Abstract
A dynamic network of metabolic adaptations, inflammatory responses, and redox homeostasis is known to drive tumor progression. A considerable overlap among these processes exists, but several of their key regulators remain unknown. To this end, here we investigated the role of the proinflammatory cytokine IL-1β in connecting these processes in glioma cells. We found that glucose starvation sensitizes glioma cells to IL-1β–induced apoptosis in a manner that depended on reactive oxygen species (ROS). Although IL-1β–induced JNK had no effect on cell viability under glucose deprivation, it mediated nuclear translocation of hexokinase 2 (HK2). This event was accompanied by increases in the levels of sirtuin 6 (SIRT6), nuclear factor erythroid 2–related factor 2 (Nrf2), and xanthine oxidoreductase (XOR). SIRT6 not only induced ROS-mediated cell death but also facilitated nuclear Nrf2–HK2 interaction. Recruitment of the Nrf2–HK2 complex to the ARE site on XOR promoter regulated its expression. Importantly, HK2 served as transcriptional coactivator of Nrf2 to regulate XOR expression, indicated by decreased XOR levels in siRNA-mediated Nrf2 and HK2 knockdown experiments. Our results highlight a non-metabolic role of HK2 as transcriptional coactivator of Nrf2 to regulate XOR expression under conditions of proinflammatory and metabolic stresses. Our insights also underscore the importance of nuclear activities of HK2 in the regulation of genes involved in redox homeostasis.
Keywords: c-Jun N-terminal kinase (JNK), glucose, hexokinase, interleukin 1 (IL-1), nuclear factor 2 (erythroid-derived 2-like factor) (NFE2L2) (Nrf2), SIRT6
Introduction
Emerging data indicate that interplay between inflammation and metabolism plays a critical role in tumor progression. In addition to their ability to extensively metabolize glucose for aiding increased energy demands, cancer cells are also under oxidative stress associated with increased production of ROS2 (1). The rapid glycolytic rate in glioblastoma (2) is concomitant with elevated levels of hexokinase 2 (HK2), which catalyzes the first step of the glycolytic pathway (3). Although HK1 is the predominant isoform in low-grade gliomas, highly up-regulated HK2 levels in glioblastoma multiforme correlates with poor prognosis (3). The subcellular localization of HK2 is sensitive to extracellular glucose with the distribution of HK2 between cytoplasm and mitochondria dynamically being regulated by glucose availability (4). HK2 regulates ROS levels (5), and glucose withdrawal increases ROS production in glioma cells (6). Also, the ability of diverse chemotherapeutic agents to induce glioma cell apoptosis through increased intracellular ROS generation is known (7–9).
Nuclear factor erythroid 2–related factor 2 (Nrf2) is a redox-sensitive transcription factor that provides cytoprotection against oxidative stress. Oxidative stress mediates activation of Nrf2 (10), which is known to regulate ROS production by mitochondria and NADPH oxidase (11). In addition to contributing toward the maintenance of redox homeostasis, Nrf2 affects the expression of metabolic genes (12, 13). Interestingly, SIRT6 not only regulates redox homeostasis by serving as an Nrf2 coactivator (14) but also affects glucose homeostasis via HIF-1α (15). Nrf2 regulates HIF-1α accumulation (16), and HIF-1α serves as a regulator of HK2 (17). Moreover, IL-1β–induced HK2 in glioma is dependent on the relative abundance of HIF-1α–dependent SIRT6 levels (18). Furthermore, HIF-1α–dependent subcellular localization of HK2 regulates cytoskeletal organization to consequently affect MHC-I clustering under inflammatory conditions in glioma cells (19).
Disrupting glycolytic flux serves as a trigger for inflammation and cell death (20). Interestingly, glycolytic inhibitors and metabolic conditions that affect hexokinase function and localization induce inflammasome activation involved in IL-1β secretion (21). Xanthine oxidoreductase (XOR), involved in catalyzing purines to uric acid, regulates IL-1β secretion upon NLRP3 inflammasome activation (22). Moreover, XOR is also known to regulate HIF-1α through ROS in glioma cells (23). Given that inflammation rewires energy metabolism in the tumor microenvironment (24), the prospect of targeting altered metabolism in inflammation has been suggested to have substantial therapeutic promise (25). As there is a link among inflammation, metabolic status, and redox homeostasis, we investigated the effect of inflammation and glucose deprivation on oxidative stress in glioma cells.
Results
Increased death upon glucose deprivation in IL-1β–treated glioma cells is ROS-dependent
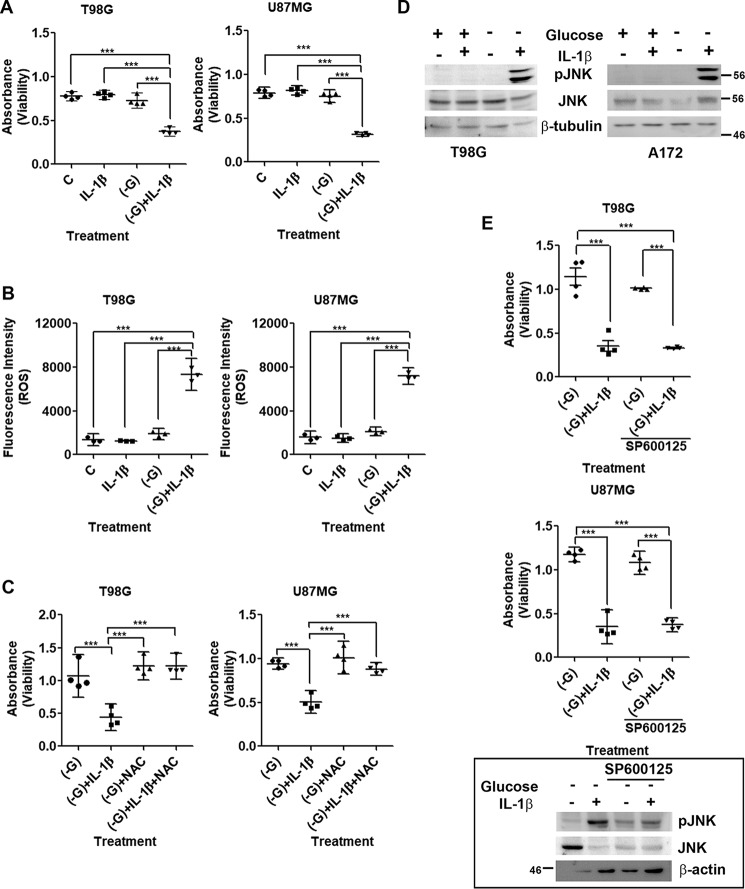
Although treatment with IL-1β or glucose deprivation alone had no effect on glioma cell viability, IL-1β triggered a significant decrease in cell viability under low-glucose conditions within 3 h (Fig. 1A). Death was accompanied by alteration in the expression of molecules associated with cell cycle progression (Fig. S1A) and an increase in cytochrome c and Bcl2 levels (Fig. S1B). Cancer cells exhibit glucose metabolism–dependent inhibition of cytochrome c–mediated apoptosis (26), and glucose deprivation promotes ROS generation and death (6). An increase in ROS generation was observed in glucose-deprived cells treated with IL-1β as compared with those exposed to low-glucose condition (Fig. 1B). As an elevated ROS level induces glioma cell apoptosis upon inhibition of glucose metabolism (7), we evaluated the role of elevated ROS in affecting glioma cell viability. The ability of ROS inhibitor N-acetyl-l-cysteine (NAC) to rescue glucose-deprived cells from IL-1β–induced death suggested that apoptosis triggered by IL-1β under glucose starvation is ROS-mediated (Fig. 1C). However, pretreatment of cells with PEGylated catalase (an H2O2-decomposing enzyme) was unable to rescue death, indicating that H2O2 is not the major ROS species contributing to cell death (Fig. S1C). This was concomitant with no change in H2O2 levels in cells treated with IL-1β under glucose deprivation (Fig. S1D).
Figure 1.
Glucose starvation sensitizes glioma cells to IL-1β–induced apoptosis in a ROS-dependent manner. A, IL-1β induces glioma cell death under glucose deprivation. B, increase in DHE fluorescence intensity depicting heightened ROS generation in glucose-deprived IL-1β–treated cells. C, increase in absorbance representing rescue of cell death by the ROS inhibitor NAC. D, Western blots demonstrating increased pJNK levels in glioma cells treated with IL-1β in the absence of glucose. E, MTS assay showing JNK-independent cell death. The inset shows the efficacy of JNK inhibitor. The graphs represent scatter plots with each data point representing average absorbance values depicting glioma cell viability (A, C, and E) and average fluorescence intensities depicting ROS levels (B). −G denotes glucose-free DMEM. SP600125 is a JNK inhibitor. One-way ANOVA (Bonferroni's multiple comparison test) was used for statistical analysis. Error bars represent S.E. (n = 4 in A, C, and E; n = 3 in B). ***, p < 0.001.
IL-1β–induced death upon glucose deprivation is JNK-independent
We have previously demonstrated the importance of ROS-induced JNK activation in triggering glioma cell apoptosis (7). On investigating the status of JNK in IL-1β–treated glioma cells in the presence and absence of glucose, an increase in JNK phosphorylation was observed only in cells treated with IL-1β in the absence of glucose (Fig. 1D). However, JNK inhibitor SP600125 failed to rescue death in cells treated with IL-1β under conditions of glucose deprivation (Fig. 1E), suggesting non-involvement of JNK in the process. The induction of cell death in both U87MG (p53 wildtype) and T98G (p53 mutant) cell lines in response to inflammation and glucose deprivation suggested that induction of death under this condition is independent of p53 status.
Increased SIRT6 expression regulates cell death
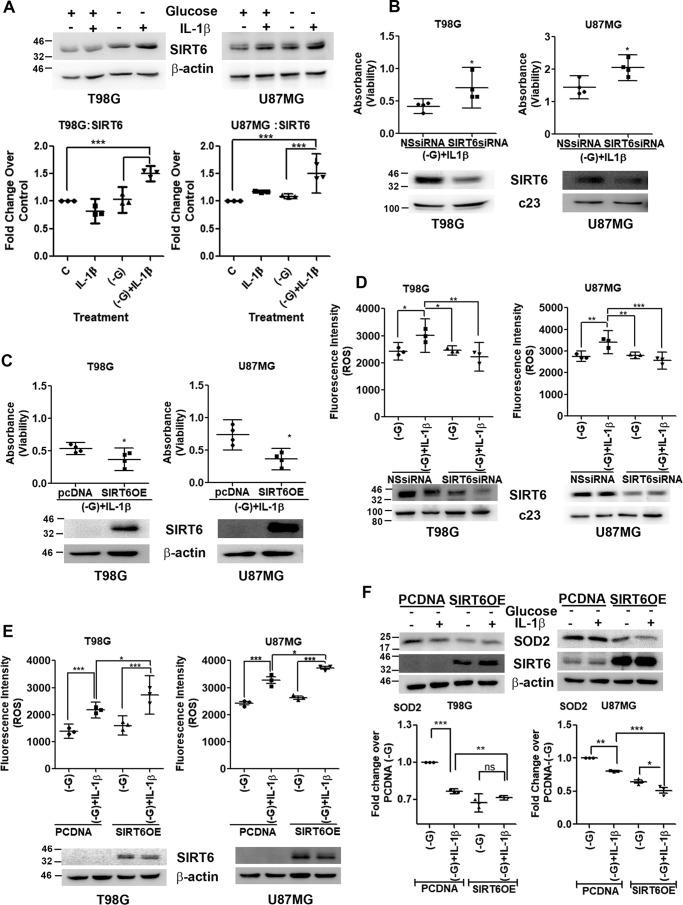
In addition to its function as a regulator of ROS generation (27), SIRT6 also plays a dominant role in affecting energy balance through control of glucose homeostasis (15). In addition, IL-1β increases SIRT6 levels in glioma cells (18). An increase in SIRT6 levels was observed upon IL-1β treatment under glucose deprivation (Fig. 2A). On evaluating the contribution of elevated SIRT6 levels to IL-1β–induced death under glucose-deprived condition, viability of cells upon SIRT6 overexpression and depletion was determined. Although siRNA-mediated SIRT6 knockdown increased viability of IL-1β–treated cells under glucose deprivation (Fig. 2B), ectopic SIRT6 expression increased cell death (Fig. 2C). Given the known function of SIRT6 as a regulator of ROS generation (27) and the ability of SIRT6 to affect viability of IL-1β–treated glucose-deprived cells, ROS levels upon SIRT6 knockdown as well as overexpression were investigated. A decrease in ROS levels upon SIRT6 knockdown (Fig. 2D) and an increase upon ectopic SIRT6 expression (Fig. 2E) were observed. The increase in ROS levels was concomitant with a decrease in the expression of mitochondrial superoxide dismutase (SOD2), a critical detoxifier of mitochondrial ROS (Fig. 2F). This decrease in SOD2 levels in glucose-deprived cells treated with IL-1β was further diminished upon SIRT6 overexpression (Fig. 2F).
Figure 2.
SIRT6 affects cell viability through regulation of redox homeostasis. A, Western blot demonstrating SIRT6 levels in IL-1β–treated glioma cells in the presence or absence of glucose. Blots are representative images of three independent experiments showing similar results. Blots were reprobed for β-actin to establish equivalent loading. Densitometry data depicting -fold change in SIRT6 expression over control under the indicated treatment conditions normalized to corresponding loading controls are shown. siRNA-mediated knockdown of SIRT6 increases (B) and SIRT6 overexpression decreases (C) viability of glioma cells deprived of glucose and treated with IL-1β as determined by MTS assay. Insets in B and C show knockdown efficiency of SIRT6 siRNA and increased SIRT6 expression upon transfection with SIRT6 overexpression construct. D and E, DHE fluorescence intensity showing that ROS generation in glucose-deprived IL-1β–treated cells is SIRT6-dependent. F, SIRT6 regulates SOD2 expression under a combination of inflammatory and metabolic stresses. Densitometry data depict -fold change in SOD2 expression over control under the indicated treatment conditions normalized to corresponding loading controls. Each data point in the scatter plots represents average absorbance values depicting glioma cell viability (n = 4) (B and C) and fluorescence values depicting ROS levels under the indicated treatment conditions from independent experiments (n = 3) (D and E). −G denotes glucose-free DMEM. NSsiRNA, nonspecific siRNA; SIRT6OE, SIRT6 overexpression. Two-tailed paired Student's t test (B and C) and one-way ANOVA (Bonferroni's multiple comparison test) (A, D, E, and F) were used for statistical analysis. Error bars represent S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant.
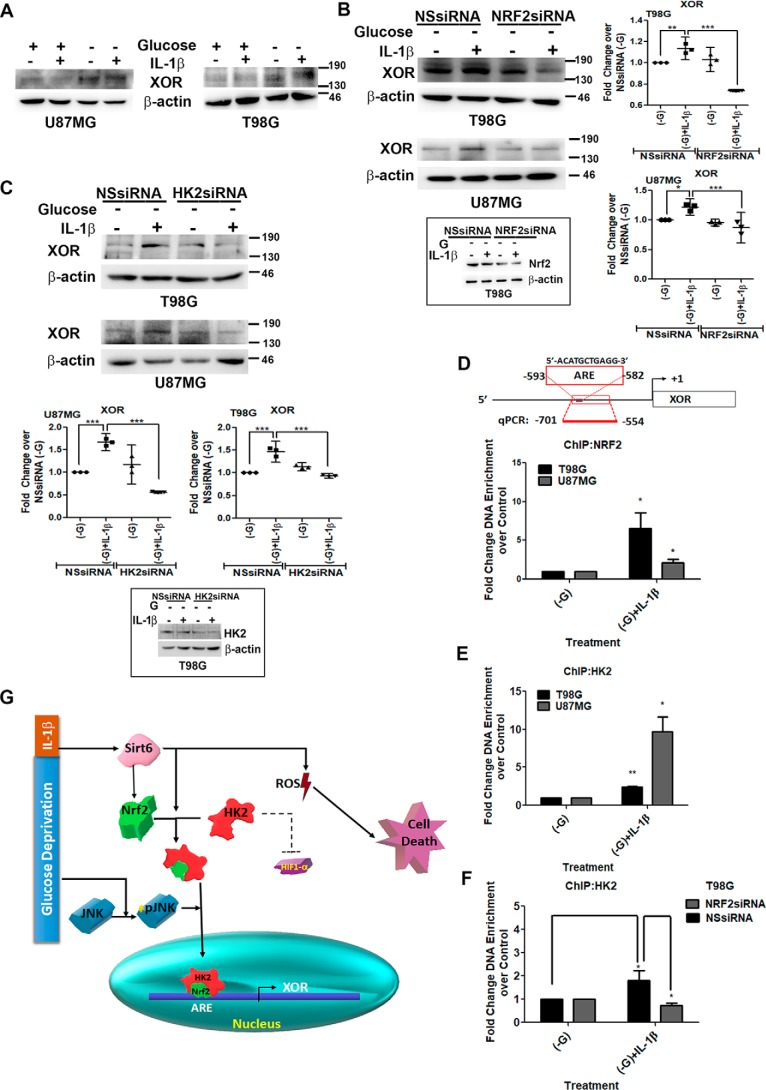
IL-1β increases nuclear localization of HK2 upon glucose deprivation
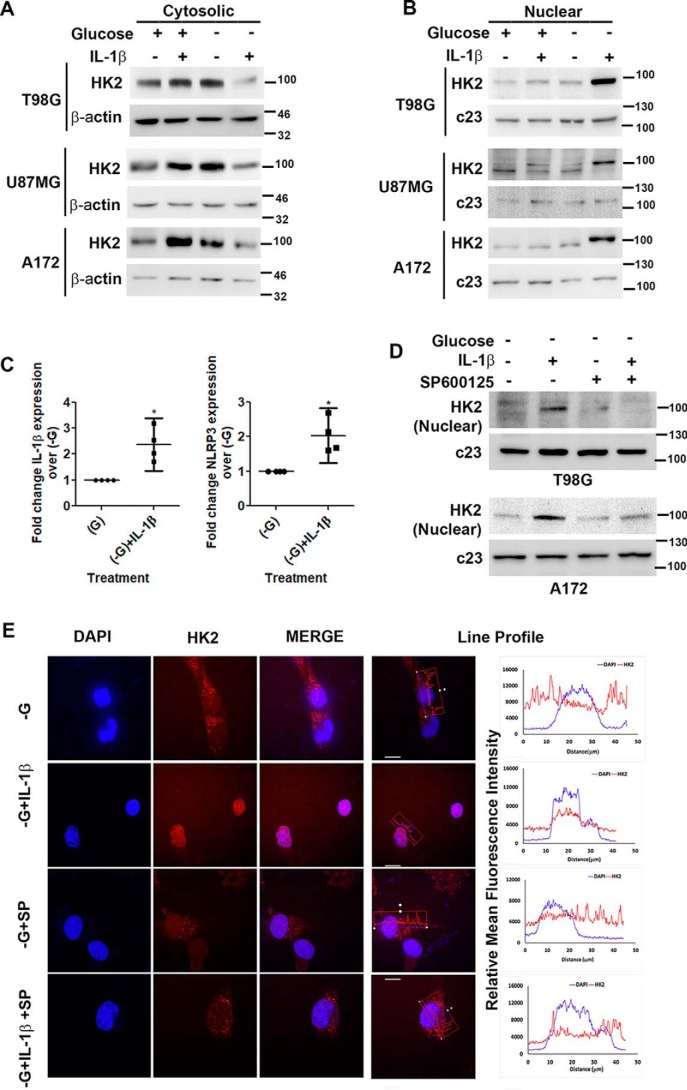
Inhibiting HK2 activity by 2-deoxyglucose, which competitively inhibits cellular uptake and utilization of glucose, increases its nuclear localization (28). Because both JNK (7) and SIRT6 (18) regulate HK2 expression in glioma cells, we investigated the status of HK2 in IL-1β–treated glucose-deprived cells expressing elevated pJNK and SIRT6 levels. Western blot analysis revealed a decrease in cytosolic (Fig. 3A) and an increase in nuclear (Fig. 3B) HK2 levels in IL-1β–treated cells upon glucose deprivation. SIRT6 enhances the expression of proinflammatory cyto-/chemokines (29), and HK2 dissociation from mitochondria triggers NLRP3 inflammasome activation and IL-1β production (21). On investigating the status of NLRP3 and IL-1β in cells exhibiting heightened SIRT6 and nuclear HK2 levels, an increase in the mRNA expression of both was observed upon glucose deprivation only in the presence of IL-1β (Fig. 3C). As we have previously reported JNK-dependent regulation of HK2 in glioma cells (7) and because JNK activation was dramatically elevated under conditions of increased nuclear HK2 translocation, the role of JNK in this shuttling was investigated. The IL-1β–induced increase in nuclear HK2 localization upon glucose deprivation was JNK-dependent as SP600125 prevented IL-1β–mediated HK2 accumulation in the nucleus under this condition (Fig. 3, D and E).
Figure 3.
IL-1β induces JNK-dependent nuclear localization of HK2. Shown are Western blots demonstrating cytosolic (A) and nuclear (B) HK2 levels in glioma cells treated with IL-1β in the presence or absence of glucose. C, qRT-PCR analysis shows increased IL-1β and NLRP3 mRNA levels in cells treated with IL-1β under glucose deprivation. Each data point in the scatter plots represents -fold change with respect to glucose-free DMEM (−G) from independent experiments (n = 4). D, Western blot demonstrating nuclear HK2 levels in cells treated with or without IL-1β or SP600125 in the presence or absence of glucose. Western blots are representative images of three independent experiments showing similar results. Blots were reprobed for β-actin or c23 to establish equivalent loading. E, JNK regulates nuclear localization of HK2. Immunofluorescence microscopy revealed nuclear HK2 localization in glucose-deprived cells in the presence of IL-1β. Treatment with JNK inhibitor (SP600125) prevented HK2 localization to the nucleus. Cells were immunostained with anti-HK2 (HK2; red). The nucleus is marked with DAPI (blue). Merged images (Merge) are shown. Representative images of 63× magnification from three independent experiments are shown for the indicated conditions. SP denotes JNK inhibitor (SP600125). Adjacent line profiles show mean fluorescence intensities of HK2 and DAPI measured by ZEN lite 2.3 software (scale bars, 10 μm). Error bars represent S.E. *, p < 0.05.
HK2 has no role in cell death but negatively regulates HIF-1α activation
HK2 determines cellular fate by affecting both cytoprotection and apoptosis induction based on the metabolic state (30). To investigate the involvement of altered HK2 localization in affecting cell death, the viability of cells upon siRNA-mediated HK2 knockdown was determined. HK2 knockdown failed to rescue cell death (Fig. S2A), suggesting the non-involvement of HK2 in IL-1β–triggered cell death under glucose starvation. Although HK2 induces apoptosis upon glucose depletion, it is also known to protect cells from death during hypoxia (30). Given the non-involvement of HK2 in death induction, we investigated whether its inability to rescue death could have stemmed from changes in HIF-1α levels. Interestingly, HK2 knockdown further heightened HIF-1α activation in IL-1β–treated cells deprived of glucose (Fig. S2B). This further confirmed our previous observation that a negative correlation exists between HK2 and HIF-1α levels in glioma cells (18).
SIRT6 regulates Nrf2
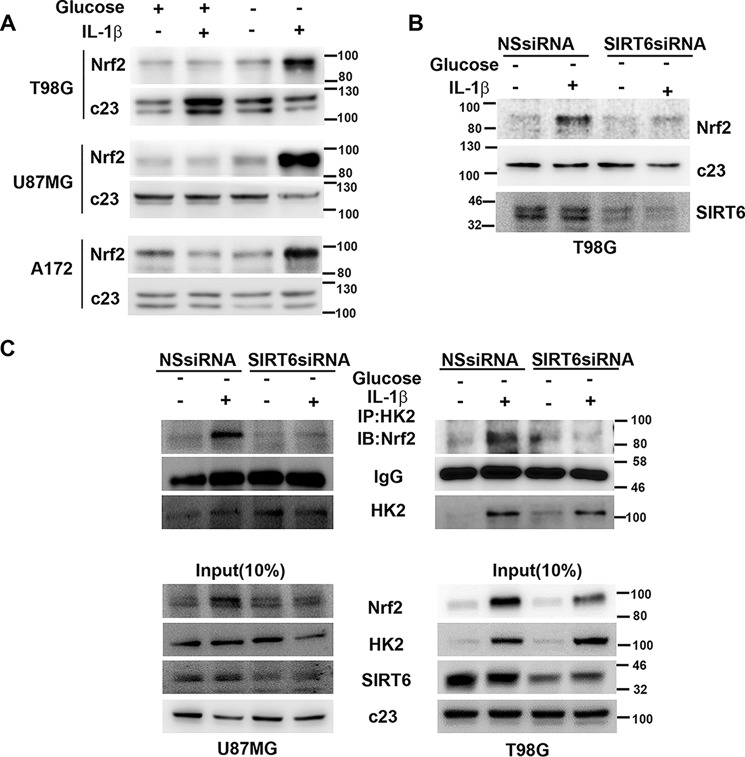
SIRT6 regulates oxidative stress responses by serving as an Nrf2 coactivator (14), and Nrf2 accumulation in cancer cells offers protection against oxidative stress (31). Importantly, Nrf2 is translocated to the nucleus in response to pro-oxidant stimuli (32). On determining Nrf2 levels in glucose-deprived IL-1β–treated cells exhibiting heightened ROS, an increase in nuclear Nrf2 levels was observed (Fig. 4A). As the increased nuclear Nrf2 level was concomitant with elevated SIRT6 levels, the role of SIRT6 in regulating Nrf2 levels was investigated. The ability of siRNA-mediated SIRT6 knockdown to decrease the nuclear Nrf2 level in glucose-deprived IL-1β–treated cells (Fig. 4B) further indicated the role of SIRT6 as a regulator of Nrf2.
Figure 4.
SIRT6 regulates Nrf2–HK2 association. A, Western blot depicting elevated nuclear Nrf2 expression in IL-1β–treated glioma cells under glucose starvation. B, siRNA-mediated knockdown of SIRT6 prevents nuclear Nrf2 accumulation in cells treated with IL-1β under glucose starvation as demonstrated by Western blot analysis. Blots were reprobed for c23 to establish equivalent loading. C, SIRT6 positively regulates nuclear HK2–Nrf2 interaction in the presence of IL-1β in glucose-starved glioma cells. Coimmunoprecipitation shows decreased HK2–Nrf2 association in IL-1β–treated glucose-deprived cells upon siRNA-mediated knockdown of SIRT6. A confounding effect of unequal precipitation on the interaction studies was ruled out by almost equal IgG levels in each condition. Blots are representative images of three independent experiments showing similar results. NSsiRNA, nonspecific siRNA; IP, immunoprecipitation; IB, immunoblotting.
HK2 interacts with Nrf2 in a SIRT6-dependent manner
As the increased nuclear Nrf2 level was concomitant with increased nuclear HK2 accumulation, we investigated the association between the two. Immunoprecipitation revealed increased association between Nrf2 and HK2 under glucose-deprived conditions only in the presence of IL-1β (Fig. 4C). Moreover, not only did siRNA-mediated SIRT6 knockdown decrease the nuclear Nrf2 level in glucose-deprived IL-1β–treated cells, but it also abrogated Nrf2–HK2 interaction (Fig. 4C). These results provide strong evidence that Nrf2 is an interacting partner of HK2 and that SIRT6 is crucial in facilitating the interaction.
XOR is a target of HK2 and Nrf2
Nuclear translocation of Nrf2 in response to oxidative stress triggers a transcriptional program through its binding to the antioxidant response element (ARE) of antioxidant genes associated with maintenance of cellular redox balance (33). HK2 serves as an intracellular glucose sensor of yeast cells to affect gene regulation (34) whereby it functions through a variety of structurally unrelated factors to sustain transcriptional repression at the SUC2 gene (34). In addition, dissociation of HK2 from mitochondria activates the NLRP3 inflammasome (21), and XOR-dependent IL-1β secretion upon NLRP3 inflammasome activation has been shown (21). Given the involvement of XOR in regulating cellular redox homeostasis through ROS generation (23), the status of XOR in glucose-deprived IL-1β–treated cells was determined. An increase in XOR expression was observed in IL-1β–treated glucose-deprived cells, which also exhibited elevated ROS, IL-1β, and NLRP3 levels, as compared with cells treated with IL-1β or glucose-deprived medium alone (Fig. 5A). This increase was accompanied by elevated XOR mRNA levels (Fig. S2E) as well as increased enzyme activity (Fig. S2C). As Nrf2 was found to interact with HK2, the role of HK2 and Nrf2 in regulating XOR expression was determined. The increase in XOR expression was found to be dependent on both HK2 and Nrf2 as siRNA-mediated knockdown of Nrf2 (Fig. 5B) and HK2 (Fig. 5C) decreased IL-1β–induced XOR expression under glucose deprivation. Also, RNA polymerase II was found to be part of this nuclear Nrf2–HK2 complex (Fig. S2D).
Figure 5.
HK2 serves as a coactivator of Nrf2 in regulating XOR expression. A, IL-1β increases XOR levels under conditions of glucose deprivation. siRNA-mediated knockdown of either Nrf2 (B) or HK2 (C) prevents IL-1β–induced XOR expression in glucose-deprived cells as depicted by Western blot analysis. Insets show knockdown efficiency of Nrf2 and HK2 siRNAs. Western blot images are representation of three independent experiments showing similar results. Blots were reprobed for β-actin to establish equivalent loading. Densitometry data of -fold change in XOR expression over control under different treatment conditions normalized to corresponding loading controls are shown. Each data point in the scatter plot denotes -fold change with respect to control from independent experiments (n = 3). D and E, ChIP performed on the region containing the ARE site on XOR promoter in T98G and U87MG glioma cells indicates increased Nrf2 and HK2 binding at the ARE site in IL-1β–treated glucose-deprived cells. F, ChIP assay depicting decreased HK2 binding at XOR promoter upon siRNA-mediated Nrf2 knockdown in IL-1β–treated glucose-deprived cells. Diluted input (5%) was used as a positive control. Relative enrichment was calculated with respect to control levels after correction for background signals. Graphs are representative data of three independent experiments. −G denotes glucose-deprived DMEM. One-way ANOVA (Bonferroni's multiple comparison test) was used for statistical analysis. Error bars represent S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001. G, schematic depiction of the importance of HK2 as a coactivator of Nrf2 in regulation of XOR under inflammatory and metabolic stresses. NSsiRNA, nonspecific siRNA; qPCR, quantitative PCR.
To investigate whether other known targets of Nrf2 require HK2 in their regulation, the expression of known Nrf2 target heme oxygenase 1 (HMOX1) (35) was determined under HK2 knockdown condition in cells treated with IL-1β under glucose deprivation. Although siRNA-mediated knockdown of HK2 abrogated the IL-1β– and glucose deprivation stress–induced increase in XOR mRNA levels, HK2 had no effect on HMOX1 expression (Fig. S2E). Thus, the ability of HK2–Nrf2 complex to regulate Nrf2-dependent genes was found to be target-specific, and not all Nrf2 target genes follow HK2-dependent regulation.
Nrf2 knockdown abolishes binding of HK2–Nrf2 complex to ARE site on XOR promoter
Several studies have reported nuclear shuttling of HK2 (34, 36), and in the context of SUC2 promoter HK2 functions as a transcriptional repressor (34). In view of our observations that XOR expression is regulated by both Nrf2 and HK2, the effect of occupancy of Nrf2–HK2 complex at the ARE site of XOR promoter on its expression was investigated. A chromatin immunoprecipitation (ChIP) assay revealed increased enrichment of Nrf2 (Fig. 5D) as well as HK2 (Fig. 5E) on the ARE site of XOR promoter in glucose-deprived IL-1β–treated cells. This raises the possibility that HK2 could directly bind DNA or may bind to ARE through its interaction with Nrf2. However, enhanced binding of HK2 in glucose-deprived IL-1β–treated cells was abrogated upon Nrf2 knockdown (Fig. 5F). Taken together, the results suggest that abundantly available nuclear HK2 in glucose-deprived IL-1β–treated cells serves as a coactivator of Nrf2 in XOR transcriptional regulation by facilitating increased binding of HK2–Nrf2 complex to the ARE site on XOR promoter (Fig. 5G).
Discussion
Non-canonical functions of glycolytic enzymes in gene regulation are just beginning to be understood with nuclear shuttling of such enzymes being involved in the regulation of transcriptional events in glioma (38). Several studies have reported nuclear shuttling of HK2 in yeast as well as in cancer cells (28, 36, 39, 40) with nuclear localization of HK2 being regulated by glucose (39). Although the presence of glucose reduces HK2 nuclear translocation (28), glucose deprivation in itself was not sufficient to affect nuclear HK2 translocation in glioma cells. An inflammatory stimulus is necessary for prompting HK2 cytoplasmic–nuclear shuttle under glucose-deprived conditions. As IL-1β is known to induce a prolonged hypoglycemia in the brain (42), it is possible that the nuclear HK2 level becomes pronounced under glucose deprivation only in the presence of IL-1β. Although HK2 had no involvement in cell death, it served as a negative regulator of HIF-1α. This is of interest as HIF-1α–dependent subcellular localization of HK2 is known to regulate inflammation-mediated cytoskeletal organization that influences immune-related outcome (19).
Importantly, SIRT6 served as a negative regulator of cell death under metabolically compromised inflammatory conditions. This ability of SIRT6 to affect death could be attributed to its ability to regulate ROS levels (27). As HIF-1α–dependent SIRT6 abundance regulates HK2 levels in IL-1β–treated glioma cells (18), this study underscores SIRT6's role as a crucial factor in the orchestration of redox-regulatory responses under inflammatory conditions. In addition to regulating cell death, SIRT6-dependent nuclear Nrf2 accumulation and subsequent formation of Nrf2–HK2 complex were found to be indispensable for XOR expression. Disruption of glycolytic flux–mediated induction of mitochondrial ROS accumulation (43, 44) as well as dissociation of hexokinase from mitochondria is known to activate the NLRP3 inflammasome (21). As XOR inhibition attenuates NLRP3 inflammasome activation by impairing IL-1β secretion (22), it is possible that increased XOR is crucial for regulating inflammatory cell responses under diminished glycolytic flux. Our findings indicate that inhibiting glucose flux alone is not sufficient to induce apoptosis in the absence of an inflammatory trigger. It is the unique state of a cell exhibiting limited glycolytic flux, inflammation, and oxidative stress that sets the stage for SIRT6-mediated death.
Compared with the relatively well known metabolic functions of HK2 in tumor progression, the present study highlights a previously unknown non-metabolic role of HK2 in regulating transcription of genes associated with redox regulation by serving as a coactivator of Nrf2. Although the nuclear translocation of HK2 and its ability to serve as a coactivator of Nrf2 are a prerequisite for facilitating transcription of XOR, the expression of the Nrf2 target HMOX1 was found to be independent of HK2. Thus, Nrf2–HK2 complex appears to be specific for regulation of XOR and is not a general feature for regulating other known Nrf2 targets. However, it is possible that the coactivator function of HK2 may not be limited only to transactivation of Nrf2. It will therefore be interesting to identify other HK2-binding partners to reveal the non-canonical function of HK2 in a broad range of cellular functions. As there is considerable overlap between metabolic and inflammatory responses in glioma cells, understanding mechanisms through which metabolic genes subserve non-canonical functions to affect growth in metabolically stressed inflammatory conditions atypical of the tumor microenvironment would provide new insights into glioma biology.
Materials and methods
Cell culture and treatment
Glioblastoma cell lines A172, U87MG, and T98G obtained from American Type Culture Collection (Manassas, VA) were cultured in DMEM (Life Technologies) supplemented with 10% FBS (Gibco, Life Technologies). On attaining semiconfluence, cells were switched to serum-free medium, and after 6 h cells were glucose-starved in glucose-free DMEM (Life Technologies) in the presence or absence of 10 ng/ml IL-1β (R&D Systems). Treatment with the ROS inhibitor NAC (2.5 mm), JNK inhibitor SP600125 (10 μm), and H2O2-decomposing enzyme PEGylated catalase (20 μm) was performed according to experimental requirements.
Determination of cell viability
The viability of cells treated with IL-1β in the presence or absence of glucose-free DMEM was assessed using the MTS assay (Promega, Madison, WI) as described (8). Similarly, the viability of cells treated with the ROS inhibitor NAC or H2O2 inhibitor PEGylated catalase or transfected with HK2 siRNA, SIRT6 siRNA, or SIRT6 overexpression construct and treated with IL-1β in the presence or absence of glucose was determined. Values were expressed as average absorbance of technical replicates for every treatment condition.
Measurement of ROS
Intracellular ROS generation in cells treated with IL-1β in the presence and absence of glucose and ROS inhibitor or upon siRNA-mediated SIRT6 knockdown as well as SIRT6 overexpression was assessed using fluorescent dye dihydroethidium (DHE) as described previously (8). Briefly, cells were stained with 1 μm DHE in serum-free medium for 45 min at 37 °C and then washed twice with 1× PBS. Fluorescence intensity was measured at 535-nm wavelength using an Infinite M200PRO (Tecan) microplate plate reader.
Measurement of hydrogen peroxide
The AmplexTM Red Hydrogen Peroxide/Peroxidase Assay was used to determine the relative levels of H2O2 in cells deprived of glucose in the presence or absence of IL-1β. Briefly, cells were collected in 1× reaction buffer, and 50 μl of Amplex Red/HRP working solution was added to the 50-μl cell suspension according to the manufacturer's instructions. The samples were incubated in the dark at room temperature for 30 min in a black 96-well plate. Fluorescence intensity was measured at an excitation wavelength of 530 nm and an emission wavelength of 590 nm using an Infinite M200PRO microplate plate reader.
Western blot analysis
Western blot analysis was performed on protein lysates isolated from control cells or cells treated with IL-1β in the presence or absence of glucose and/or transfected with different constructs or siRNAs as described previously (37) using antibodies against Nrf2 (catalog number ab31163), SIRT6 (catalog number ab62739), p27 (catalog number ab32034), SOD2 (catalog number ab13533) (Abcam), pJNK (catalog number 4668), JNK (catalog number 9252), HK2 (catalog number 2867) (Cell Signaling Technology), RNA polymerase II (catalog number 39097) (Active Motif), p21 (catalog number 05-345) (Upstate), cyclin E (catalog number sc247), cyclin A (catalog number sc239), Bcl2 (catalog number sc7382), cytochrome c (catalog number sc13560), and XOR (catalog number sc20991) (Santa Cruz Biotechnology). Secondary antibodies were purchased from Vector Laboratories Inc. (Burlingame, CA). The blots were stripped and reprobed with anti-β-actin (catalog number A3854) (Sigma), anti-β-tubulin (catalog number sc9104), or anti-c23 (catalog number sc55486) (Santa Cruz Biotechnology) to determine equivalent loading (41). Images were photographed using ECL (Millipore) on a Syngene G:Box system (Cambridge, UK) using Gene-Sys software.
Transfection
5 × 103 cells were seeded in 96-well plates, and 2 h prior to transfection cell medium was replaced with Opti-MEM (Gibco, Life Technologies). Transfection with 70 nm duplex HK2, 50 nm SIRT6, and Nrf2 or nonspecific siRNA (Thermo Fischer Scientific) was carried out using Lipofectamine RNAiMAX reagent (Life Technologies-Invitrogen) as described previously (37). Similarly, transfection with either 10 ng of Renilla luciferase expression vector (pRL-TK) or 0.3 μg of HIF-1α luciferase construct was performed using Lipofectamine 2000 (Life Technologies), and luciferase activity was measured using the Dual-Luciferase assay kit according to the manufacturer's protocol (Promega) using a GloMax 96 microplate luminometer (Promega) as described previously (37).
Confocal microscopy
For immunofluorescence staining, cells were grown in a 4-well chamber glass slide system (Nunc Lab-Tek) and treated with JNK inhibitor (SP600125) before depriving cells of glucose and treating with IL-1β. After washing with 1× PBS, the cells were fixed in 4% paraformaldehyde for 20 min and subsequently permeabilized by 0.1% Triton-X and 0.1% BSA in 1× PBS for 10 min at room temperature. Cells were blocked with 2% BSA and 3% normal donkey serum in 1× PBS, incubated with anti-HK2 antibody at a dilution of 1:500, washed three times with 1× PBS, and incubated with Alexa Fluor 594–conjugated anti-rabbit antiserum (Invitrogen catalog number 21207) at a dilution of 1:500 for 1 h at room temperature. Mounting was done with DAPI, and the slides were visualized using a spinning disk confocal microscope (Zeiss Observer.Z1). Multiple images were taken from different fields at 63× magnification. Image analysis was carried using ZEN lite 2.3 (Carl Zeiss) software to generate a line profile of the mean fluorescence intensity.
Coimmunoprecipitation
Endogenous HK2 was immunoprecipitated with anti-HK2 antibody from nuclear extracts obtained from treated and/or transfected cells. Briefly, nuclear lysates were incubated with 3 μg of the indicated antibody for 16 h and subsequently incubated for 4 h with a mixture of protein G/A-Sepharose beads (GE Healthcare). The immunoprecipitated samples were resolved by 8–10% SDS-PAGE after washing the beads five times in immunoprecipitation buffer. 10% input was also resolved. Western blot analysis was performed with the immunoprecipitates and inputs with specific antibodies.
Xanthine oxidoreductase activity assay
The Amplex Red Xanthine/Xanthine Oxidase Assay kit (Thermo Fisher catalog number a22182) was used for detecting xanthine oxidase activity in the cell lysates according to the manufacturer's instructions.
ChIP and ChIP-qPCR assay
ChIP was performed by enzymatic DNA shearing (ChIP-IT Enzymatic kit, Active Motif) to investigate Nrf2 binding on the ARE site (−593 to −582 bp) of XOR promoter as described previously (37). Cells were deprived of glucose in the presence or absence of IL-1β for 2–4 h and fixed in 1% formaldehyde at room temperature for 8 min. Isolated nuclei were lysed and then enzymatically sheared with the enzymatic shearing kit (Active Motif). Antibodies against Nrf2 or HK2 were used for immunoprecipitation. Following reverse cross-linking and DNA purification, DNA from input (1:10 diluted) or immunoprecipitated samples were assayed by qRT-PCR. The primers spanning the ARE site on XOR promoter were as follows: XOR ChIP primer forward, 5′-TTTACAAGGCACTCCCAAAA-3′; and reverse, 5′-TGAACCTGACTCAAATCCTG-3′.
Quantitative real-time PCR
To analyze mRNA levels of different genes in cells treated with IL-1β in the presence or absence of glucose or HK2 knockdown, real-time PCR was performed as described previously (13) using a ViiA7 Real Time thermocycler (Applied Biosystems Inc.), and results were plotted as -fold change over control. All samples were normalized with their respective 18S rRNA Ct values. qRT-PCR primers used are listed in Table 1.
Table 1.
qRT-PCR primers
| Gene | Primer |
|---|---|
| XOR | Forward, 5′-GGACAGTTGTGGCTCTTGAGGT-3′ |
| Reverse, 5′-GGAAGGTTGGTTTTGCACAGCC-3′ | |
| IL-1β | Forward, 5′-GACCTTCCAGGAGAATGACC-3′ |
| Reverse, 5′-GGCTTATCATCTTTCAACACG-3′ | |
| NLRP3 | Forward, 5′-CTTCTCTGATGACCGCCAAG-3′ |
| Reverse, 5′-GCAGCAAACTGGAAAGGAAG-3′ | |
| HMOX1 | Forward, 5′-GGGTGATAGAAGAGGCCAAGA-3′ |
| Reverse, 5′-AGCTCCTGCAACTCCTCAAA-3′ | |
| HK2 | Forward, 5′-TCGCATCTGCTTGCCTACTTC-3′ |
| Reverse, 5′-CTTCTGGAGCCCATTGTCCGT-3′ | |
| 18S rRNA | Forward, 5′-CAGCCACCCGAGATTGAGCA-3′ |
| Reverse, 5′-TAGTAGCGACGGGCGGTGTG-3′ |
Statistical analysis
All comparisons between groups were performed either by two-tailed paired Student's t test or one-way ANOVA (Bonferroni's multiple comparison test) for multiple comparisons between more than two groups. All p values less than 0.05 were taken as significant.
Author contributions
T. S., P. G., and E. S. conceived and designed the study. T. S., P. G., and P. G. acquired data. T. S., P. G., P. G., and S. P. analyzed and interpreted data (e.g. statistical analysis). T. S. and E. S. wrote, reviewed, and revised the manuscript. E. S. supervised the study.
Supplementary Material
Acknowledgments
The technical assistance of Shanker Datt Joshi and Rajesh Kumar Kumawat is acknowledged.
The work was supported by Department of Biotechnology (Government of India) Research Grant BT/MED/30/SP11016/2015 (to E. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- ROS
- reactive oxygen species
- HK
- hexokinase
- SIRT6
- sirtuin 6
- Nrf2
- nuclear factor erythroid 2–related factor 2
- XOR
- xanthine oxidoreductase
- NAC
- N-acetyl-l-cysteine
- NLRP3
- NOD-like receptor family pyrin domain–containing protein 3
- HIF
- hypoxia-inducible factor
- SOD2
- mitochondrial superoxide dismutase
- pJNK
- phosphorylated JNK
- ARE
- antioxidant response element
- HMOX1
- heme oxygenase 1
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- DHE
- dihydroethidium
- qRT-PCR
- quantitative RT-PCR
- ANOVA
- analysis of variance.
References
- 1. Szatrowski T. P., and Nathan C. F. (1991) Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 51, 794–798 [PubMed] [Google Scholar]
- 2. Zhou Y., Zhou Y., Shingu T., Feng L., Chen Z., Ogasawara M., Keating M. J., Kondo S., and Huang P. (2011) Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J. Biol. Chem. 286, 32843–32853 10.1074/jbc.M111.260935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wolf A., Agnihotri S., Micallef J., Mukherjee J., Sabha N., Cairns R., Hawkins C., and Guha A. (2011) Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 208, 313–326 10.1084/jem.20101470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. John S., Weiss J. N., and Ribalet B. (2011) Subcellular localization of hexokinases I and II directs the metabolic fate of glucose. PLoS One 6, e17674 10.1371/journal.pone.0017674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu R., Wyatt E., Chawla K., Tran M., Ghanefar M., Laakso M., Epting C. L., and Ardehali H. (2012) Hexokinase II knockdown results in exaggerated cardiac hypertrophy via increased ROS production. EMBO Mol. Med. 4, 633–646 10.1002/emmm.201200240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Graham N. A., Tahmasian M., Kohli B., Komisopoulou E., Zhu M., Vivanco I., Teitell M. A., Wu H., Ribas A., Lo R. S., Mellinghoff I. K., Mischel P. S., and Graeber T. G. (2012) Glucose deprivation activates a metabolic and signaling amplification loop leading to cell death. Mol. Syst. Biol. 8, 589 10.1038/msb.2012.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dixit D., Ghildiyal R., Anto N. P., and Sen E. (2014) Chaetocin-induced ROS-mediated apoptosis involves ATM-YAP1 axis and JNK-dependent inhibition of glucose metabolism. Cell Death Dis. 5, e1212 10.1038/cddis.2014.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dixit D., Sharma V., Ghosh S., Koul N., Mishra P. K., and Sen E. (2009) Manumycin inhibits STAT3, telomerase activity, and growth of glioma cells by elevating intracellular reactive oxygen species generation. Free Radic. Biol. Med. 47, 364–374 10.1016/j.freeradbiomed.2009.04.031 [DOI] [PubMed] [Google Scholar]
- 9. Sharma V., Joseph C., Ghosh S., Agarwal A., Mishra M. K., and Sen E. (2007) Kaempferol induces apoptosis in glioblastoma cells through oxidative stress. Mol. Cancer Ther. 6, 2544–2553 10.1158/1535-7163.MCT-06-0788 [DOI] [PubMed] [Google Scholar]
- 10. Nguyen T., Nioi P., and Pickett C. B. (2009) The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 284, 13291–13295 10.1074/jbc.R900010200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kovac S., Angelova P. R., Holmström K. M., Zhang Y., Dinkova-Kostova A. T., and Abramov A. Y. (2015) Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim. Biophys. Acta 1850, 794–801 10.1016/j.bbagen.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., and Motohashi H. (2012) Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 10.1016/j.ccr.2012.05.016 [DOI] [PubMed] [Google Scholar]
- 13. Ahmad F., Dixit D., Sharma V., Kumar A., Joshi S. D., Sarkar C., and Sen E. (2016) Nrf2-driven TERT regulates pentose phosphate pathway in glioblastoma. Cell Death Dis. 7, e2213 10.1038/cddis.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pan H., Guan D., Liu X., Li J., Wang L., Wu J., Zhou J., Zhang W., Ren R., Zhang W., Li Y., Yang J., Hao Y., Yuan T., Yuan G., et al. (2016) SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 26, 190–205 10.1038/cr.2016.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R. E., Vadysirisack D. D., Guimaraes A., Marinelli B., Wikstrom J. D., Nir T., Clish C. B., Vaitheesvaran B., Iliopoulos O., Kurland I., Dor Y., et al. (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 10.1016/j.cell.2009.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim T. H., Hur E. G., Kang S. J., Kim J. A., Thapa D., Lee Y. M., Ku S. K., Jung Y., and Kwak M. K. (2011) NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1α. Cancer Res. 71, 2260–2275 10.1158/0008-5472.CAN-10-3007 [DOI] [PubMed] [Google Scholar]
- 17. Mathupala S. P., Rempel A., and Pedersen P. L. (2001) Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J. Biol. Chem. 276, 43407–43412 10.1074/jbc.M108181200 [DOI] [PubMed] [Google Scholar]
- 18. Gupta P., Sheikh T., and Sen E. (2017) SIRT6 regulated nucleosomal occupancy affects Hexokinase 2 expression. Exp. Cell Res. 357, 98–106 10.1016/j.yexcr.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 19. Ghosh S., Gupta P., and Sen E. (2016) TNFα driven HIF-1α-hexokinase II axis regulates MHC-I cluster stability through actin cytoskeleton. Exp. Cell Res. 340, 116–124 10.1016/j.yexcr.2015.11.016 [DOI] [PubMed] [Google Scholar]
- 20. Sanman L. E., Qian Y., Eisele N. A., Ng T. M., van der Linden W. A., Monack D. M., Weerapana E., and Bogyo M. (2016) Disruption of glycolytic flux is a signal for inflammasome signaling and pyroptotic cell death. eLife 5, e13663 10.7554/eLife.13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolf A. J., Reyes C. N., Liang W., Becker C., Shimada K., Wheeler M. L., Cho H. C., Popescu N. I., Coggeshall K. M., Arditi M., and Underhill D. M. (2016) Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell 166, 624–636 10.1016/j.cell.2016.05.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ives A., Nomura J., Martinon F., Roger T., LeRoy D., Miner J. N., Simon G., Busso N., and So A. (2015) Xanthine oxidoreductase regulates macrophage IL1β secretion upon NLRP3 inflammasome activation. Nat. Commun. 6, 6555 10.1038/ncomms7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Griguer C. E., Oliva C. R., Kelley E. E., Giles G. I., Lancaster J. R. Jr, and Gillespie G. Y. (2006) Xanthine oxidase-dependent regulation of hypoxia-inducible factor in cancer cells. Cancer Res. 66, 2257–2263 10.1158/0008-5472.CAN-05-3364 [DOI] [PubMed] [Google Scholar]
- 24. Martinez-Outschoorn U. E., Curry J. M., Ko Y. H., Lin Z., Tuluc M., Cognetti D., Birbe R. C., Pribitkin E., Bombonati A., Pestell R. G., Howell A., Sotgia F., and Lisanti M. P. (2013) Oncogenes and inflammation rewire host energy metabolism in the tumor microenvironment: RAS and NFκB target stromal MCT4. Cell Cycle 12, 2580–2597 10.4161/cc.25510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Neill L. A., and Hardie D. G. (2013) Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 493, 346–355 10.1038/nature11862 [DOI] [PubMed] [Google Scholar]
- 26. Huber H. J., Dussmann H., Kilbride S. M., Rehm M., and Prehn J. H. (2011) Glucose metabolism determines resistance of cancer cells to bioenergetic crisis after cytochrome-c release. Mol. Syst. Biol. 7, 470 10.1038/msb.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maksin-Matveev A., Kanfi Y., Hochhauser E., Isak A., Cohen H. Y., and Shainberg A. (2015) Sirtuin 6 protects the heart from hypoxic damage. Exp. Cell Res. 330, 81–90 10.1016/j.yexcr.2014.07.013 [DOI] [PubMed] [Google Scholar]
- 28. Neary C. L., and Pastorino J. G. (2010) Nucleocytoplasmic shuttling of hexokinase II in a cancer cell. Biochem. Biophys. Res. Commun. 394, 1075–1081 10.1016/j.bbrc.2010.03.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bauer I., Grozio A., Lasigliè D., Basile G., Sturla L., Magnone M., Sociali G., Soncini D., Caffa I., Poggi A., Zoppoli G., Cea M., Feldmann G., Mostoslavsky R., Ballestrero A., et al. (2012) The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J. Biol. Chem. 287, 40924–40937 10.1074/jbc.M112.405837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mergenthaler P., Kahl A., Kamitz A., van Laak V., Stohlmann K., Thomsen S., Klawitter H., Przesdzing I., Neeb L., Freyer D., Priller J., Collins T. J., Megow D., Dirnagl U., Andrews D. W., et al. (2012) Mitochondrial hexokinase II (HKII) and phosphoprotein enriched in astrocytes (PEA15) form a molecular switch governing cellular fate depending on the metabolic state. Proc. Natl. Acad. Sci. U.S.A. 109, 1518–1523 10.1073/pnas.1108225109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jaramillo M. C., and Zhang D. D. (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191 10.1101/gad.225680.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Theodore M., Kawai Y., Yang J., Kleshchenko Y., Reddy S. P., Villalta F., and Arinze I. J. (2008) Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J. Biol. Chem. 283, 8984–8994 10.1074/jbc.M709040200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T. W., Wasserman W. W., and Biswal S. (2010) Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 38, 5718–5734 10.1093/nar/gkq212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vega M., Riera A., Fernández-Cid A., Herrero P., and Moreno F. (2016) Hexokinase 2 is an intracellular glucose sensor of yeast cells that maintains the structure and activity of Mig1 repressor complex. J. Biol. Chem. 291, 7267–7285 10.1074/jbc.M115.711408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reichard J. F., Motz G. T., and Puga A. (2007) Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 35, 7074–7086 10.1093/nar/gkm638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Herrero P., Martínez-Campa C., and Moreno F. (1998) The hexokinase 2 protein participates in regulatory DNA-protein complexes necessary for glucose repression of the SUC2 gene in Saccharomyces cerevisiae. FEBS Lett. 434, 71–76 10.1016/S0014-5793(98)00872-2 [DOI] [PubMed] [Google Scholar]
- 37. Ghosh S., Paul A., and Sen E. (2013) Tumor necrosis factor α-induced hypoxia-inducible factor 1α-β-catenin axis regulates major histocompatibility complex class I gene activation through chromatin remodeling. Mol. Cell. Biol. 33, 2718–2731 10.1128/MCB.01254-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W., Gao X., Aldape K., and Lu Z. (2011) Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480, 118–122 10.1038/nature10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahuatzi D., Herrero P., de la Cera T., and Moreno F. (2004) The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 279, 14440–14446 10.1074/jbc.M313431200 [DOI] [PubMed] [Google Scholar]
- 40. Peláez R., Herrero P., and Moreno F. (2009) Nuclear export of the yeast hexokinase 2 protein requires the Xpo1 (Crm1)-dependent pathway. J. Biol. Chem. 284, 20548–20555 10.1074/jbc.M109.013730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharma V., Dixit D., Koul N., Mehta V. S., and Sen E. (2011) Ras regulates interleukin-1β-induced HIF-1α transcriptional activity in glioblastoma. J. Mol. Med. 89, 123–136 10.1007/s00109-010-0683-5 [DOI] [PubMed] [Google Scholar]
- 42. Del Rey A., Roggero E., Randolf A., Mahuad C., McCann S., Rettori V., and Besedovsky H. O. (2006) IL-1 resets glucose homeostasis at central levels. Proc. Natl. Acad. Sci. U.S.A. 103, 16039–16044 10.1073/pnas.0607076103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schroder K., Zhou R., and Tschopp J. (2010) The NLRP3 inflammasome: a sensor for metabolic danger? Science 327, 296–300 10.1126/science.1184003 [DOI] [PubMed] [Google Scholar]
- 44. Zhou R., Yazdi A. S., Menu P., and Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.