Abstract
Cytochrome c oxidase (COX) was initially purified more than 70 years ago. A tremendous amount of insight into its structure and function has since been gleaned from biochemical, biophysical, genetic, and molecular studies. As a result, we now appreciate that COX relies on its redox-active metal centers (heme a and a3, CuA and CuB) to reduce oxygen and pump protons in a reaction essential for most eukaryotic life. Questions persist, however, about how individual structural subunits are assembled into a functional holoenzyme. Here, we focus on what is known and what remains to be learned about the accessory proteins that facilitate CuA site maturation.
Keywords: chaperone, copper, cytochrome c oxidase (complex IV), mitochondria, protein assembly, COX assembly factors, CuA site formation
Cytochrome c oxidase is a multisubunit enzyme of dual genetic origin
COX2 is a member of the A1 subgroup of a diverse superfamily of heme-copper oxidases. Embedded in the inner mitochondrial membrane, it is a multimeric protein complex composed of structural subunits that are encoded by two distinct genomes. The three largest of these, COX1–3, are mitochondrially encoded and form the catalytic core of the enzyme. COX1 contains the two heme (a and a3) moieties and a mononuclear CuB center, all of which are buried within the lipid bilayer in the fully assembled holoenzyme. COX2 harbors a mixed valence, binuclear CuA site within a cupredoxin fold that is localized to the intermembrane space (IMS) and is solvent-exposed. The CuA site accepts electrons from cytochrome c, and subsequent electron transfer steps to the heme a and then the heme a3–CuB metal centers of COX1 ultimately allow COX to convert molecular oxygen to water. Four protons are pumped across the membrane during each catalytic cycle and contribute to the electrochemical gradient that is required for aerobic ATP production. The catalytic core is surrounded by a variable number of nuclear-encoded structural subunits (8 in yeast and 11 in humans), which function collectively to stabilize the holoenzyme, provide sites for the allosteric modulation of its catalytic activity, and facilitate its organization into higher order structures termed supercomplexes or respirasomes (1, 2). High resolution structures of mammalian COX (3) and of eight mammalian respirasomes that contain COX (4) have been invaluable to advancing our understanding of how inter-subunit interactions impinge upon enzyme activity and dimerization and the integration of COX into higher order structures.
COX is assembled in a modular fashion
COX assembly is a very complex process that requires the stoichiometric expression of its nuclear- and mitochondrially-encoded subunits and the ordered incorporation of its heme and copper prosthetic groups. The basic blueprint that defines the steps and mechanisms that facilitate COX assembly within the inner mitochondrial membrane has been derived almost exclusively from studies of mutant yeast strains and cell lines from patients who presented with an isolated COX deficiency. Elegant work done by Nitjmans et al. (5) in the late 1990s established that the individual structural subunits of COX are matured and assembled in modules, with the formation of three assembly intermediates (S1–S3) preceding the biogenesis of the mature holoenzyme (S4). Subsequent studies identified the existence of unique maturation modules specific to COX1, COX2, and COX3 (6, 7) and challenged the exact subunit composition of individual modules and the order in which they are added to the assembling holoenzyme (8). However, the original concept that COX is built using a modular blueprint remains intact.
Maturation of individual modules and progression through the various stages of holoenzyme assembly requires a surprisingly large number of accessory proteins, termed COX assembly factors. Pioneering studies of yeast nuclear petite (PET) mutant collections identified more than 30 different complementation groups that encode for gene products with unique functions in holoenzyme biogenesis (9, 10), many of which are conserved in humans (11). At least six of these complementation groups encode for accessory proteins crucial to the maturation of COX2 and the metallation of its CuA site (Table 1). Additionally, evolutionarily conserved COX assembly factors with poorly characterized functions like COA5 (12, 13) and CMC2 (14, 15) may also function in this pathway. Pathogenic mutations in COX2 (16–18), COX20 (19, 20), COA6 (21–23), SCO1 (24–26), and SCO2 (27, 28) all cause severe, early onset forms of fatal disease because of an isolated COX deficiency (Table 1). Characterization of human cell lines in which COX20 (29), COA6 (23), SCO1 (30), or SCO2 (31, 32) function is impaired indicates stalling of COX assembly always occurs at the S2 stage, when fully matured COX2 is normally inserted into the assembling holoenzyme. Loss of function mutations in the remaining COX assembly factors with putative (OXA1L) or established (COX17 and COX18) roles in COX2 maturation have yet to be described in COX-deficient patients (Table 1), despite candidate gene screening (33, 34) and the advent of next generation sequencing technologies, suggesting that such variants may be embryonic lethal in humans. Consistent with this idea, Cox17 deletion in the mouse results in a severe COX deficiency at embryonic day 6.5 and lethality shortly thereafter (35). The clinically diverse forms of fatal disease caused by mutations that compromise the function of any member of the COX2 assembly module (Table 1) means that there is great interest from a biomedical perspective in defining the precise roles of the individual COX assembly factors, and identifying how they collaborate with one another at various stages as COX2 matures. Such insight, once garnered, will be equally fascinating from a basic biological point of view, for it will clarify how each step in COX2 maturation ultimately leads to the metallation of its CuA site.
Table 1.
COX2 and COX assembly factors required for polypeptide maturation and CuA site formation
| Gene | Function | Clinical course(s) of disease |
|---|---|---|
| COX2 | Catalytic core | Myopathy (16), encephalopathy (17), multisystem disorder (18) |
| OXA1L | N-terminal insertase? | NAa (33) |
| COX18 | C-terminal insertase | NA (34) |
| COX20 | Chaperone | Dystonia and ataxia (19) |
| COX17 | IMS Cu trafficking | NA (35) |
| COA6 | CuA site maturation | Hypertrophic cardiomyopathy (21, 22) |
| SCO1 | CuA site maturation | Hypertrophic cardiomyopathy and encephalopathy (24), hepatopathy (25), encephalopathy (26) |
| SCO2 | CuA site maturation | Hypertrophic cardiomyopathy and encephalopathy (27, 28) |
a NA means not applicable.
Co-translational insertion of apo-COX2 into the inner membrane is paired with the translocation of its N terminus into the IMS
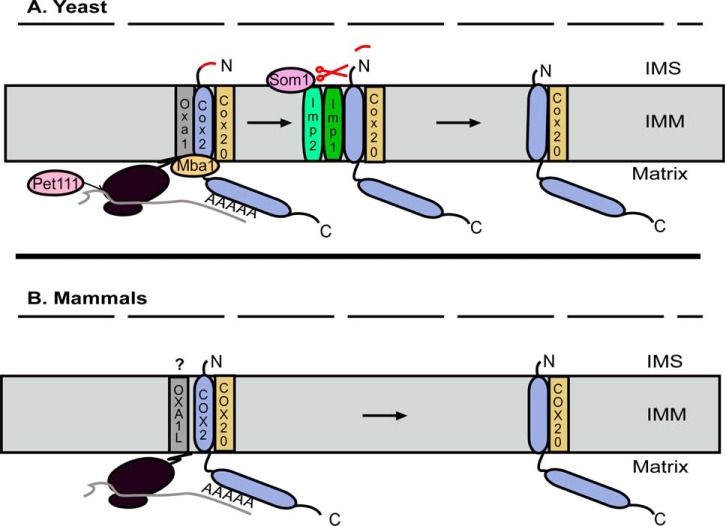
COX2 is a polytopic protein with two transmembrane domains whose N and C termini protrude into the IMS. Because of its inherent hydrophobicity, COX2 insertion into the inner membrane is always coupled to mRNA translation. In yeast, a long 5′-untranslated leader sequence is recognized by the translational activator Pet111, which recruits COX2 mRNA to specific ribosomes (Fig. 1A). Mammalian COX2 mRNA is essentially devoid of a 5′-untranslated leader sequence, and searches for the ortholog of Pet111 based on amino acid identity have proved fruitless. However, it has long been postulated that mammalian mitochondrial mRNAs contain recognition motifs within their coding sequences that are similarly vital to their efficient translation, and this is now known to be true at least for COX1 (36), raising the intriguing possibility that a mammalian-specific COX2 translational activator remains to be discovered.
Figure 1.
Co-translational insertion of COX2 and export of its N terminus. A, in yeast, Pet111 binds to Cox2 mRNA to stimulate its translation. Translation is coupled with polypeptide insertion into the inner mitochondrial membrane (IMM), in a process that is facilitated by both Oxa1 and Mba1. The C terminus of Oxa1 interacts with the large ribosomal subunit, and its interaction with Mba1 positions it near the exit tunnel where Oxa1 promotes Cox2 insertion and export of its N terminus into the IMS. Cox20 interacts with Mba1 and Cox2 to stabilize and prevent degradation of the pre-protein. The N terminus of the pre-protein is then proteolytically processed to yield mature Cox2 by a peptidase complex composed of Imp1, Imp2, and Som1. B, in mammals, the hydrophilic N terminus of COX2 is much shorter than that of yeast Cox2, and it does not require proteolytic processing upon membrane insertion. Although OXA1L physically interacts with mitochondrial ribosomes and functions as an insertase, questions remain about whether it promotes co-translational insertion of COX2 into the IMM or whether this function is fulfilled by COX20. The outer mitochondrial membrane is represented by the dashed line in both panels.
Co-translational insertion of yeast Cox2 into the membrane also requires Oxa1, a member of the conserved YidC/Alb3/Oxa1 family of insertases (37). The C terminus of Oxa1 interacts with the large ribosomal subunit in close proximity to the polypeptide exit tunnel where it promotes protein insertion into the inner membrane (38, 39). Oxa1 function in this regard is aided by Mba1, which acts to tether ribosomes to the inner membrane on the matrix side of the leaflet (Fig. 1A) (40). The human homolog of Oxa1, OXA1L, also physically interacts with mitochondrial ribosomes via its C-terminal tail (41, 42) and is able to restore COX assembly in an OXA1 null yeast strain, arguing that Oxa1-dependent insertion of the N-terminal transmembrane domain of Cox2 and the simultaneous translocation of its N terminus into the IMS is an evolutionarily conserved function (Fig. 1B) (43). However, OXA1L knockdown in HEK293 cells leads to a diminution in the abundance of complexes I and V rather than COX (44). This has led to the suggestion that in mammals COX20 may be sufficient for insertion of the N-terminal transmembrane domain of COX2, given that its hydrophilic N terminus is shorter than that of yeast Cox2 and does not require proteolytic processing upon membrane insertion (45). Further studies of OXA1L are required to discriminate between these distinct mechanistic possibilities, but they are challenged by its broad function as an insertase of subunits of multiple complexes of oxidative phosphorylation.
COX20 stabilizes apo-COX2 in the inner membrane to protect it from degradation and promote the export of its C terminus into the IMS by COX18
Although the mechanisms that facilitate export of the N-terminal tail of mammalian COX2 remain unclear, recent studies emphasize that COX20 fulfills an essential, evolutionarily conserved role during the early stages of COX2 maturation (Figs. 1 and 2). COX20 contains two transmembrane domains with N and C termini that also protrude into the IMS (29) and serves as a chaperone, stabilizing the newly synthesized polypeptide and protecting it from degradation (29, 46). Consistent with this idea, deletion of COX20 in either yeast (46) or human (29) cells results in rapid COX2 turnover. In yeast, Cox20 interacts with Cox2 and Mba1 at the ribosome (Fig. 1A) (47). Cox20 then presents the pre-protein to an inner membrane peptidase complex, which cleaves the N-terminal yeast leader sequence within the IMS to yield mature apo-Cox2 (Fig. 1A) (48).
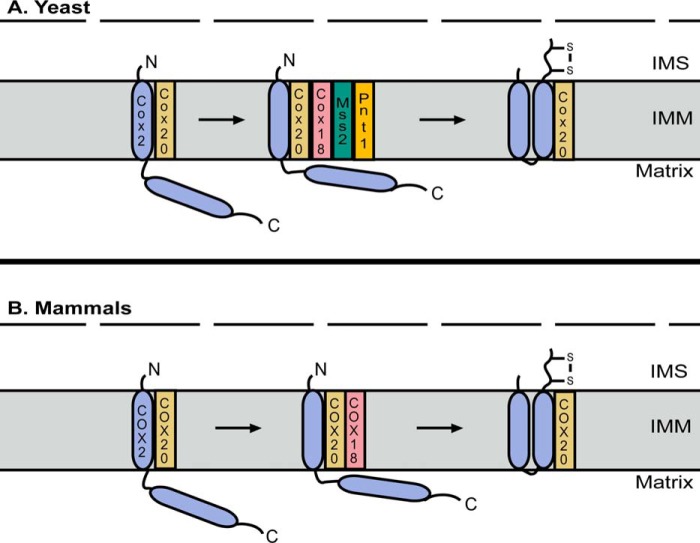
Figure 2.
Insertion and export of the C terminus of COX2. A, in yeast, insertion and export of the C-terminal transmembrane domain of Cox2 requires recruitment of Cox18 to the Cox2–Cox20 complex. The insertase function of Cox18 then depends on its physical association with Mss2 and Pnt1. B, in mammals, COX18 is similarly recruited to the COX2–COX20 complex where it is thought to function alone to promote insertion of the C-terminal transmembrane domain of COX2 and export of its C terminus into the IMS. The outer mitochondrial membrane is represented by the dashed line in both panels. IMM, inner mitochondrial membrane.
Cleavage of the N-terminal leader sequence results in Mba1 dissociation from the Cox20–Cox2 complex (47) and the subsequent recruitment of Cox18, which promotes the insertion of the C-terminal transmembrane domain and concomitant export of the long soluble C-terminal tail of Cox2 (Fig. 2A) (49). In yeast, deletion of COX18 stalls Cox2 maturation and leads to the accumulation of a species in which the N-terminal transmembrane domain is inserted into the membrane, and the leader sequence has been cleaved (50). Deletion of COX18 in human cells similarly leads to the accumulation of a stalled COX2–COX20 complex (45). Although COX18 belongs to the OXA1 protein family (50), COX2 is its only known substrate in both yeast and mammals (45). Yeast experiments in which Oxa1 was overexpressed in a COX18 null strain failed to restore growth on a non-fermentable carbon source even though some export of the C-terminal tail of Cox2 was observed, emphasizing that Cox18 translocates the tail across the membrane and delivers it in a state competent for metallation of its CuA site (51). In yeast, Cox18 depends on a physical association with two additional factors, Mss2 and Pnt1 (50, 52), for its function, although it is thought that COX18 acts alone in mammals to promote insertion of the C-terminal transmembrane domain of COX2 and the export of its tail into the IMS (Fig. 2B) (45).
COA6, SCO1, and SCO2 form a metallochaperone module that interacts with the COX20–COX2 complex to metallate the CuA site
Following the export of its C-terminal tail into the IMS, COX2 remains bound to COX20 and is competent for CuA site metallation (29, 45). Physical interaction studies indicate that dissociation of COX18 from the COX20–COX2 complex coincides with the recruitment of a metallochaperone module composed of SCO1, SCO2, and COA6 (45). This trio of proteins then functions as a collective to deliver and insert copper into the CuA site (Fig. 3), the ligands for which are two conserved cysteines, two histidines, a glutamate, and a methionine contained within a cupredoxin fold (53).
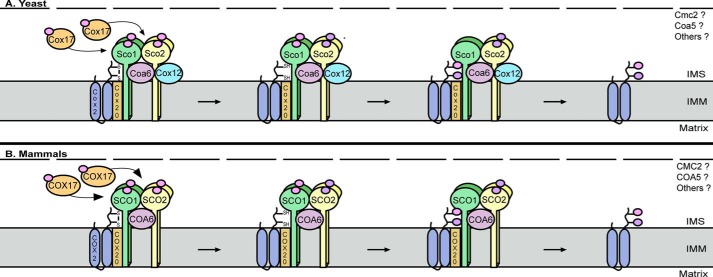
Figure 3.
CuA site maturation of COX2. In yeast (A) and mammals (B), Cox17 and COX17, respectively, transfer Cu(I) ions to Sco1/SCO1 and Sco2/SCO2 to load the metallochaperone module. Because SCO1 and SCO2 are homodimers (30), this requires two successive rounds of COX17-dependent Cu(I) delivery to fully metallate each protein. In vitro studies support a model whereby copper-loaded SCO2 functions as a thiol-disulfide oxidoreductase to reduce the cysteinyl sulfurs of COX2 (66), in a reaction that converts Cu(I) (pink circle) to Cu(II) (purple circle) and requires COA6 function in vivo. In yeast, reduction of the cysteinyl sulfurs of Cox2 also requires Coa6 and Sco2, as well as Cox12 (69). Whether the human homolog of Cox12, COX6B, is similarly important for CuA site maturation is unclear. Upon priming of the cysteinyl sulfurs of Cox2/COX2, the CuA site is metallated by Sco1/SCO1. Whether other evolutionarily conserved COX assembly factors with poorly defined roles in COX assembly also participate in CuA site maturation remains unknown. The outer mitochondrial membrane is represented by the dashed line in both panels. 1MM, inner mitochondrial membrane.
Metallation of the CuA site of COX2 by its metallochaperone module requires a dedicated upstream copper donor within the IMS, and COX17 fulfills this role in both yeast and humans (Fig. 3). COX17 was the first member to be identified from a surprisingly large family of soluble proteins with twin CX9C motifs localized to the IMS (54). The cysteinyl sulfurs of these twin CX9C motifs are recognized by the MIA40–ERV1 import pathway within the IMS, which oxidizes them to form a pair of disulfide bonds that convert an intrinsically disordered protein into one with a helical hairpin structure (55–58). Mutagenesis studies in yeast revealed that two additional conserved cysteines at the N terminus of Cox17 are responsible for the coordination of Cu(I) ions (59, 60). Results from a comprehensive suite of functional genetic experiments argue that COX17 receives its copper from an evolutionarily conserved, labile Cu(I) pool that is housed in the matrix (61). Although Cobine and co-workers (62, 63) have provided some important insight into how Cu(I) is trafficked to and within mitochondria, the mechanisms that regulate IMS copper translocation and transfer to COX17 have yet to be identified. It is clear, however, that both yeast Cox17 and human COX17 transfer copper to both SCO1 and SCO2 (Fig. 3). Elegant work from Winge and co-workers (64) used in vitro and in vivo copper transfer studies to demonstrate that Cox17 requires a cysteine within one of its twin CX9C motifs, Cys57, to be competent to transfer copper to Sco1. Equally elegant work by Bertini and co-workers (65) used NMR to directly demonstrate that COX17 transfers Cu(I) to both SCO1 and SCO2 and that this copper transfer step to SCO1 may be coupled to electron transfer, which serves to reduce the copper-binding site and promote ligand exchange.
SCO1 and SCO2 are paralogs that must be able to bind copper to promote CuA site maturation (30, 31, 66). Both proteins were originally identified in yeast as high copy suppressors of a COX17 point mutant strain (54). Subsequent studies, however, found that only deletion of SCO1 resulted in an inability to grow on a non-fermentable carbon source (67, 68), and until recently, the function of yeast Sco2 remained unknown (see below) (69). Both SCO proteins bind Cu(I) via a conserved CXXXC motif and a conserved histidine (70, 71) and rely on an additional conserved aspartate residue to coordinate Cu(II) (71). One copper atom is bound per monomer, and mutations that impair Cu(I) or Cu(II) binding result in a non-functional SCO protein, arguing that this property is crucial to their roles in COX assembly (71). The importance of copper binding to SCO1 function is emphasized by two parallel studies that both found the pathogenic P174L substitution in SCO1 severely compromises COX17-dependent copper transfer (72, 73). Whether mutations in SCO2 also impair copper transfer from COX17 remains unclear. However, the most common pathogenic variant of SCO2, SCO2 E140K, rescues the COX deficiency in SCO2 patients when overexpressed, arguing that it is competent for metallation (31).
Although SCO1 and SCO2 are part of the same metallochaperone module, each protein physically interacts with COA6 (23, 69, 74) to fulfill a distinct function during the metallation of the CuA site of COX2 (Fig. 3) (30, 31, 66). Yeast studies were the first to show that Sco1 physically interacts with Cox2 (75) and map the functional interaction between the two proteins to a conserved sequence motif within loop 8 of Sco1 (76), which is solvent-exposed and undergoes structural rearrangement when Sco1 transitions from an apo- to a metallated conformer (77). Mutations in residues within this loop 8 motif do not affect physical interactions with Cox17 or copper binding, arguing that Sco1 uses distinct interfaces to interact with Cox17 and Cox2 (76). Pulse-chase labeling of mitochondrial translation products in human cells established that impaired SCO1 function did not affect COX2 synthesis but resulted in accelerated turnover of the newly synthesized protein (73). In contrast, COX2 synthesis is greatly reduced in SCO2 patient cells, yet the residual protein that is made is much more stable than in control cells (31). These results led Shoubridge and co-workers (31) to propose that SCO2 acts upstream of SCO1 to stabilize newly synthesized COX2, and that the subsequent maturation of COX2 is contingent upon the formation of a ternary complex containing both SCO proteins and COX2. However, it is now clear that another small, soluble twin CX9C motif-containing protein, COA6, is also an essential part of the metallochaperone module that associates with the COX20–COX2 complex to catalyze copper insertion into the CuA site of COX2 (Fig. 3) (23, 45, 69, 74, 78).
The fine mechanistic details of CuA site metallation in higher eukaryotes remain to be collected, but recent years have yielded a wealth of new information that brings much sharper focus to how the metallochaperone module functions as a unit. Elegant work from Vila and co-workers (66) has established that SCO1 and SCO2 are sufficient for CuA site maturation in vitro, with SCO2 functioning as a thiol-disulfide oxidoreductase to reduce the cysteinyl sulfurs of COX2 and SCO1 acting as the copper donor. However, in this system SCO2 performs its redox function in a copper-bound state rather than through a simple disulfide-exchange reaction (66), and questions remain with respect to the fate of its Cu(II) ion after SCO2 acts as an electron donor (Fig. 3). The cysteines of the CXXXC motif of SCO1 also exist as a mixed population composed of both oxidized disulfides and reduced thiols in vivo (26, 31), and how the cysteinyl sulfurs of SCO1 are oxidized to drive copper insertion into the CuA site in the IMS is also unclear. Excitingly, the findings from a flurry of recent studies strongly suggest that COA6 might in fact bridge these distinct aspects of SCO1 and SCO2 function to directly support CuA site maturation. COA6 is reported to be a copper-binding protein (23, 74), raising the intriguing possibility that it might transition from an apo- to a copper-loaded conformer upon SCO2-dependent reduction of the cysteinyl sulfurs of the CuA site. Consistent with this idea, human COA6 and SCO2 form a complex that is essential for COX2 maturation and COX assembly (74) and a yeast COA6/SCO2 double mutant is less fit than the single mutants when grown on a non-fermentable carbon source, arguing that SCO2 and COA6 fulfill overlapping functions in CuA site metallation (69). Yeast Coa6 and human COA6 also physically interact with both Cox2/COX2 and Sco1/SCO1, and these interactions are crucial for the stability of newly synthesized polypeptide (23, 69).
Future directions
Our current understanding of mammalian COX2 maturation and CuA site metallation involves at least four distinct stages (Figs. 1–3). First, the N-terminal tail of COX2 is exported into the IMS as its N-terminal transmembrane domain is inserted into the inner membrane, in a translocation reaction that may involve OXA1L and/or COX20 but that has yet to be fully defined (1, 45). Second, COX20 associates with COX2 as it is co-translationally inserted into the membrane to stabilize the polypeptide and prevent its degradation. COX18 then promotes the simultaneous insertion of the C-terminal transmembrane domain of COX2 and translocation of its C-terminal tail into the IMS. Third, COX18 release from the COX20–COX2 complex coincides with the recruitment of the metallochaperone module composed of SCO1, SCO2, and COA6. Finally, the metallochaperone module functions collectively to reduce the cysteinyl sulfurs of COX2, with a SCO1 homodimer (30) inserting two copper ions into COX2 that it originally received from COX17 to form the CuA site. Whether COA6 oxidizes the CXXXC motif of SCO1 to promote insertion of copper into the CuA site, thereby priming its cysteinyl sulfurs to accept copper from SCO2, and whether this copper is in turn transferred to apo-SCO1 to prime SCO1 for a subsequent round of metallation are important, open, and exciting questions.
Despite the tremendous recent progress, many other questions remain with respect to the maturation of COX2 and the biogenesis of its CuA site. The first, and most obvious, question is whether the full complement of proteins that participates in this process has been identified. The twin CX9C motif-containing family of proteins contains a surprisingly large number of soluble factors that are essential for COX assembly in yeast (14), and several of these, including Cmc1 (79), Cox19 (80), and Cox23 (81), are conserved in humans and have been shown to play a role in mitochondrial copper metabolism. However, recent studies indicate that all three of these factors fulfill important functions in delivering copper to COX1 rather than COX2 (82–84). Other family members with poorly understood but evolutionarily conserved roles in COX assembly, such as CMC2 and COA5, nonetheless remain and may facilitate CuA site maturation directly or indirectly by regulating IMS copper trafficking (Fig. 3). It is equally plausible that unrelated or novel mammalian-specific factors relevant to this process have yet to be identified.
A second critical question is whether the relative timing and composition of protein constituents that make up the various subassembly modules critical to COX2 maturation and CuA site formation are constant. For example, it remains to be seen whether the metallochaperone module is homogeneous in its composition or whether there is dynamic exchange of proteins in and out of this module as COX2 is matured. This is an important consideration because there is some discord in the literature with respect to the COX assembly factors that have been identified within this module using affinity purification methods (Ref. 26 versus Ref. 78). However, it is important to note that some of these discrepancies may be explained by inter-lab variation in methodology, which may favor the enrichment and therefore capture of one complex over another (e.g. 35S-pulse-chase labeling protocols (45)). The mitochondrial proteome also differs across tissues (85, 86), and it is therefore worth considering the underexplored possibility that the constituents of the COX2 assembly module may in fact be specific to different cell types and/or species (11).
Other, more nuanced but equally important mechanistic questions also persist, independent of the complement of proteins that catalyzes CuA site maturation and its relative conservation across cell types. Why, for example, do both SCO proteins need to be able to coordinate Cu(I) and Cu(II), and how does this relate to the biogenesis of the mixed valence, binuclear CuA site? Redox functions have also been proposed for both human SCO1 (87) and SCO2 (31) based on in vivo and structural data, yet teasing apart the functional significance of their redox and copper-binding properties to CuA site maturation has yet to be realized. In fact, our general knowledge of redox homeostasis in the IMS and its impact on the redox state of cysteinyl sulfurs critical to protein function remains limited (88, 89). Beautiful work from Riemer and co-workers (88) has established that the IMS is not an oxidizing environment as was once thought and that its glutathione pool in fact exhibits the same redox potential as that of the cytosol. However, they have since demonstrated that thiol oxidation occurs within the reducing environment of the IMS because glutaredoxin levels are rate-limiting (89). The potential interplay between glutaredoxin and the metallochaperone module has obvious but as yet poorly understood consequences for maturation of the CuA site. More specifically, it remains unclear whether glutaredoxin abundance may be altered within the IMS and, if it can be, how this may impinge upon the redox state of cysteines within the CXXXC motif of SCO proteins and those within the twin CX9C motifs of COA6 to modulate the function of the metallochaperone module. We impatiently await the mechanistic insight that will be afforded by future studies that tackle these big and small questions.
Acknowledgments
We recognize the outstanding contributions of those who have investigated bacterial aa3-type oxidases and apologize for our inability to cite these and other unrelated studies due to space limitations. We thank Drs. Dennis Winge (University of Utah) and Paul Cobine (Auburn University, Auburn, AL) for critical feedback on an earlier version of this manuscript.
This is the fifth article in the Thematic Minireview series “Metals in Biology 2018: Copper homeostasis and utilization in redox enzymes.” The authors declare that they have no conflicts of interest with the contents of this article.
- COX
- cytochrome c oxidase
- IMS
- intermembrane space.
References
- 1. Timon-Gomez A., Nyvltova E., Abriata L. A., Vila A. J., Hosler J., and Barrientos A. (2017) Mitochondrial cytochrome c oxidase biogenesis: recent developments. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2017.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sinkler C. A., Kalpage H., Shay J., Lee I., Malek M. H., Grossman L. I., and Huttemann M. (2017) Tissue- and condition-specific isoforms of mammalian cytochrome c oxidase subunits: from function to human disease. Oxid. Med. Cell. Longev. 2017, 10.1155/2017/1534056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., and Yoshikawa S. (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 10.1126/science.272.5265.1136 [DOI] [PubMed] [Google Scholar]
- 4. Wu M., Gu J., Guo R., Huang Y., and Yang M. (2016) Structure of mammalian respiratory supercomplex I1III2IV1. Cell 167, 1598–1609.e1510 10.1016/j.cell.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 5. Nijtmans L. G., Taanman J. W., Muijsers A. O., Speijer D., and Van den Bogert C. (1998) Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem 254, 389–394 10.1046/j.1432-1327.1998.2540389.x [DOI] [PubMed] [Google Scholar]
- 6. Su C.-H., McStay G. P., and Tzagoloff A. (2014) The Cox3p assembly module of yeast cytochrome oxidase. Mol. Biol. Cell 25, 965–976 10.1091/mbc.E13-10-0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soto I. C., Fontanesi F., Liu J., and Barrientos A. (2012) Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta 1817, 883–897 10.1016/j.bbabio.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vidoni S., Harbour M. E., Guerrero-Castillo S., Signes A., Ding S., Fearnley I. M., Taylor R. W., Tiranti V., Arnold S., Fernandez-Vizarra E., and Zeviani M. (2017) MR-1S interacts with PET100 and PET117 in module-based assembly of human cytochrome c oxidase. Cell Rep. 18, 1727–1738 10.1016/j.celrep.2017.01.044 [DOI] [PubMed] [Google Scholar]
- 9. McEwen J. E., Ko C., Kloeckner-Gruissem B., and Poyton R. O. (1986) Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J. Biol. Chem. 261, 11872–11879 [PubMed] [Google Scholar]
- 10. Tzagoloff A., and Dieckmann C. L. (1990) PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shoubridge E. A. (2001) Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106, 46–52 10.1002/ajmg.1378 [DOI] [PubMed] [Google Scholar]
- 12. Khalimonchuk O., Rigby K., Bestwick M., Pierrel F., Cobine P. A., and Winge D. R. (2008) Pet191 is a cytochrome c oxidase assembly factor in Saccharomyces cerevisiae. Eukaryot. Cell 7, 1427–1431 10.1128/EC.00132-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huigsloot M., Nijtmans L. G., Szklarczyk R., Baars M. J., van den Brand M. A., Hendriksfranssen M. G., van den Heuvel L. P., Smeitink J. A., Huynen M. A., and Rodenburg R. J. (2011) A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am. J. Hum. Genet. 88, 488–493 10.1016/j.ajhg.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Longen S., Bien M., Bihlmaier K., Kloeppel C., Kauff F., Hammermeister M., Westermann B., Herrmann J. M., and Riemer J. (2009) Systematic analysis of the twin cx(9)c protein family. J. Mol. Biol. 393, 356–368 10.1016/j.jmb.2009.08.041 [DOI] [PubMed] [Google Scholar]
- 15. Horn D., Zhou W., Trevisson E., Al-Ali H., Harris T. K., Salviati L., and Barrientos A. (2010) The conserved mitochondrial twin Cx9C protein Cmc2 is a Cmc1 homologue essential for cytochrome c oxidase biogenesis. J. Biol. Chem. 285, 15088–15099 10.1074/jbc.M110.104786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rahman S., Taanman J. W., Cooper J. M., Nelson I., Hargreaves I., Meunier B., Hanna M. G., García J. J., Capaldi R. A., Lake B. D., Leonard J. V., and Schapira A. H. (1999) A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am. J. Hum. Genet. 65, 1030–1039 10.1086/302590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clark K. M., Taylor R. W., Johnson M. A., Chinnery P. F., Chrzanowska-Lightowlers Z. M., Andrews R. M., Nelson I. P., Wood N. W., Lamont P. J., Hanna M. G., Lightowlers R. N., and Turnbull D. M. (1999) An mtDNA mutation in the initiation codon of the cytochrome c oxidase subunit II gene results in lower levels of the protein and a mitochondrial encephalomyopathy. Am. J. Hum. Genet. 64, 1330–1339 10.1086/302361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Horváth R., Schoser B. G., Müller-Höcker J., Völpel M., Jaksch M., and Lochmüller H. (2005) Mutations in mtDNA-encoded cytochrome c oxidase subunit genes causing isolated myopathy or severe encephalomyopathy. Neuromuscul. Disord. 15, 851–857 10.1016/j.nmd.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 19. Szklarczyk R., Wanschers B. F., Nijtmans L. G., Rodenburg R. J., Zschocke J., Dikow N., van den Brand M. A., Hendriks-Franssen M. G., Gilissen C., Veltman J. A., Nooteboom M., Koopman W. J., Willems P. H., Smeitink J. A., Huynen M. A., and van den Heuvel L. P. (2013) A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum. Mol. Genet. 22, 656–667 10.1093/hmg/dds473 [DOI] [PubMed] [Google Scholar]
- 20. Doss S., Lohmann K., Seibler P., Arns B., Klopstock T., Zühlke C., Freimann K., Winkler S., Lohnau T., Drungowski M., Nürnberg P., Wiegers K., Lohmann E., Naz S., Kasten M., et al. (2014) Recessive dystonia-ataxia syndrome in a Turkish family caused by a COX20 (FAM36A) mutation. J. Neurol. 261, 207–212 10.1007/s00415-013-7177-7 [DOI] [PubMed] [Google Scholar]
- 21. Calvo S. E., Compton A. G., Hershman S. G., Lim S. C., Lieber D. S., Tucker E. J., Laskowski A., Garone C., Liu S., Jaffe D. B., Christodoulou J., Fletcher J. M., Bruno D. L., Goldblatt J., Dimauro S., et al. (2012) Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci. Transl. Med. 4, 118ra110 10.1126/scitranslmed.3003310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baertling F., A. M. van den Brand M., Hertecant J. L., Al-Shamsi A., P. van den Heuvel L., Distelmaier F., Mayatepek E., Smeitink J. A., Nijtmans L. G., and Rodenburg R. J. (2015) Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Hum. Mutat. 36, 34–38 10.1002/humu.22715 [DOI] [PubMed] [Google Scholar]
- 23. Stroud D. A., Maher M. J., Lindau C., Vögtle F. N., Frazier A. E., Surgenor E., Mountford H., Singh A. P., Bonas M., Oeljeklaus S., Warscheid B., Meisinger C., Thorburn D. R., and Ryan M. T. (2015) COA6 is a mitochondrial complex IV assembly factor critical for biogenesis of mtDNA-encoded COX2. Hum. Mol. Genet. 24, 5404–5415 10.1093/hmg/ddv265 [DOI] [PubMed] [Google Scholar]
- 24. Stiburek L., Vesela K., Hansikova H., Hulkova H., and Zeman J. (2009) Loss of function of Sco1 and its interaction with cytochrome c oxidase. Am. J. Physiol. Cell Physiol 296, C1218–C1226 10.1152/ajpcell.00564.2008 [DOI] [PubMed] [Google Scholar]
- 25. Valnot I., Osmond S., Gigarel N., Mehaye B., Amiel J., Cormier-Daire V., Munnich A., Bonnefont J. P., Rustin P., and Rötig A. (2000) Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet. 67, 1104–1109 10.1016/S0002-9297(07)62940-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leary S. C., Antonicka H., Sasarman F., Weraarpachai W., Cobine P. A., Pan M., Brown G. K., Brown R., Majewski J., Ha K. C., Rahman S., and Shoubridge E. A. (2013) Novel mutations in SCO1 as a cause of fatal infantile encephalopathy and lactic acidosis. Hum. Mutat 34, 1366–1370 10.1002/humu.22385 [DOI] [PubMed] [Google Scholar]
- 27. Jaksch M., Ogilvie I., Yao J., Kortenhaus G., Bresser H. G., Gerbitz K. D., and Shoubridge E. A. (2000) Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 9, 795–801 10.1093/hmg/9.5.795 [DOI] [PubMed] [Google Scholar]
- 28. Papadopoulou L. C., Sue C. M., Davidson M. M., Tanji K., Nishino I., Sadlock J. E., Krishna S., Walker W., Selby J., Glerum D. M., Coster R. V., Lyon G., Scalais E., Lebel R., Kaplan P., et al. (1999) Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 23, 333–337 10.1038/15513 [DOI] [PubMed] [Google Scholar]
- 29. Bourens M., Boulet A., Leary S. C., and Barrientos A. (2014) Human COX20 cooperates with SCO1 and SCO2 to mature COX2 and promote the assembly of cytochrome c oxidase. Hum. Mol. Genet. 23, 2901–2913 10.1093/hmg/ddu003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leary S. C., Kaufman B. A., Pellecchia G., Guercin G. H., Mattman A., Jaksch M., and Shoubridge E. A. (2004) Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 13, 1839–1848 10.1093/hmg/ddh197 [DOI] [PubMed] [Google Scholar]
- 31. Leary S. C., Sasarman F., Nishimura T., and Shoubridge E. A. (2009) Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1. Hum. Mol. Genet. 18, 2230–2240 10.1093/hmg/ddp158 [DOI] [PubMed] [Google Scholar]
- 32. Stiburek L., Vesela K., Hansikova H., Pecina P., Tesarova M., Cerna L., Houstek J., and Zeman J. (2005) Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 392, 625–632 10.1042/BJ20050807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coenen M. J., Smeitink J. A., Smeets R., Trijbels F. J., and van den Heuvel L. P. (2005) Mutation detection in four candidate genes (OXA1L, MRS2L, YME1L and MIPEP) for combined deficiencies in the oxidative phosphorylation system. J. Inherit. Metab. Dis. 28, 1091–1097 10.1007/s10545-005-4483-y [DOI] [PubMed] [Google Scholar]
- 34. Sacconi S., Salviati L., and Trevisson E. (2009) Mutation analysis of COX18 in 29 patients with isolated cytochrome c oxidase deficiency. J. Hum. Genet. 54, 419–421 10.1038/jhg.2009.36 [DOI] [PubMed] [Google Scholar]
- 35. Takahashi Y., Kako K., Kashiwabara S., Takehara A., Inada Y., Arai H., Nakada K., Kodama H., Hayashi J., Baba T., and Munekata E. (2002) Mammalian copper chaperone Cox17p has an essential role in activation of cytochrome c oxidase and embryonic development. Mol. Cell. Biol. 22, 7614–7621 10.1128/MCB.22.21.7614-7621.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J. E., Lochmüller H., Chevrette M., Kaufman B. A., Horvath R., and Shoubridge E. A. (2009) Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 41, 833–837 10.1038/ng.390 [DOI] [PubMed] [Google Scholar]
- 37. Bonnefoy N., Fiumera H. L., Dujardin G., and Fox T. D. (2009) Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim. Biophys. Acta 1793, 60–70 10.1016/j.bbamcr.2008.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jia L., Dienhart M., Schramp M., McCauley M., Hell K., and Stuart R. A. (2003) Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22, 6438–6447 10.1093/emboj/cdg624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szyrach G., Ott M., Bonnefoy N., Neupert W., and Herrmann J. M. (2003) Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 22, 6448–6457 10.1093/emboj/cdg623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ott M., Prestele M., Bauerschmitt H., Funes S., Bonnefoy N., and Herrmann J. M. (2006) Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 25, 1603–1610 10.1038/sj.emboj.7601070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haque M. E., Elmore K. B., Tripathy A., Koc H., Koc E. C., and Spremulli L. L. (2010) Properties of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L and its interactions with mammalian mitochondrial ribosomes. J. Biol. Chem. 285, 28353–28362 10.1074/jbc.M110.148262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haque M. E., Spremulli L. L., and Fecko C. J. (2010) Identification of protein-protein and protein-ribosome interacting regions of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L. J. Biol. Chem. 285, 34991–34998 10.1074/jbc.M110.163808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bonnefoy N., Kermorgant M., Groudinsky O., Minet M., Slonimski P. P., and Dujardin G. (1994) Cloning of a human gene involved in cytochrome oxidase assembly by functional complementation of an oxa1− mutation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 91, 11978–11982 10.1073/pnas.91.25.11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stiburek L., Fornuskova D., Wenchich L., Pejznochova M., Hansikova H., and Zeman J. (2007) Knockdown of human Oxa1l impairs the biogenesis of F1Fo-ATP synthase and NADH:ubiquinone oxidoreductase. J. Mol. Biol. 374, 506–516 10.1016/j.jmb.2007.09.044 [DOI] [PubMed] [Google Scholar]
- 45. Bourens M., and Barrientos A. (2017) Human mitochondrial cytochrome c oxidase assembly factor COX18 acts transiently as a membrane insertase within the subunit 2 maturation module. J. Biol. Chem. 292, 7774–7783 10.1074/jbc.M117.778514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Elliott L. E., Saracco S. A., and Fox T. D. (2012) Multiple roles of the Cox20 chaperone in assembly of Saccharomyces cerevisiae cytochrome c oxidase. Genetics 190, 559–567 10.1534/genetics.111.135665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lorenzi I., Oeljeklaus S., Ronsor C., Bareth B., Warscheid B., Rehling P., and Dennerlein S. (2016) The ribosome-associated Mba1 escorts Cox2 from insertion machinery to maturing assembly intermediates. Mol. Cell. Biol. MCB.00361–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hell K., Tzagoloff A., Neupert W., and Stuart R. A. (2000) Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 275, 4571–4578 10.1074/jbc.275.7.4571 [DOI] [PubMed] [Google Scholar]
- 49. Fiumera H. L., Broadley S. A., and Fox T. D. (2007) Translocation of mitochondrially synthesized Cox2 domains from the matrix to the intermembrane space. Mol. Cell. Biol. 27, 4664–4673 10.1128/MCB.01955-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saracco S. A., and Fox T. D. (2002) Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell 13, 1122–1131 10.1091/mbc.01-12-0580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fiumera H. L., Dunham M. J., Saracco S. A., Butler C. A., Kelly J. A., and Fox T. D. (2009) Translocation and assembly of mitochondrially coded Saccharomyces cerevisiae cytochrome c oxidase subunit Cox2 by Oxa1 and Yme1 in the absence of Cox18. Genetics 182, 519–528 10.1534/genetics.109.101196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Broadley S. A., Demlow C. M., and Fox T. D. (2001) Peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C tail in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 7663–7672 10.1128/MCB.21.22.7663-7672.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Speno H., Taheri M. R., Sieburth D., and Martin C. T. (1995) Identification of essential amino acids within the proposed CuA binding site in subunit II of cytochrome c oxidase. J. Biol. Chem. 270, 25363–25369 10.1074/jbc.270.43.25363 [DOI] [PubMed] [Google Scholar]
- 54. Glerum D. M., Shtanko A., and Tzagoloff A. (1996) Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271, 14504–14509 10.1074/jbc.271.24.14504 [DOI] [PubMed] [Google Scholar]
- 55. Chacinska A., Pfannschmidt S., Wiedemann N., Kozjak V., Sanjuán Szklarz L. K., Schulze-Specking A., Truscott K. N., Guiard B., Meisinger C., and Pfanner N. (2004) Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 10.1038/sj.emboj.7600389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., and Herrmann J. M. (2005) A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 10.1016/j.cell.2005.04.011 [DOI] [PubMed] [Google Scholar]
- 57. Banci L., Bertini I., Cefaro C., Cenacchi L., Ciofi-Baffoni S., Felli I. C., Gallo A., Gonnelli L., Luchinat E., Sideris D., and Tokatlidis K. (2010) Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc. Natl. Acad. Sci. U.S.A. 107, 20190–20195 10.1073/pnas.1010095107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sideris D. P., Petrakis N., Katrakili N., Mikropoulou D., Gallo A., Ciofi-Baffoni S., Banci L., Bertini I., and Tokatlidis K. (2009) A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol. 187, 1007–1022 10.1083/jcb.200905134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heaton D., Nittis T., Srinivasan C., and Winge D. R. (2000) Mutational analysis of the mitochondrial copper metallochaperone Cox17. J. Biol. Chem. 275, 37582–37587 10.1074/jbc.M006639200 [DOI] [PubMed] [Google Scholar]
- 60. Punter F. A., and Glerum D. M. (2003) Mutagenesis reveals a specific role for Cox17p in copper transport to cytochrome oxidase. J. Biol. Chem. 278, 30875–30880 10.1074/jbc.M302358200 [DOI] [PubMed] [Google Scholar]
- 61. Cobine P. A., Ojeda L. D., Rigby K. M., and Winge D. R. (2004) Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem. 279, 14447–14455 10.1074/jbc.M312693200 [DOI] [PubMed] [Google Scholar]
- 62. Vest K. E., Leary S. C., Winge D. R., and Cobine P. A. (2013) Copper import into the mitochondrial matrix in Saccharomyces cerevisiae is mediated by Pic2, a mitochondrial carrier family protein. J. Biol. Chem. 288, 23884–23892 10.1074/jbc.M113.470674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vest K. E., Wang J., Gammon M. G., Maynard M. K., White O. L., Cobine J. A., Mahone W. K., and Cobine P. A. (2016) Overlap of copper and iron uptake systems in mitochondria in Saccharomyces cerevisiae. Open Biol. 6, 150223 10.1098/rsob.150223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Horng Y. C., Cobine P. A., Maxfield A. B., Carr H. S., and Winge D. R. (2004) Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome c oxidase. J. Biol. Chem. 279, 35334–35340 10.1074/jbc.M404747200 [DOI] [PubMed] [Google Scholar]
- 65. Banci L., Bertini I., Ciofi-Baffoni S., Hadjiloi T., Martinelli M., and Palumaa P. (2008) Mitochondrial copper(I) transfer from Cox17 to Sco1 is coupled to electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 6803–6808 10.1073/pnas.0800019105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Morgada M. N., Abriata L. A., Cefaro C., Gajda K., Banci L., and Vila A. J. (2015) Loop recognition and copper-mediated disulfide reduction underpin metal site assembly of CuA in human cytochrome oxidase. Proc. Natl. Acad. Sci. U.S.A. 112, 11771–11776 10.1073/pnas.1505056112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schulze M., and Rödel G. (1988) SCO1, a yeast nuclear gene essential for accumulation of mitochondrial cytochrome c oxidase subunit II. Mol. Gen. Genet. 211, 492–498 10.1007/BF00425706 [DOI] [PubMed] [Google Scholar]
- 68. Glerum D. M., Shtanko A., and Tzagoloff A. (1996) SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271, 20531–20535 10.1074/jbc.271.34.20531 [DOI] [PubMed] [Google Scholar]
- 69. Ghosh A., Pratt A. T., Soma S., Theriault S. G., Griffin A. T., Trivedi P. P., and Gohil V. M. (2016) Mitochondrial disease genes COA6, COX6B and SCO2 have overlapping roles in COX2 biogenesis. Hum. Mol. Genet. 25, 660–671 10.1093/hmg/ddv503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nittis T., George G. N., and Winge D. R. (2001) Yeast Sco1, a protein essential for cytochrome c oxidase function is a Cu(I)-binding protein. J. Biol. Chem. 276, 42520–42526 10.1074/jbc.M107077200 [DOI] [PubMed] [Google Scholar]
- 71. Horng Y. C., Leary S. C., Cobine P. A., Young F. B., George G. N., Shoubridge E. A., and Winge D. R. (2005) Human Sco1 and Sco2 function as copper-binding proteins. J. Biol. Chem. 280, 34113–34122 10.1074/jbc.M506801200 [DOI] [PubMed] [Google Scholar]
- 72. Banci L., Bertini I., Ciofi-Baffoni S., Leontari I., Martinelli M., Palumaa P., Sillard R., and Wang S. (2007) Human Sco1 functional studies and pathological implications of the P174L mutant. Proc. Natl. Acad. Sci. U.S.A. 104, 15–20 10.1073/pnas.0606189103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cobine P. A., Pierrel F., Leary S. C., Sasarman F., Horng Y. C., Shoubridge E. A., and Winge D. R. (2006) The P174L mutation in human Sco1 severely compromises Cox17-dependent metallation but does not impair copper binding. J. Biol. Chem. 281, 12270–12276 10.1074/jbc.M600496200 [DOI] [PubMed] [Google Scholar]
- 74. Pacheu-Grau D., Bareth B., Dudek J., Juris L., Vögtle F. N., Wissel M., Leary S. C., Dennerlein S., Rehling P., and Deckers M. (2015) Cooperation between COA6 and SCO2 in COX2 maturation during cytochrome c oxidase assembly links two mitochondrial cardiomyopathies. Cell Metab. 21, 823–833 10.1016/j.cmet.2015.04.012 [DOI] [PubMed] [Google Scholar]
- 75. Lode A., Kuschel M., Paret C., and Rödel G. (2000) Mitochondrial copper metabolism in yeast: interaction between Sco1p and Cox2p. FEBS Lett. 485, 19–24 10.1016/S0014-5793(00)02176-1 [DOI] [PubMed] [Google Scholar]
- 76. Rigby K., Cobine P. A., Khalimonchuk O., and Winge D. R. (2008) Mapping the functional interaction of Sco1 and Cox2 in cytochrome oxidase biogenesis. J. Biol. Chem. 283, 15015–15022 10.1074/jbc.M710072200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Banci L., Bertini I., Calderone V., Ciofi-Baffoni S., Mangani S., Martinelli M., Palumaa P., and Wang S. (2006) A hint for the function of human Sco1 from different structures. Proc. Natl. Acad. Sci. U.S.A. 103, 8595–8600 10.1073/pnas.0601375103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ghosh A., Trivedi P. P., Timbalia S. A., Griffin A. T., Rahn J. J., Chan S. S., and Gohil V. M. (2014) Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum. Mol. Genet. 23, 3596–3606 10.1093/hmg/ddu069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Horn D., Al-Ali H., and Barrientos A. (2008) Cmc1p is a conserved mitochondrial twin CX9C protein involved in cytochrome c oxidase biogenesis. Mol. Cell. Biol. 28, 4354–4364 10.1128/MCB.01920-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nobrega M. P., Bandeira S. C., Beers J., and Tzagoloff A. (2002) Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J. Biol. Chem. 277, 40206–40211 10.1074/jbc.M207348200 [DOI] [PubMed] [Google Scholar]
- 81. Barros M. H., Johnson A., and Tzagoloff A. (2004) COX23, a homologue of COX17, is required for cytochrome oxidase assembly. J. Biol. Chem. 279, 31943–31947 10.1074/jbc.M405014200 [DOI] [PubMed] [Google Scholar]
- 82. Bourens M., and Barrientos A. (2017) A CMC1-knockout reveals translation-independent control of human mitochondrial complex IV biogenesis. EMBO Rep. 18, 477–494 10.15252/embr.201643103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bode M., Woellhaf M. W., Bohnert M., van der Laan M., Sommer F., Jung M., Zimmermann R., Schroda M., and Herrmann J. M. (2015) Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase. Mol. Biol. Cell 26, 2385–2401 10.1091/mbc.E14-11-1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dela Cruz R., Jeong M. Y., and Winge D. R. (2016) Cox1 mutation abrogates need for Cox23 in cytochrome c oxidase biogenesis. Microb. Cell 3, 275–284 10.15698/mic2016.07.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., and Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 10.1016/S0092-8674(03)00926-7 [DOI] [PubMed] [Google Scholar]
- 86. Forner F., Foster L. J., Campanaro S., Valle G., and Mann M. (2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics 5, 608–619 10.1074/mcp.M500298-MCP200 [DOI] [PubMed] [Google Scholar]
- 87. Williams J. C., Sue C., Banting G. S., Yang H., Glerum D. M., Hendrickson W. A., and Schon E. A. (2005) Crystal structure of human SCO1: implications for redox signaling by a mitochondrial cytochrome c oxidase “assembly” protein. J. Biol. Chem. 280, 15202–15211 10.1074/jbc.M410705200 [DOI] [PubMed] [Google Scholar]
- 88. Kojer K., Bien M., Gangel H., Morgan B., Dick T. P., and Riemer J. (2012) Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J. 15;31(14):3160–82 10.1038/emboj.2012.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kojer K., Peleh V., Calabrese G., Herrmann J. M., and Riemer J. (2015) Kinetic control by limiting glutaredoxin amounts enables thiol oxidation in the reducing mitochondrial intermembrane space. Mol. Biol. Cell 26, 195–204 10.1091/mbc.E14-10-1422 [DOI] [PMC free article] [PubMed] [Google Scholar]