper-only SOD at the host–pathogen interface where it can react with superoxide from host NOX (macrophages or neutrophils) or from fungal NOX. The H2O2 generated may be used in signaling as has been shown for C. albicans where copper-only SOD5 and the FRE8 NOX promote morphogenesis of the fungus (78).
SOD5-like protein domains in animals
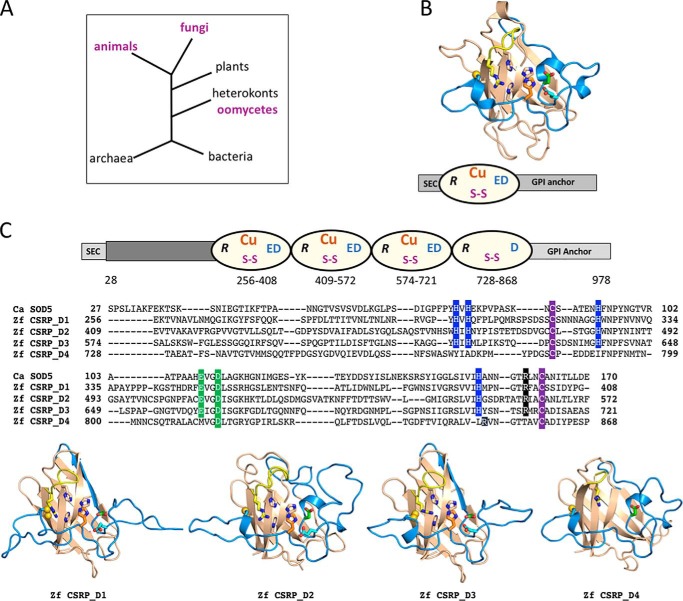
We have searched for SOD5-like proteins outside of the fungal kingdom. Our definition of a SOD5-like protein is one with the predicted Greek key β-barrel fold of the Cu,Zn-SOD family, including the copper site, disulfide, and active-site arginine but lacking sequences for zinc binding and the ESL and retaining SOD5 equivalents to Glu-110 and Asp-113. The only non-fungal organisms that express ≈20–30-kDa (predicted molecular mass of mature protein) SOD5-like SODs are oomycetes, a line of heterokont eukaryotes distantly removed from the fungal kingdom (Fig. 3A). As with fungi, the oomycete proteins are predicted to be secreted GPI-anchored extracellular SODs (70). Oomycetes are derived from photosynthetic microbes and are thought have acquired genetic material through horizontal gene transfer from a fungal ancestor (79–81). Copper-only SODs were apparently carried over as part of this genetic transfer.
Figure 3.
Evolution of copper-only SOD domains. A, phylogenetic tree of the distribution of copper-only SOD5-like domains in animals, fungi, and oomycetes (purple). B, three-dimensional structure of C. albicans apo-SOD5 is shown above the schematic of the full-length native protein where the N- and C-terminal sequences for secretion and GPI anchorage are in light gray and the Greek key β-barrel domain is depicted as an oval with active-site arginine (R), copper site, disulfide cysteines (S–S), and active-site Glu-110 and Asp-113 (ED). C, top, schematic of the predicted full-length D. rerio zebrafish (Zf) CSRP (XP_001343650.5) where the individual SOD5-like domains and key features are highlighted using the same scheme as for C. albicans SOD5 in B, bottom. D. rerio CSRP contains additional sequences at the N terminus (dark gray) of unknown nature. The ω-site for the GPI anchor is predicted to be at residue 955. C, middle, alignment of the individual SOD5-like domains of D. rerio CSRP against C. albicans SOD5. The copper-binding histidines are in blue; disulfide is in purple; active-site arginine is in black; and positions equivalent to SOD5 Glu-110 and Asp-113 are in green. The overall amino acid identity and similarity compared with SOD5 is as follows: CSRP_D1, 33% identity and 51% similarity; CSRP_D2, 30 and 47%; CSRP_D3, 31 and 56%; and CSRP_D4, 27 and 43%. C, bottom, each of the four SOD5-like domains were modeled onto C. albicans SOD5 using the MPI bioinformatics toolkit (93, 94). The most C-terminal repeat (CSRP_D4) is lacking a copper site but retains the predicted overall fold as well as other hallmark features of copper-only SOD5-like domains, as indicated.
Interestingly, SOD5-like protein sequences are also found in specific classes of animals but not in any plants, protists, archaea, or eubacteria we could identify (Fig. 3A). As with fungi and oomycetes, the animal proteins are largely predicted to be extracellular with GPI anchors and to exhibit the signatures of SOD5-like SODs defined above. However, in animals the SOD5-like protein sequences are not 20–30-kDa SOD enzymes but rather protein domains in much larger polypeptides of ≈100 kDa we define as CSRP (copper-only SOD repeat proteins). An example is illustrated in Fig. 3C with CSRP of the zebrafish Danio rerio (XP_001343650.5). The protein is predicted to contain four tandem repeats of SOD5-like SOD domains separated by very short linkers. Each domain contains the Greek key β-barrel fold structure, disulfide cysteines, active-site arginine, and equivalents to SOD5 Glu/Gln-110 and Asp-113, except for domain four, which has a methionine instead of the Glu-110 equivalent. All but the fourth domain retain the four copper-binding histidines (Fig. 3C, top and middle). The ESL is missing, as is the zinc site in all four domains of the zebrafish CSRP. Modeling of zebrafish CSRP shows how similar each domain is to the C. albicans SOD5 prototype, including the positioning of the predicted copper site in domains 1–3 (Fig. 3B and 3C, bottom). It is curious that the fourth domain is very similar to SOD5 in overall fold and positioning of the disulfide and active-site arginine but has no copper site (Fig. 3C, middle and bottom). This identical pattern of three copper-binding repeats followed by a fourth non-copper–binding domain appears preserved within the class of bony fishes/Osteichthyes, e.g. CSRPs from red piranha (XP_017575212.1), common carp (KTF72519.1), and Atlantic salmon (XP_014036762.1).
CSRPs occur throughout the animal kingdom from the unicellular Capsaspora owczarzaki (KJE90024.1) to diverse marine invertebrates and insecta to vertebrate teleosts. In fact, at least one study looking at the evolution of SODs remarks on the presence of repeated SOD domains in Anopheles gambiae (82). These CSRPs have been annotated widely as Cu,Zn-SODs, but they are clearly more related to fungal copper-only SODs. Expression of the transcripts have been analyzed, e.g. the Pacific oyster Crassostrea gigas CSRP (83) (EKC41617.1), and the proteins are produced as has been shown in proteomic analysis of placozoans (XP_002114624.1, Uniprot. No. B3S3A9) (84). Interestingly, CSRPs are not uniformly distributed in animals, and to date all CSRPs we have identified are in aquatic organisms and winged insects. We have yet to identify CSRP in lunged animals, e.g. avians, reptiles, and mammals. The significance of this distribution is currently not understood as the function of these curious SOD-like repeat proteins remains a mystery. Do these multidomain CSRPs function similarly to their smaller, single-domain fungal counterparts in disproportionating superoxide, or have they evolved with an entirely distinct activity in animals? The possible function of animal CSRPs in the metabolism or sensing of reactive oxygen species and/or metals is worthy of investigation.
Concluding remarks
The eukaryotic copper-only SOD protein is not just a single unit SOD enzyme, but a protein domain conserved in evolution since the split of animals and fungi ≈1.5 billion years ago (Fig. 3A). In virtually all cases examined so far, including fungi, oomycetes, and animals, the copper-only SOD-like protein is predicted to be outside the cell, therefore serving in some capacity involving the environment. The fungal copper-only SOD enzyme is as fast as its Cu,Zn-SOD sister and can protect fungal pathogens from host oxidative insults as well as operate in signaling processes involving fungus-derived superoxide. The function of animal CSRPs is currently unknown, but there is precedence for diversification of small copper-binding proteins. For example, copper-binding ATX1/ATOX functions as either a single-domain ≈8-kDa copper chaperone (85, 86) or as one of three domains in the copper chaperone CCS (87, 88) or as repeated protein domains in copper-transporting ATPases (89–92). Similarly, the SOD5-like copper-binding domain may have been diversified in evolution to function in numerous capacities for metal and redox homeostasis.
Acknowledgments
We thank P. John Hart and Sabrina Schatzman for helpful discussions.
This work was supported by National Institutes of Health Grant GM50016 (to V. C. C.). This is the fourth article in the Thematic Minireview series “Metals in Biology 2018: Copper homeostasis and utilization in redox enzymes.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SOD
- superoxide dismutase
- ESL
- electrostatic loop
- ALS
- amyotrophic lateral sclerosis
- MtSodC
- M. tuberculosis copper-only superoxide dismutase C
- CSRP
- copper-SOD repeat protein
- ecSOD
- extracellular Cu,Zn-SOD
- NOX
- NADPH oxidase
- GPI
- glycosylphosphatidylinositol.
References
- 1. McCord J. M., and Fridovich I. (1969) Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055 [PubMed] [Google Scholar]
- 2. Carrico R. J., and Deutsch H. F. (1970) The presence of zinc in human cytocuprein and some properties of the apoprotein. J. Biol. Chem. 245, 723–727 [PubMed] [Google Scholar]
- 3. Sheng Y., Abreu I. A., Cabelli D. E., Maroney M. J., Miller A. F., Teixeira M., and Valentine J. S. (2014) Superoxide dismutases and superoxide reductases. Chem. Rev. 114, 3854–3918 10.1021/cr4005296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fridovich I. (1983) Superoxide radical: an endogenous toxicant. Annu. Rev. Pharmacol. Toxicol. 23, 239–257 10.1146/annurev.pa.23.040183.001323 [DOI] [PubMed] [Google Scholar]
- 5. Cadenas E., and Davies K. J. (2000) Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med. 29, 222–230 10.1016/S0891-5849(00)00317-8 [DOI] [PubMed] [Google Scholar]
- 6. Juarez J. C., Manuia M., Burnett M. E., Betancourt O., Boivin B., Shaw D. E., Tonks N. K., Mazar A. P., and Doñate F. (2008) Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 7147–7152 10.1073/pnas.0709451105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reddi A. R., and Culotta V. C. (2013) SOD1 integrates signals from oxygen and glucose to repress respiration. Cell 152, 224–235 10.1016/j.cell.2012.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fattman C. L., Schaefer L. M., and Oury T. D. (2003) Extracellular superoxide dismutase in biology and medicine. Free Radic. Biol. Med. 35, 236–256 10.1016/S0891-5849(03)00275-2 [DOI] [PubMed] [Google Scholar]
- 9. Broxton C. N., and Culotta V. C. (2016) SOD enzymes and microbial pathogens: surviving the oxidative storm of infection. PLoS Pathog. 12, e1005295 10.1371/journal.ppat.1005295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fenlon L. A., and Slauch J. M. (2014) Phagocyte roulette in Salmonella killing. Cell Host Microbe 15, 7–8 10.1016/j.chom.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banci L., Bertini I., Cabelli D. E., Hallewell R. A., Tung J. W., and Viezzoli M. S. (1991) A characterization of copper/zinc superoxide dismutase mutants at position 124. Zinc-deficient proteins. Eur. J. Biochem. 196, 123–128 10.1111/j.1432-1033.1991.tb15794.x [DOI] [PubMed] [Google Scholar]
- 12. Ellerby L. M., Cabelli D. E., Graden J. A., and Valentine J. S. (1996) Copper-zinc superoxide dismutase: why not pH-dependent? J. Am. Chem. Soc. 118, 6556–6561 10.1021/ja953845x [DOI] [Google Scholar]
- 13. Potter S. Z., Zhu H., Shaw B. F., Rodriguez J. A., Doucette P. A., Sohn S. H., Durazo A., Faull K. F., Gralla E. B., Nersissian A. M., and Valentine J. S. (2007) Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J. Am. Chem. Soc. 129, 4575–4583 10.1021/ja066690+ [DOI] [PubMed] [Google Scholar]
- 14. Roberts B. R., Tainer J. A., Getzoff E. D., Malencik D. A., Anderson S. R., Bomben V. C., Meyers K. R., Karplus P. A., and Beckman J. S. (2007) Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS. J. Mol. Biol. 373, 877–890 10.1016/j.jmb.2007.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spagnolo L., Törö I., D'Orazio M., O'Neill P., Pedersen J. Z., Carugo O., Rotilio G., Battistoni A., and Djinovic-Carugo K. (2004) Unique features of the sodC-encoded superoxide dismutase from Mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site. J. Biol. Chem. 279, 33447–33455 10.1074/jbc.M404699200 [DOI] [PubMed] [Google Scholar]
- 16. Gleason J. E., Galaleldeen A., Peterson R. L., Taylor A. B., Holloway S. P., Waninger-Saroni J., Cormack B. P., Cabelli D. E., Hart P. J., and Culotta V. C. (2014) Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc. Natl. Acad. Sci. U.S.A. 111, 5866–5871 10.1073/pnas.1400137111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goscin S. A., and Fridovich I. (1972) The purification and properties of superoxide dismutase from Saccharomyces cerevisiae. Biochim. Biophys. Acta 289, 276–283 10.1016/0005-2744(72)90078-2 [DOI] [PubMed] [Google Scholar]
- 18. Bermingham-McDonogh O., Gralla E. B., and Valentine J. S. (1988) The copper,zinc-superoxide dismutase gene of Saccharomyces cerevisiae: cloning, sequencing, and biological activity. Proc. Natl. Acad. Sci. U.S.A. 85, 4789–4793 10.1073/pnas.85.13.4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crapo J. D., Oury T., Rabouille C., Slot J. W., and Chang L. Y. (1992) Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc. Natl. Acad. Sci. U.S.A. 89, 10405–10409 10.1073/pnas.89.21.10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jaarsma D., Rognoni F., van Duijn W., Verspaget H. W., Haasdijk E. D., and Holstege J. C. (2001) CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. 102, 293–305 [DOI] [PubMed] [Google Scholar]
- 21. Okado-Matsumoto A., and Fridovich I. (2001) Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J. Biol. Chem. 276, 38388–38393 10.1074/jbc.M105395200 [DOI] [PubMed] [Google Scholar]
- 22. Sturtz L. A., Diekert K., Jensen L. T., Lill R., and Culotta V. C. (2001) A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 276, 38084–38089 [DOI] [PubMed] [Google Scholar]
- 23. Weisiger R. A., and Fridovich I. (1973) Mitochondrial superoxide simutase. Site of synthesis and intramitochondrial localization. J. Biol. Chem. 248, 4793–4796 [PubMed] [Google Scholar]
- 24. Turner B. J., Atkin J. D., Farg M. A., Zang D. W., Rembach A., Lopes E. C., Patch J. D., Hill A. F., and Cheema S. S. (2005) Impaired extracellular secretion of mutant superoxide dismutase 1 associates with neurotoxicity in familial amyotrophic lateral sclerosis. J. Neurosci. 25, 108–117 10.1523/JNEUROSCI.4253-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mondola P., Damiano S., Sasso A., and Santillo M. (2016) The Cu,Zn superoxide dismutase: not only a dismutase enzyme. Front. Physiol. 7, 594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsang C. K., Liu Y., Thomas J., Zhang Y., and Zheng X. F. (2014) Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 5, 3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wood L. K., and Thiele D. J. (2009) Transcriptional activation in yeast in response to copper deficiency involves copper-zinc superoxide dismutase. J. Biol. Chem. 284, 404–413 10.1074/jbc.M807027200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Banci L., Bertini I., Boca M., Girotto S., Martinelli M., Valentine J. S., and Vieru M. (2008) SOD1 and amyotrophic lateral sclerosis: mutations and oligomerization. PLoS ONE 3, e1677 10.1371/journal.pone.0001677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chattopadhyay M., and Valentine J. S. (2009) Aggregation of copper-zinc superoxide dismutase in familial and sporadic ALS. Antioxid. Redox Signal. 11, 1603–1614 10.1089/ars.2009.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Renton A. E., Chiò A., and Traynor B. J. (2014) State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 17, 17–23 10.1038/nn.3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sheng Y., Chattopadhyay M., Whitelegge J., and Valentine J. S. (2012) SOD1 aggregation and ALS: role of metallation states and disulfide status. Curr. Top. Med. Chem. 12, 2560–2572 [DOI] [PubMed] [Google Scholar]
- 32. Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X., et al. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- 33. Puget K., and Michelson A. M. (1974) Isolation of a new copper-containing superoxide dismutase bacteriocuprein. Biochem. Biophys. Res. Commun. 58, 830–838 10.1016/S0006-291X(74)80492-4 [DOI] [PubMed] [Google Scholar]
- 34. Benov L., Chang L. Y., Day B., and Fridovich I. (1995) Copper,zinc superoxide dismutase in Escherichia coli: periplasmic localization. Arch. Biochem. Biophys. 319, 508–511 10.1006/abbi.1995.1324 [DOI] [PubMed] [Google Scholar]
- 35. Imlay K. R., and Imlay J. A. (1996) Cloning and analysis of sodC, encoding the copper-zinc superoxide dismutase of Escherichia coli. J. Bacteriol. 178, 2564–2571 10.1128/jb.178.9.2564-2571.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marklund S. L. (1982) Human copper-containing superoxide dismutase of high molecular weight. Proc. Natl. Acad. Sci. U.S.A. 79, 7634–7638 10.1073/pnas.79.24.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Antonyuk S. V., Strange R. W., Marklund S. L., and Hasnain S. S. (2009) The structure of human extracellular copper-zinc superoxide dismutase at 1.7 A resolution: insights into heparin and collagen binding. J. Mol. Biol. 388, 310–326 10.1016/j.jmb.2009.03.026 [DOI] [PubMed] [Google Scholar]
- 38. Marklund S. L., Holme E., and Hellner L. (1982) Superoxide dismutase in extracellular fluids. Clin. Chim. Acta 126, 41–51 10.1016/0009-8981(82)90360-6 [DOI] [PubMed] [Google Scholar]
- 39. Oury T. D., Crapo J. D., Valnickova Z., and Enghild J. J. (1996) Human extracellular superoxide dismutase is a tetramer composed of two disulphide-linked dimers: a simplified, high-yield purification of extracellular superoxide dismutase. Biochem. J. 317, 51–57 10.1042/bj3170051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Strömqvist M. (1993) Characterization of recombinant human extracellular superoxide dismutase. J. Chromatogr. 621, 139–148 10.1016/0378-4347(93)80089-M [DOI] [PubMed] [Google Scholar]
- 41. Strömqvist M., Holgersson J., and Samuelsson B. (1991) Glycosylation of extracellular superoxide dismutase studied by high-performance liquid chromatography and mass spectrometry. J. Chromatogr. 548, 293–301 10.1016/S0021-9673(01)88611-8 [DOI] [PubMed] [Google Scholar]
- 42. Lambeth J. D. (2004) NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4, 181–189 10.1038/nri1312 [DOI] [PubMed] [Google Scholar]
- 43. Moris D., Spartalis M., Tzatzaki E., Spartalis E., Karachaliou G. S., Triantafyllis A. S., Karaolanis G. I., Tsilimigras D. I., and Theocharis S. (2017) The role of reactive oxygen species in myocardial redox signaling and regulation. Ann. Transl. Med. 5, 324 10.21037/atm.2017.06.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Panday A., Sahoo M. K., Osorio D., and Batra S. (2015) NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol. Immunol. 12, 5–23 10.1038/cmi.2014.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Battistoni A., Folcarelli S., Rotilio G., Capasso C., Pesce A., Bolognesi M., and Desideri A. (1996) Crystallization and preliminary X-ray analysis of the monomeric Cu,Zn superoxide dismutase from Escherichia coli. Protein Sci. 5, 2125–2127 10.1002/pro.5560051020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tainer J. A., Getzoff E. D., Beem K. M., Richardson J. S., and Richardson D. C. (1982) Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J. Mol. Biol. 160, 181–217 10.1016/0022-2836(82)90174-7 [DOI] [PubMed] [Google Scholar]
- 47. Hart P. J., Balbirnie M. M., Ogihara N. L., Nersissian A. M., Weiss M. S., Valentine J. S., and Eisenberg D. (1999) A structure-based mechanism for copper-zinc superoxide dismutase. Biochemistry 38, 2167–2178 10.1021/bi982284u [DOI] [PubMed] [Google Scholar]
- 48. Perry J. J., Shin D. S., Getzoff E. D., and Tainer J. A. (2010) The structural biochemistry of the superoxide dismutases. Biochim. Biophys. Acta 1804, 245–262 10.1016/j.bbapap.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Valentine J. S., Pantoliano M. W., McDonnell P. J., Burger A. R., and Lippard S. J. (1979) pH-dependent migration of copper(II) to the vacant zinc-binding site of zinc-free bovine erythrocyte superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 76, 4245–4249 10.1073/pnas.76.9.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seetharaman S. V., Winkler D. D., Taylor A. B., Cao X., Whitson L. J., Doucette P. A., Valentine J. S., Schirf V., Demeler B., Carroll M. C., Culotta V. C., and Hart P. J. (2010) Disrupted zinc-binding sites in structures of pathogenic SOD1 variants D124V and H80R. Biochemistry 49, 5714–5725 10.1021/bi100314n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fisher C. L., Cabelli D. E., Tainer J. A., Hallewell R. A., and Getzoff E. D. (1994) The role of arginine 143 in the electrostatics and mechanism of Cu,Zn superoxide dismutase: computational and experimental evaluation by mutational analysis. Proteins 19, 24–34 10.1002/prot.340190105 [DOI] [PubMed] [Google Scholar]
- 52. Fisher C. L., Cabelli D. E., Hallewell R. A., Beroza P., Lo T. P., Getzoff E. D., and Tainer J. A. (1997) Computational, pulse-radiolytic, and structural investigations of lysine-136 and its role in the electrostatic triad of human Cu,Zn superoxide dismutase. Proteins 29, 103–112 10.1002/(SICI)1097-0134(199709)29:1%3C103::AID-PROT8%3E3.0.CO%3B2-G [DOI] [PubMed] [Google Scholar]
- 53. Getzoff E. D., Cabelli D. E., Fisher C. L., Parge H. E., Viezzoli M. S., Banci L., and Hallewell R. A. (1992) Faster superoxide dismutase mutants designed by enhancing electrostatic guidance. Nature 358, 347–351 10.1038/358347a0 [DOI] [PubMed] [Google Scholar]
- 54. Getzoff E. D., Tainer J. A., Weiner P. K., Kollman P. A., Richardson J. S., and Richardson D. C. (1983) Electrostatic recognition between superoxide and copper,zinc superoxide dismutase. Nature 306, 287–290 10.1038/306287a0 [DOI] [PubMed] [Google Scholar]
- 55. Vandal O. H., Nathan C. F., and Ehrt S. (2009) Acid resistance in Mycobacterium tuberculosis. J. Bacteriol. 191, 4714–4721 10.1128/JB.00305-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Piddington D. L., Fang F. C., Laessig T., Cooper A. M., Orme I. M., and Buchmeier N. A. (2001) Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect. Immun. 69, 4980–4987 10.1128/IAI.69.8.4980-4987.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Battistoni A. (2003) Role of prokaryotic Cu,Zn superoxide dismutase in pathogenesis. Biochem. Soc. Trans. 31, 1326–1329 10.1042/bst0311326 [DOI] [PubMed] [Google Scholar]
- 58. Martchenko M., Alarco A. M., Harcus D., and Whiteway M. (2004) Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15, 456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. De Groot P. W., Hellingwerf K. J., and Klis F. M. (2003) Genome-wide identification of fungal GPI proteins. Yeast 20, 781–796 10.1002/yea.1007 [DOI] [PubMed] [Google Scholar]
- 60. Amorim-Vaz S., Tran Vdu T., Pradervand S., Pagni M., Coste A. T., and Sanglard D. (2015) RNA enrichment method for quantitative transcriptional analysis of pathogens in vivo applied to the fungus Candida albicans. MBio 6, e00942–00915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bruno V. M., Shetty A. C., Yano J., Fidel P. L. Jr., Noverr M. C., and Peters B. M. (2015) Transcriptomic analysis of vulvovaginal candidiasis identifies a role for the NLRP3 inflammasome. MBio. 6, e00182–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fanning S., Xu W., Solis N., Woolford C. A., Filler S. G., and Mitchell A. P. (2012) Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot. Cell 11, 896–904 10.1128/EC.00103-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frohner I. E., Bourgeois C., Yatsyk K., Majer O., and Kuchler K. (2009) Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 71, 240–252 10.1111/j.1365-2958.2008.06528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pierce J. V., Dignard D., Whiteway M., and Kumamoto C. A. (2013) Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot. Cell 12, 37–49 10.1128/EC.00236-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thewes S., Kretschmar M., Park H., Schaller M., Filler S. G., and Hube B. (2007) In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol. Microbiol. 63, 1606–1628 10.1111/j.1365-2958.2007.05614.x [DOI] [PubMed] [Google Scholar]
- 66. Nantel A., Dignard D., Bachewich C., Harcus D., Marcil A., Bouin A. P., Sensen C. W., Hogues H., van het Hoog M., Gordon P., Rigby T., Benoit F., Tessier D. C., Thomas D. Y., and Whiteway M. (2002) Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13, 3452–3465 10.1091/mbc.E02-05-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fradin C., De Groot P., MacCallum D., Schaller M., Klis F., Odds F. C., and Hube B. (2005) Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56, 397–415 10.1111/j.1365-2958.2005.04557.x [DOI] [PubMed] [Google Scholar]
- 68. Miramón P., Dunker C., Windecker H., Bohovych I. M., Brown A. J., Kurzai O., and Hube B. (2012) Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS ONE 7, e52850 10.1371/journal.pone.0052850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tscherner M., Zwolanek F., Jenull S., Sedlazeck F. J., Petryshyn A., Frohner I. E., Mavrianos J., Chauhan N., von Haeseler A., and Kuchler K. (2015) The Candida albicans histone acetyltransferase Hat1 regulates stress resistance and virulence via distinct chromatin assembly pathways. PLoS Pathog. 11, e1005218 10.1371/journal.ppat.1005218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Peterson R. L., Galaleldeen A., Villarreal J., Taylor A. B., Cabelli D. E., Hart P. J., and Culotta V. C. (2016) The phylogeny and active site design of eukaryotic copper-only superoxide dismutases. J. Biol. Chem. 291, 20911–20923 10.1074/jbc.M116.748251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Petersen S. V., Kristensen T., Petersen J. S., Ramsgaard L., Oury T. D., Crapo J. D., Nielsen N. C., and Enghild J. J. (2008) The folding of human active and inactive extracellular superoxide dismutases is an intracellular event. J. Biol. Chem. 283, 15031–15036 10.1074/jbc.M801548200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Qin Z., Itoh S., Jeney V., Ushio-Fukai M., and Fukai T. (2006) Essential role for the Menkes ATPase in activation of extracellular superoxide dismutase: implication for vascular oxidative stress. FASEB J. 20, 334–336 10.1096/fj.05-4564fje [DOI] [PubMed] [Google Scholar]
- 73. Youseff B. H., Holbrook E. D., Smolnycki K. A., and Rappleye C. A. (2012) Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 8, e1002713 10.1371/journal.ppat.1002713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tamayo D., Muñoz J. F., Lopez Á., Uran M., Herrera J., Borges C. L., Restrepo Á., Soares C. M., Taborda C. P., Almeida A. J., McEwen J. G., and Hernández O. (2016) Identification and analysis of the role of superoxide dismutases isoforms in the pathogenesis of Paracoccidioides spp. PLoS Negl. Trop. Dis. 10, e0004481 10.1371/journal.pntd.0004481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Breitenbach M., Weber M., Rinnerthaler M., Karl T., and Breitenbach-Koller L. (2015) Oxidative stress in fungi: its function in signal transduction, interaction with plant hosts, and lignocellulose degradation. Biomolecules 5, 318–342 10.3390/biom5020318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Heller J., and Tudzynski P. (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390 10.1146/annurev-phyto-072910-095355 [DOI] [PubMed] [Google Scholar]
- 77. Takemoto D., Kamakura S., Saikia S., Becker Y., Wrenn R., Tanaka A., Sumimoto H., and Scott B. (2011) Polarity proteins Bem1 and Cdc24 are components of the filamentous fungal NADPH oxidase complex. Proc. Natl. Acad. Sci. U.S.A. 108, 2861–2866 10.1073/pnas.1017309108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rossi D. C. P., Gleason J. E., Sanchez H., Schatzman S. S., Culbertson E. M., Johnson C. J., McNees C. A., Coelho C., Nett J. E., Andes D. R., Cormack B. P., and Culotta V. C. (2017) Candida albicans FRE8 encodes a member of the NADPH oxidase family that produces a burst of ROS during fungal morphogenesis. PLoS Pathog. 13, e1006763 10.1371/journal.ppat.1006763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Richards T. A., Soanes D. M., Jones M. D., Vasieva O., Leonard G., Paszkiewicz K., Foster P. G., Hall N., and Talbot N. J. (2011) Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. U.S.A. 108, 15258–15263 10.1073/pnas.1105100108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Richards T. A., and Talbot N. J. (2007) Plant parasitic oomycetes such as phytophthora species contain genes derived from three eukaryotic lineages. Plant Signal. Behav. 2, 112–114 10.4161/psb.2.2.3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Savory F., Leonard G., and Richards T. A. (2015) The role of horizontal gene transfer in the evolution of the oomycetes. PLoS Pathog. 11, e1004805 10.1371/journal.ppat.1004805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Landis G. N., and Tower J. (2005) Superoxide dismutase evolution and life span regulation. Mech. Ageing Dev. 126, 365–379 10.1016/j.mad.2004.08.012 [DOI] [PubMed] [Google Scholar]
- 83. Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., Yang P., Zhang L., Wang X., Qi H., Xiong Z., Que H., Xie Y., Holland P. W., Paps J., et al. (2012) The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54 10.1038/nature11413 [DOI] [PubMed] [Google Scholar]
- 84. Ringrose J. H., van den Toorn H. W., Eitel M., Post H., Neerincx P., Schierwater B., Altelaar A. F., and Heck A. J. (2013) Deep proteome profiling of Trichoplax adhaerens reveals remarkable features at the origin of metazoan multicellularity. Nat. Commun. 4, 1408 10.1038/ncomms2424 [DOI] [PubMed] [Google Scholar]
- 85. Lin S. J., and Culotta V. C. (1995) The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc. Natl. Acad. Sci. U.S.A. 92, 3784–3788 10.1073/pnas.92.9.3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pufahl R. A., Singer C. P., Peariso K. L., Lin S. J., Schmidt P. J., Fahrni C. J., Culotta V. C., Penner-Hahn J. E., and O'Halloran T. V. (1997) Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278, 853–856 10.1126/science.278.5339.853 [DOI] [PubMed] [Google Scholar]
- 87. Lamb A. L., Wernimont A. K., Pufahl R. A., Culotta V. C., O'Halloran T. V., and Rosenzweig A. C. (1999) Crystal structure of the copper chaperone for superoxide dismutase. Nat. Struct. Biol. 6, 724–729 10.1038/11489 [DOI] [PubMed] [Google Scholar]
- 88. Schmidt P. J., Rae T. D., Pufahl R. A., Hamma T., Strain J., O'Halloran T. V., and Culotta V. C. (1999) Multiple protein domains contribute to the action of the copper chaperone for superoxide dismutase. J. Biol. Chem. 274, 23719–23725 10.1074/jbc.274.34.23719 [DOI] [PubMed] [Google Scholar]
- 89. Barry A. N., Shinde U., and Lutsenko S. (2010) Structural organization of human Cu-transporting ATPases: learning from building blocks. J. Biol. Inorg. Chem. 15, 47–59 10.1007/s00775-009-0595-4 [DOI] [PubMed] [Google Scholar]
- 90. Chelly J., Tümer Z., Tønnesen T., Petterson A., Ishikawa-Brush Y., Tommerup N., Horn N., and Monaco A. P. (1993) Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat. Genet. 3, 14–19 10.1038/ng0193-14 [DOI] [PubMed] [Google Scholar]
- 91. Lutsenko S., Barnes N. L., Bartee M. Y., and Dmitriev O. Y. (2007) Function and regulation of human copper-transporting ATPases. Physiol. Rev. 87, 1011–1046 10.1152/physrev.00004.2006 [DOI] [PubMed] [Google Scholar]
- 92. Strausak D., La Fontaine S., Hill J., Firth S. D., Lockhart P. J., and Mercer J. F. (1999) The role of GMXCXXC metal binding sites in the copper-induced redistribution of the Menkes protein. J. Biol. Chem. 274, 11170–11177 10.1074/jbc.274.16.11170 [DOI] [PubMed] [Google Scholar]
- 93. Alva V., Nam S. Z., Söding J., and Lupas A. N. (2016) The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 44, W410–W415 10.1093/nar/gkw348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Söding J., Biegert A., and Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]