Abstract
Mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) integrates various environmental signals to regulate cell growth and metabolism. DEPTOR, also termed DEPDC6, is an endogenous inhibitor of mTORC1 and mTORC2 activities. The abundance of DEPTOR centrally orchestrates the mTOR signaling network. However, the mechanisms by which DEPTOR stability is regulated are still elusive. Here, we report that OTU domain–containing ubiquitin aldehyde-binding protein 1 (OTUB1) specifically deubiquitinates DEPTOR in a deubiquitination assay. We found that OTUB1 directly interacted with DEPTOR via its N-terminal domain, deubiquitinated DEPTOR, and thereby stabilized DEPTOR in a Cys-91–independent but Asp-88–dependent manner, suggesting that OTUB1 targets DEPTOR for deubiquitination via a deubiquitinase activity–independent non-canonical mechanism. The interaction between OTUB1 and DEPTOR was enhanced when the cells were treated with amino acids. Moreover, OTUB1 suppressed amino acid–induced activation of mTORC1 in a DEPTOR-dependent manner and thereby ultimately controlled cellular autophagy, cell proliferation, and size. Our findings reveal a mechanism that stabilizes the mTORC1 inhibitor DEPTOR via OTUB1's deubiquitinase activity. Our insights may inform research into various mTOR activity–related diseases, such as cancer, and may contribute to the identification of new diagnostic markers and therapeutic strategies for cancer treatments.
Keywords: cancer biology, cell signaling, deubiquitylation (deubiquitination), protein stability, tumor cell biology, DEPTOR, OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1), mTORC1
Introduction
The mammalian target of rapamycin complex (mTORC) signaling pathway, a central regulator of mechanistic target of rapamycin complex 1 (mTORC1),4 orchestrates both intracellular and extracellular signals to control cell metabolism, growth, proliferation, and survival (1–4). Great efforts have been made to improve our understanding of how mTORC1 integrates systemic and local nutrient signals, which constitute a regulatory circuit involving the essential activators of mTORC1, such as small GTPase proteins Rheb and Rags (5–9). The activated mTORC1 phosphorylates substrates, such as S6K, 4EBP1, ULK1, and TFEB, and thus regulates cell growth, autophagy, and cell metabolism (10, 11). Deregulated mTORC1 signaling is closely associated with a wide range of diseases, including various cancers, metabolic diseases, and developmental disorders (1, 5, 11, 12).
DEPTOR, also known as DEPDC6, plays an important role in both mTORC1 and mTORC2 signaling pathways. DEPTOR was degraded rapidly and promoted the activity of mTOR kinase when cells were shifted from unfavorable to favorable environments (4). It was also found that DEPTOR suppressed mTOR activity by interacting with the FAT domain of mTOR via its C-terminal PDZ domain. The N terminus of DEPTOR contains two DEP domains with largely unknown function. Moreover, DEPTOR is an unstable protein and can be rapidly degraded under growth factor stimulation (4, 13). Importantly, previous studies have described an mTOR-dependent phosphorylation-driven pathway underlying which DEPTOR was degraded by SCFβTrCP (4, 13–15), suggesting a plausible existence of potential DUBs that contribute to DEPTOR stability.
Ubiquitination is an essential modification during the post-translational process (16). Ubiquitin (Ub) is a small polypeptide and is attached to the amino group of an acceptor, which is usually on the lysine residues of substrates (16). Ubiquitination is initiated by the E1 enzyme–dependent activation of ubiquitin followed by its transfer to an E2 ubiquitin-conjugating enzyme. The resultant thioester linkage of Ub to E2 enzyme acts in concert with a ubiquitin ligase (E3) to conjugate ubiquitin on substrates (17, 18). This cycle can be repeated to form various ubiquitin chains (Lys-48, Lys-63, Lys-33, Lys-29, Lys-27, Lys-6, or Lys-11) (19, 20). The type of ubiquitin chain linkage determines the destination of the corresponding ubiquitination. For example, Lys-48–linked Ub chains usually target proteins for proteasome degradation. In contrast, Lys-63–linked Ub chains are non-degradable and often orchestrate protein complex in signaling pathways (20–22). Selection of the substrate is dependent on the E3, whereas the E2 can determine the specific type of ubiquitin chain (23).
DUBs are cysteine proteases (with the exception of the JAMM family DUBs, which are metalloproteases) (24, 25). They can catalyze the removal of Ub from Ub-modified proteins and for the processing of tandemly linked Ub precursors that are nascently translated (24, 26, 27). Based on the structure of the active site and the mechanism of catalysis, DUBs are divided into five groups: UCHs, USPs, MJDs, OTUs, and JAMMs (24). The release of Ub is achieved either by the removal of the entire Ub chain from the protein or by the removal of individual or multiple ubiquitins from the chain. Conversely, different DUBs exhibit specific preferences for mono-/poly-Ub chains (28). There are also some DUBs that target a wide range of distinct ubiquitinated proteins and display a diverse array of biological functions (29).
In this study, we identified ovarian tumor (OTU) domain–containing ubiquitin aldehyde-binding protein 1 (OTUB1) as a DEPTOR-interacting DUB. OTUB1 belongs to the OTU protein superfamily (30, 31) and is implicated in the NF-κβ and TGF-β signaling pathways (32). It can stabilize c-IAP2 (33), phosphorylated SMAD2/3 (32), and TRAF3/6 (34). OTUB1 is also involved in the functional regulation of some proteins that are critical for tumorigenesis, such as p53 (35, 36), estrogen receptor α (37, 38) FOXM1 (39), and SMAD2/3 (32, 40), indicating that OTUB1 may play an important role in tumorigenesis. Consistently, recent studies have reported that OTUB1 is overexpressed in colorectal and prostate cancers (41–43).
In this study, we identified OTUB1 as a DEPTOR-interacting protein that deubiquitinates and stabilizes DEPTOR in an Asp-88–dependent manner. Moreover, OTUB1 also negatively regulates the activation of mTORC1 by stabilizing DEPTOR protein under the changing conditions of nutrient cues, which ultimately involves in cellular autophagy, cell size alteration, and tumor growth.
Results
OTUB1 is a potential DUB of DEPTOR
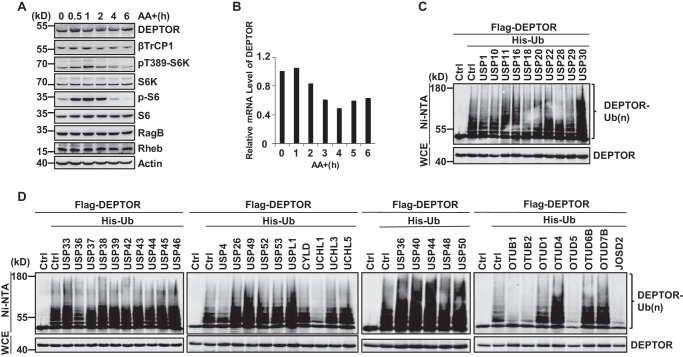
mTORC1 signaling is activated by short-term amino acid treatment (44). We were interested in studying whether and how mTORC1 activation is shut off by long-term amino acid treatment. Consistent with a previous study, treatment with amino acids results in mTORC1 activation in 1 h as determined by pS6K and pS6 (44). Interestingly, we found that under long-term amino acid stimulation up to 6 h, the activation of mTORC1 signaling was significantly decreased (Fig. 1A), suggesting that a negative feedback might be operating to control such amino acid–dependent mTORC1 signaling pathway.
Figure 1.
Screening DUBs that deubiquitinate DEPTOR. A, in HEK293T cells, mTORC1 signaling significantly declined under long-term amino acid stimulation up to 6 h as determined by pS6K and pS6. The DEPTOR protein level increased in response to long-term amino acid stimulation. HEK293T cells were starved under amino acid deprivation conditions for up to 6 h. Cells were lysed, and the protein levels of DEPTOR were detected by immunoblotting using anti-DEPTOR antibody. The expression levels of other mTOR-related proteins were detected with their specific antibodies, respectively. B, the mRNA levels of DEPTOR at different time points were detected using quantitative PCR. C and D, screening DUBs that specifically deubiquitinate DEPTOR. The ubiquitination level of DEPTOR was analyzed in HEK293T cells after transfection of different DUB plasmids. The ubiquitinated proteins were pulled down under denaturing conditions using Ni-NTA-agarose beads and detected by Western blotting. AA, amino acids; Ctrl, control; WCE, whole-cell extract.
To uncover the mechanism by which mTORC1 is inactivated under long-term amino acid treatment, we examined the expression levels of the corresponding mTORC1 components in human embryonic kidney 293T (HEK293T) cells. Interestingly, we found that the protein levels of DEPTOR, but not other proteins, were increased in response to long-term amino acid stimulation (Fig. 1A). However, we did not detect up-regulation of DEPTOR at the mRNA level (Fig. 1B), suggesting that DEPTOR levels may be regulated at the post-translational level. It has been reported that SCFβTrCP targets DEPTOR for ubiquitination-dependent degradation (4, 13–15). However, it is not clear whether DEPTOR is regulated by deubiquitination as well. To explore the potential relevance of DUBs for DEPTOR stability, we screened a library of different DUBs by examining the ubiquitination levels of DEPTOR. Our data showed that OTUB1, OTUB2, OTUD5, UCHL1, and JOSD2, but not other DUBs, significantly removed the ubiquitination chain from DEPTOR (Fig. 1, C and D).
OTUB1 interacts with DEPTOR via its N-terminal domain
To further identify which DUB is involved in the DEPTOR deubiquitination, we examined the interaction between the above-mentioned DUBs and DEPTOR by a coimmunoprecipitation assay. Our data showed that OTUB1, OTUB2, OTUD5, UCHL1, and JOSD2, but not other DUBs, could interact with DEPTOR (Fig. 2, A and B). The interaction between exogenous or endogenous DEPTOR and OTUB1 was further confirmed by a coimmunoprecipitation assay in HEK293T cells (Fig. 2, C–E). These data suggest that OTUB1 may directly interact with DEPTOR.
Figure 2.
OTUB1 directly interacts with DEPTOR, and their interaction is regulated by amino acid signaling. A, FLAG-OTUB1, FLAG-OTUB2, FLAG-OTUD5, or FLAG-JOSD2 and HA-DEPTOR were cotransfected into HEK293T cells, and then cells were lysed. DEPTOR was immunoprecipitated using anti-HA or anti-FLAG antibody. Coimmunoprecipitated OTUB1, OTUB2, OTUD5, and JOSD2 were detected using anti-FLAG or -HA antibody, respectively. B, HA-OTUB1, HA-UCHL1, and FLAG-DEPTOR were cotransfected into HEK293T cells and detected by Western blotting. C, HA-OTUB1 and FLAG-DEPTOR were cotransfected into HEK293T cells, and cells were lysed. DEPTOR was immunoprecipitated using anti-FLAG antibody. Coimmunoprecipitated OTUB1 was detected using anti HA-antibody. D, transfected cells were cotransfected with FLAG-OTUB1 and HA-DEPTOR, and the cell lysates were immunoprecipitated using anti-HA or FLAG antibody, respectively. E, endogenous DEPTOR was immunoprecipitated from HeLa cells using rabbit DEPTOR polyclonal antibody or control rabbit IgG antibody. Coimmunoprecipitated endogenous OTUB1 was analyzed by Western blotting. F, domain architecture of OTUB1 and OTUB2. G, the N-terminal region of OTUB1 is critical for the interaction between OTUB1 and DEPTOR. H, the recombinant protein N+OTUB2 interacts with DEPTOR in the same way as OTUB1. I and J, the interaction between OTUB1 and DEPTOR is regulated by amino acid signaling. AA, amino acids; Ctrl, control; WCE, whole-cell extract; IP, immunoprecipitation.
OTUB1 differs from OTUB2 primarily by an N-terminal extension of 46 amino acid residues that is absent from OTUB2 (Fig. 2F). We therefore tested whether the interaction between DEPTOR and OTUB1 depends on the N terminus of OTUB1. DEPTOR was coexpressed with the wildtype or a mutant of HA-OTUB1 lacking its N-terminal extension (HA-OTUB1ΔN). As shown in Fig. 2G, our data showed that the mutant, HA-OTUB1ΔN, lost the ability to interact with DEPTOR, indicating an important role of the N terminus of OTUB1 in its interaction with DEPTOR. To further confirm this finding, we generated an OTUB1-(N-term)-OTUB2 (N+OTUB2) chimera construct and examined its interaction with DEPTOR. We found that the binding of DEPTOR was rescued by the fusion of the N terminus of OTUB1 to OTUB2 (Fig. 2H). These results together indicate that OTUB1 requires its N terminus to interact with DEPTOR.
Because the cellular protein levels of DEPTOR were increased in response to long-term amino acid stimulation as shown in Fig. 1, we hypothesized that the binding of OTUB1 and DEPTOR is mediated by amino acid signals. To this end, we examined whether the interaction between DEPTOR and OTUB1 is regulated by amino acid stimulation. As shown in Fig. 2I, the binding of OTUB1 to DEPTOR was markedly increased under amino acid treatment. The interaction between endogenous DEPTOR and OTUB1 was also strengthened under amino acid treatment (Fig. 2J), suggesting that OTUB1 may be involved in the modulation of DEPTOR stability induced by amino acid signaling.
OTUB1 stabilizes DEPTOR
We next examined whether OTUB1 is involved in DEPTOR stabilization. Our data showed that protein levels of DEPTOR were dramatically increased by coexpression of OTUB1 (Fig. 3A). We also examined whether OTUB1 affects the stability of DEPTOR using a cycloheximide (CHX) chase assay. To this end, FLAG-DEPTOR was coexpressed with or without FLAG-OTUB1 in HEK293T cells. The half-life time of FLAG-DEPTOR was analyzed. Our data showed that OTUB1 significantly prolonged the half-life of FLAG-DEPTOR (Fig. 3, B and C). The E3 ubiquitin ligase SCFβTrCP has been reported to induce DEPTOR degradation (49). Our data showed that expression of OTUB1 markedly prevented the degradation of DEPTOR induced by βTrCP (Fig. 3D).
Figure 3.
OTUB1 regulates the stability of DEPTOR. A, FLAG-DEPTOR was cotransfected with empty vector or with increasing amounts of FLAG-OTUB1 constructs in HEK293T cells. B, overexpression of OTUB1 prolongs the half-life of DEPTOR. HEK293T cells were transfected with OTUB1 and DEPTOR followed by treatment with 50 μg/ml CHX, and then cells were harvested at the indicated times. C, the protein band intensities of DEPTOR were quantified by densitometry. Error bars represent S.D. D, OTUB1 prevents βTrCP-mediated DEPTOR degradation. E, endogenous DEPTOR level was analyzed in OTUB1-deficient HeLa cell lines. F, HeLa cells were transfected with specific OTUB1 siRNA (siOTUB1). After 72 h, cells were deprived of amino acids for up to 6 h. The levels of DEPTOR were detected by immunoblotting using anti-DEPTOR antibody. Ctrl, control; AA, amino acids; siNC, nonspecific control siRNA.
Given the fact that overexpression of OTUB1 can stabilize DEPTOR protein, we tested whether endogenous OTUB1 can also regulate DEPTOR stability. To this end, we established OTUB1-deficient HeLa cell lines using the CRISPR-Cas9 system. Our data showed that expression levels of DEPTOR were obviously decreased in OTUB1-deficient HeLa cells compared with that of their parent wildtype HeLa cells (Fig. 3E). Importantly, the enhanced protein levels of DEPTOR induced by long-term amino acid treatment were abolished by OTUB1-specific siRNAs (Fig. 3F). Collectively, these data indicate that OTUB1 is a deubiquitinase of DEPTOR that can stabilize DEPTOR.
Asp-88 is required for the activity of OTUB1 toward DEPTOR
We have confirmed that OTUB1 can specifically bind to and stabilize DEPTOR. OTUB1 is a deubiquitinating enzyme that specifically removes Lys-48–linked ubiquitin chains (45). We therefore examined whether the deubiquitination of DEPTOR by OTUB1 was dependent on its catalytic activity. To this end, we generated the OTUB1 C91S mutant in which the catalytic cysteine 91 was replaced by serine and thereby lost its catalytic activity (Fig. 4A). To our surprise, our data showed that the OTUB1 C91S mutant could still abrogate the ubiquitination of DEPTOR as efficiently as the wildtype OTUB1 (Fig. 4B), indicating that Cys-91 is not required for OTUB1 to deubiquitinate DEPTOR.
Figure 4.
Asp-88 is critical for OTUB1 to suppress DEPTOR ubiquitination. A, domain architecture of OTUB1 and OTUB2. Mutations at amino acid residues used in the study are shown. B, mutant OTUB1 D88A, but not mutant OTUB1 C91S, failed to suppress DEPTOR ubiquitination. HEK293T cells were transfected with different combinations of the indicated plasmids. A ubiquitination assay was conducted using Ni-NTA pulldown under denaturing conditions followed by Western blotting in vivo. C, mutant OTUB1 D88A cannot stabilize DEPTOR in HEK293T cells. D and E, overexpression of OTUB1 D88A cannot prolong the half-life of DEPTOR in HEK293T cells. F, DEPTOR exhibits a stronger interaction with OTUB1 D88A than with wildtype OTUB1 in HEK293T cells. IP, immunoprecipitation; WCE, whole-cell extract; Ctrl, control.
OTUB1 can also stabilize proteins via an Asp-88–dependent non-canonical mechanism (36). We therefore examined whether OTUB1 stabilized DEPTOR by a non-canonical mechanism. We showed that OTUB1 requires its N terminus to interact with DEPTOR. OTUB1 C23S mutant in which the reactive cysteine in the OTUB1 N-terminal region is disrupted also behaved like the wild-type OTUB1 (Fig. 4B), indicating that Cys-23 is not required for its activity toward DEPTOR. In contrast, we found that mutation of Asp-88 to Ala in OTUB1 (D88A) dramatically abolished its deubiquitinase activity toward DEPTOR (Fig. 4B). Consistently, OTUB1 D88A lost the ability to stabilize DEPTOR (Fig. 4, C, D, and E). These results indicate that Asp-88 is required for the deubiquitinase activity of OTUB1 to stabilize DEPTOR in cells. Intriguingly, the capability of OTUB1 D88A to interact with DEPTOR was much stronger than that of wildtype OTUB1 (Fig. 4F), which can be explained by a “substrate trapping” mechanism. Together, these results reveal that Asp-88 is critical for OTUB1 to suppress DEPTOR ubiquitination in cells.
OTUB1 regulates the mTORC1 activity in amino acid signaling pathway
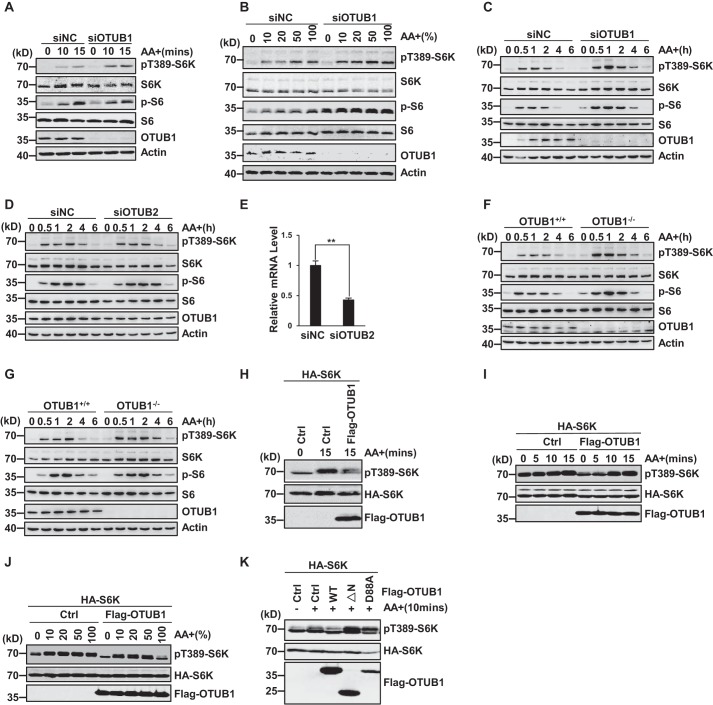
Because the interaction between OTUB1 and DEPTOR was regulated by amino acid signals (Fig. 2, I and J), we were particularly interested in exploring whether OTUB1 has any effect on mTORC1 activation induced by amino acid treatment. To this end, endogenous OTUB1 was depleted in H1299 cells using its specific siRNA, and the cells were treated with amino acid at different time points as indicated. Activation of mTORC1 was monitored by detecting the levels of phosphorylation of S6K1 at Thr-389, which is dependent on mTORC1 activation (10, 11). We found that depletion of OTUB1 dramatically increased the amino acid–induced S6K1 phosphorylation at Thr-389 at different time points as indicated (Fig. 5, A and C). Moreover, knockdown of OTUB1 markedly increased the phosphorylation level of S6K1 at Thr-389 in H1299 cells within a broad range of amino acid concentrations (Fig. 5B). In addition, the phosphorylation of S6, a target of S6K, was intimately correlated with the phosphorylation status of S6K1 (Fig. 5, A–C), indicating that OTUB1 depletion also promotes cellular S6K1 kinase activities. This phenotype is specific to OTUB1 but not OTUB2 (Fig. 5, D and E). We also confirmed this result in OTUB1-deficient HeLa cells as well as in HCT116 cells (Fig. 5, F and G). Taken together, these data indicate that OTUB1 is an endogenous negative regulator of mTORC1 activation in response to amino acid signals.
Figure 5.
OTUB1 is a negative regulator of amino acid–induced mTORC1 activation. A–C, knockdown of OTUB1 enhances amino acid–dependent mTORC1 signaling in H1299 cells. H1299 cells were treated with nonspecific control siRNA (siNC) or OTUB1-targeting siRNA (siOTUB1). The cells were treated with amino acids at the indicated concentrations (B) and durations (A and C). D and E, knockdown of OTUB2 has no effect on mTORC1 signaling. Error bars represent S.D. (**, p < 0.01). F and G, the deficiency of OTUB1 enhances amino acid–dependent mTORC1 signaling in HCT116 (F) and OTUB1-KO HeLa cells (G). H–J, OTUB1 inhibits mTORC1 signaling. HEK293T cells expressing the indicated proteins were treated with amino acids for the indicated durations (H and I) and at different concentrations (J). K, neither OTUB1ΔN nor OTUB1 D88A inhibits mTORC1 signaling. Ctrl, control; AA, amino acids.
In accordance with the negative effect of OTUB1 on mTORC1 signaling, we found that ectopic expression of OTUB1 significantly suppressed mTORC1 activation induced by amino acid treatment (Fig. 5H). Such effect was also observed with amino acid stimulation in both time- and gradient-dependent manners within a broad range of amino acid concentrations (Fig. 5, I and J). This inhibition required the N terminus of OTUB1 and its Asp-88 residue (Fig. 5K). Taken together, our data demonstrate that OTUB1 is a negative regulator of amino acid–induced mTORC1 activation, which is dependent on the Asp-88 site of OTUB1 and its interaction with DEPTOR.
OTUB1 regulates cell proliferation, cell size, and autophagy
It has been demonstrated that amino acid deprivation can induce autophagy due to inhibition of mTORC1 activities (46, 47). Given that OTUB1 negatively regulates mTORC1 activation, we then examined whether OTUB1 is involved in regulation of autophagy. Our data showed that the autophagy induced by amino acid starvation was markedly promoted by OTUB1 overexpression as detected by the production of GFP-LC3II puncta (Fig. 6, A and B). On the contrary, OTUB1 depletion significantly decreased the autophagy induced by amino acid starvation (Fig. 6, C and D). Together, these data indicate that OTUB1 is a negative regulator of amino acid starvation–induced autophagy.
Figure 6.
OTUB1 regulates cell growth, proliferation, and cell size through DEPTOR. A–D, OTUB1 induces the formation of LC3 puncta in HeLa cells. HeLa cells were transfected with OTUB1 (A and B) or with the OTUB1 siRNA (C and D). The numbers of LC3 puncta in vector-, OTUB1-, or OTUB1 siRNA–transfected cells were counted. Error bars represent S.D. (**, p < 0.01). E, H1299 cells were transfected with siRNAs as indicated, and cell proliferation was analyzed using an MTT assay. The cell proliferation rate was normalized to that of the control group. Error bars represent S.D. (**, p < 0.01). F, H1299 cells were transfected with control or OTUB1 siRNAs (siOTUB1), and FACS analysis was performed to determine cell size. G, FACS analysis was performed to determine cell size of OTUB1-KO HeLa cells. siNC, nonspecific control siRNA; FCS, forward cell scatter.
Previous studies have shown that mTORC1 is involved in cell growth (47). Given that OTUB1 mediates the stability of DEPTOR, we examined whether OTUB1 affects cell proliferation using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. As shown in Fig. 6E, knockdown of OTUB1 by its specific siRNA significantly increased the proliferation rate of HeLa cells. To further determine whether the effects of OTUB1 on mTORC1 signaling are physiologically significant, we measured the cell size of OTUB1-deficient cells. Our data showed that cells depleted of OTUB1 by siRNA were larger than those of parallel control cells (Fig. 6F). Similar results were obtained in OTUB1-knockout HeLa cells (Fig. 6G). Collectively, these data illustrate that OTUB1 regulates cell proliferation, cell size, and autophagy.
Discussion
OTUB1 is a member of the OTU domain–containing cysteine proteases and catalyzes the cleavage of Lys-48–linked poly-Ub in vitro (45). In this study, we found that OTUB1 directly interacts with DEPTOR in cells and in vitro, resulting in stabilization of DEPTOR. DEPTOR is an endogenous inhibitor of both mTORC1 and mTORC2 (4). We found that overexpression of OTUB1 negatively regulates mTORC1 activation in response to amino acid signals. Thus, we have identified OTUB1 as a new regulator for stabilizing DEPTOR.
In contrast to OTUB1, an N-terminal extension of 46 amino acid residues is absent from OTUB2 (48). Although both can deubiquitinate DEPTOR, OTUB2 cannot interact with DEPTOR. We have proven that the N-terminal region of OTUB1 is necessary for the binding of OTUB1 to DEPTOR as there was no interaction between OTUB1ΔN and DEPTOR. It is worthwhile to mention that such binding also exists between N+OTUB2 and DEPTOR, although whether N+OTUB2 possesses the ability to stabilize DEPTOR is still unknown.
Cys-91 is an essential catalytic site for OTUB1 enzymatic activities toward poly-Ub chains (48). However, mutation of the catalytic Cys-91 did not abolish the ability of OTUB1 to deubiquitinate and stabilize DEPTOR. Interestingly, our data showed that overexpression of OTUB1 D88A or abrogation of endogenous OTUB1 by siRNA markedly impaired DEPTOR stability and mTORC1 activities in response to amino acid signals. It has been reported that the Asp-88 site of OTUB1 is critical for its binding to E2 and suppresses its Ub-conjugating activity in vitro (36). Thus, it appears that OTUB1 reduces cellular DEPTOR ubiquitination primarily via non-canonical inhibition of UbcH5 or other E2, although we cannot exclude the possibility that OTUB1 also directly inhibits βTrCP E3 activity. This observation is consistent with a non-canonical mechanism by which OTUB1 suppresses the chromatin ubiquitination induced by DNA damage (48).
By screening deubiquitinase enzymes of DEPTOR, we found that, in addition to OTUB1, OTUB2, OTUB5, UCHL1, and JOSD2 also can deubiquitinate DEPTOR, although they do not possess the same interaction with DEPTOR as OTUB1. Our data also excluded the possibility that OTUB2 and OTUD5 deubiquitinate DEPTOR via forming a heterodimer with OTUB1 (data not shown). Therefore, their functional roles in DEPTOR deubiquitination await further investigation. In summary, our study reveals a novel role of OTUB1 in regulation of DEPTOR stability and mTORC1 activities.
Experimental procedures
Cell culture and transfection
All cell lines were received from the Chinese Academy of Sciences Committee Type Culture Collection Cell Bank (Shanghai, China) and authenticated by the cell banks with short tandem repeat analysis. Both HeLa and HEK293T cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37 °C in the presence of 5% CO2. H1299 cells were cultured in RPMI 1640 medium with 10% heat-inactivated FBS. H1299 and HeLa cells were transfected with Lipofectamine 2000 following the manufacturer's protocol. HEK293T cells were transfected using a calcium phosphate-DNA coprecipitation method.
Plasmids and RNA interference (RNAi)
OTUB1 and its mutants were cloned into pCDNA3.1 vector with a FLAG or HA tag at its N terminus using standard cloning methods. HA-S6K was kindly provided by Dr. Kunliang Guan. His-Ub expression plasmids were constructed as described previously (9). siRNA oligonucleotides were transfected using Lipofectamine 2000. The sequences of siRNAs against OTUB1 were as follows: siRNA 1, 5′-CCGACUACCUUGUGGUCUA-3′; and siRNA 2, 5′-TGGATGACAGCAAGGAGTT-3′.
Reagents and antibodies
Anti-FLAG, anti-HA, and secondary antibodies were purchased from Sigma. The polyclonal anti-GFP antibody and mouse monoclonal anti-ubiquitin (P4D1,sc-8017) (P4D1, sc-8017) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The antibodies against OTUB1 (3783S), DEPTOR (11816S), pS6K (9234S/L), pS6 (4858S), mTOR (2983S), S6K (9202S), S6 (2217S), RagB (D18F3), and βTrCP1 (D13F10) were purchased from Cell Signaling Technology. MG132 was from Sigma, and Ni-NTA-agarose (30210) was from Qiagen. DMEM (amino acid-free) was purchased from Genetimes Technology, and amino acids (50×) were purchased from Gibco.
RNA isolation and real-time quantitative PCR
Total mRNA was extracted using TRIzol (Invitrogen), and 500 ng RNA was used to synthesize cDNA using the Prime ScriptTM RT reagent kit (Takara, DRR037A) according to the manufacturer's instructions.
Coimmunoprecipitation and Western blotting
Coimmunoprecipitation and Western blotting were performed as described previously (9). The cells were lysed in CHAPS lysis buffer (10 mm glycerophosphate, 0.3% CHAPS, 1 mm EDTA, 40 mm HEPES, pH 7.4, 120 mm NaCl plus a mixture of proteinase inhibitors). After sonication for 10 min, the soluble part of cell lysates was centrifuged at 12,000 rpm in a frozen microcentrifuge for 15 min. Then the cell lysates were centrifuged to discard the cell debris and incubated with HA or M2 beads (Abmart and Sigma) for 3 h. After washing three times with CHAPS buffer, the beads were boiled, resolved using SDS-PAGE, and analyzed by Western blotting. Results were measured and analyzed using an Odyssey system.
Deubiquitination assay
To perform the ubiquitination assay, moderate-density cells were cotransfected with His-Ub, and the transfected cells were lysed using metamorphic Buffer A (0.1 m Na2HPO4/NaH2PO4, 6 m guanidine HCl, 10 mm imidazole, pH 8.0). Then the ubiquitinated proteins were refined using Ni-NTA beads (Qiagen) for 4 h at room temperature. The beads were washed with Buffer B (0.01 m Tris-HCl, pH 8.0, 10 mm β-mercaptoethanol, 8 m urea, 0.1 m Na2HPO4/NaH2PO4, pH 8.0), Buffer C (8 m urea, 0.1 m Na2HPO4/NaH2PO4, pH 6.3, 10 mm β-mercaptoethanol, 0.01 m Tris-HCl, pH 6.3) with 0.2% Triton X-100, and Buffer C. After the above steps, beads were incubated with 30 μl of elution buffer (0.72 m β-mercaptoethanol, 30% glycerol, 200 mm imidazole, 0.15 m Tris-HCl, pH 6.7, 5% SDS) at room temperature for 30 min. Samples were analyzed by immunoblotting.
Amino acid starvation and stimulation
All cells were washed and incubated in amino acid-free RPMI 1640 medium for the appropriate time and then stimulated with amino acids (50×)(Gibco) for the indicated times. The final concentration of amino acids in the medium was the same as that in RPMI 1640 medium.
MTT assay and CHX chase assay
Cell proliferation was detected by MTT assay. H1299 cells were seeded into 96-well plates at a density of 700 cells/well. After 24, 48, 72, 96, and 120 h, 200 μl of MTT (5 mg/ml) was mixed into each well. The samples were incubated for 4 h at 37 °C and then subcultured in medium with 200 μl of dimethyl sulfoxide (DMSO). Three independent wells were analyzed at 490-nm wavelength using a spectrophotometer. For the CHX assay, CHX (50 μg/ml) was mixed into the cell culture medium, and the cells were harvested at 0–6 h. The transfected cells were lysed and analyzed by Western blotting.
Immunofluorescence
HeLa cells were seeded onto fibronectin-coated glass coverslips in 24-well plates. After transfection or other treatment, cells were deprived of amino acids for 1 h and then treated with amino acids for the indicated time periods. Afterward, the HeLa cells were washed three times with PBS and fixed for 30 min with 4% paraformaldehyde on ice. After fixing, cells were permeabilized with 0.1% Triton X-100 and washed three times with PBS with Triton X-100. The coverslips were blocked for 1 h with blocking buffer (0.3% BSA in PBS) at 4 °C and incubated with primary antibody in blocking buffer overnight at 4 °C with mixing, washed three times with PBS with Triton X-100. The secondary antibodies were incubated for 1 h at room temperature in the dark. The nucleus was stained with DAPI. The results were observed using a Zeiss LSM 510 Meta confocal system.
Autophagy analysis and cell size analysis
In the autophagy analysis assay, HeLa cells were transfected with GFP-LC3 and the plasmid as indicated in the figure. Those cells were deprived of amino acids for 3 h to induce the formation of GFP-LC3–containing puncta detected by immunofluorescence staining.
To detect cell size, HeLa cells with OTUB1 deficiency or HeLa cells were transfected with OTUB1 siRNAs. Cells were cultured in a 12-well plate until reaching moderate density and then were passaged into a 10-cm dish 24 h after transfection. Then the cells were cultured at 37 °C for 48 h. The medium was changed every day. Cells were harvested and subjected to FACS analysis to measure cell size (7).
Author contributions
L. Z., X. W., and P. W. conceived the project. L. Z., X. W., Y. Y., L. D., L. C., X. P., C. J., G. G., X. T., and W. P. performed experiments. L. Z., X. W., Y. Y., L. D., L. C., X. P., C. J., X. T., W. P., and P. W. analyzed the data. L. Z., X. W., L. D., X. G., and P. W. wrote the manuscript.
Acknowledgment
We thank Dr. Kunliang Guan for kindly providing reagents.
This work was supported by National Key Research and Development Program of China Grant 2016YFC0902102; National Natural Science Foundation of China Grants 81625019, 91440104, 91519322, and 81402417; Science Technology Commission of Shanghai Municipality Grant 16JC1404500; and Fundamental Research Funds for the Central Universities Grants 1501219106, 1500219121, and 1500219133. The authors declare that they have no conflicts of interest with the contents of this article.
- mTORC
- mechanistic target of rapamycin complex
- OTUB
- OTU domain–containing ubiquitin aldehyde-binding protein
- TFEB
- transcription factor EB
- FAT
- focal adhesion–targeting
- OTU
- ovarian tumor
- DUB
- deubiquitinating enzyme
- Ub
- ubiquitin
- pS6K
- phosphorylated S6K
- pS6
- phosphorylated S6
- CHX
- cycloheximide
- CRISPR
- clustered regularly interspaced short palindromic repeats
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- N+OTUB2
- OTUB1-(N-term)-OTUB2
- Ni-NTA
- nickel-nitrilotriacetic acid
- UCH
- UBQ C-terminal hydrolase
- USP
- ubiquitin-specific protease
- MJD
- Machado-Joseph disease protease.
References
- 1. Saxton R. A., and Sabatini D. M. (2017) mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sengupta S., Peterson T. R., and Sabatini D. M. (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 10.1016/j.molcel.2010.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dibble C. C., and Manning B. D. (2013) Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 15, 555–564 10.1038/ncb2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peterson T. R., Laplante M., Thoreen C. C., Sancak Y., Kang S. A., Kuehl W. M., Gray N. S., and Sabatini D. M. (2009) DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873–886 10.1016/j.cell.2009.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inoki K., Corradetti M. N., and Guan K. L. (2005) Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 37, 19–24 10.1038/ng1494 [DOI] [PubMed] [Google Scholar]
- 6. Dibble C. C., Elis W., Menon S., Qin W., Klekota J., Asara J. M., Finan P. M., Kwiatkowski D. J., Murphy L. O., and Manning B. D. (2012) TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 47, 535–546 10.1016/j.molcel.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inoki K., Zhu T., and Guan K. L. (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- 8. Li Y., Corradetti M. N., Inoki K., and Guan K. L. (2004) TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29, 32–38 10.1016/j.tibs.2003.11.007 [DOI] [PubMed] [Google Scholar]
- 9. Liu N., Li H., Li S., Shen M., Xiao N., Chen Y., Wang Y., Wang W., Wang R., Wang Q., Sun J., and Wang P. (2010) The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J. Biol. Chem. 285, 18858–18867 10.1074/jbc.M109.099440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sarbassov D. D., Guertin D. A., Ali S. M., and Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 10.1126/science.1106148 [DOI] [PubMed] [Google Scholar]
- 11. Laplante M., and Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Z., and Ming X. F. (2012) mTOR signalling: the molecular interface connecting metabolic stress, aging and cardiovascular diseases. Obes. Rev. 13, Suppl. 2, 58–68 10.1111/j.1467-789X.2012.01038.x [DOI] [PubMed] [Google Scholar]
- 13. Gao D., Inuzuka H., Tan M. K., Fukushima H., Locasale J. W., Liu P., Wan L., Zhai B., Chin Y. R., Shaik S., Lyssiotis C. A., Gygi S. P., Toker A., Cantley L. C., Asara J. M., et al. (2011) mTOR drives its own activation via SCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 44, 290–303 10.1016/j.molcel.2011.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duan S., Skaar J. R., Kuchay S., Toschi A., Kanarek N., Ben-Neriah Y., and Pagano M. (2011) mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol. Cell 44, 317–324 10.1016/j.molcel.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao Y., Xiong X., and Sun Y. (2011) DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell 44, 304–316 10.1016/j.molcel.2011.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 17. Rajalingam K., and Dikic I. (2016) SnapShot: expanding the ubiquitin code. Cell 164, 1074–1074.e1 10.1016/j.cell.2016.02.019 [DOI] [PubMed] [Google Scholar]
- 18. Schulman B. A., and Harper J. W. (2009) Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 10.1038/nrm2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., and Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- 20. Pickart C. M., and Fushman D. (2004) Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 10.1016/j.cbpa.2004.09.009 [DOI] [PubMed] [Google Scholar]
- 21. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 22. Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., and Goldberg A. L. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 10.1074/jbc.M609659200 [DOI] [PubMed] [Google Scholar]
- 23. Deshaies R. J., and Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 24. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., and Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 10.1016/j.cell.2005.11.007 [DOI] [PubMed] [Google Scholar]
- 25. Komander D., Clague M. J., and Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- 26. Ramanathan H. N., and Ye Y. (2012) Cellular strategies for making monoubiquitin signals. Crit. Rev. Biochem. Mol. Biol. 47, 17–28 10.3109/10409238.2011.620943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wing S. S. (2003) Deubiquitinating enzymes—the importance of driving in reverse along the ubiquitin-proteasome pathway. Int. J. Biochem. Cell Biol. 35, 590–605 10.1016/S1357-2725(02)00392-8 [DOI] [PubMed] [Google Scholar]
- 28. Kim J. H., Park K. C., Chung S. S., Bang O., and Chung C. H. (2003) Deubiquitinating enzymes as cellular regulators. J. Biochem. 134, 9–18 10.1093/jb/mvg107 [DOI] [PubMed] [Google Scholar]
- 29. Ventii K. H., and Wilkinson K. D. (2008) Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161–175 10.1042/BJ20080798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiener R., DiBello A. T., Lombardi P. M., Guzzo C. M., Zhang X., Matunis M. J., and Wolberger C. (2013) E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 20, 1033–1039 10.1038/nsmb.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soares L., Seroogy C., Skrenta H., Anandasabapathy N., Lovelace P., Chung C. D., Engleman E., and Fathman C. G. (2004) Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat. Immunol. 5, 45–54 10.1038/ni1017 [DOI] [PubMed] [Google Scholar]
- 32. Herhaus L., Al-Salihi M., Macartney T., Weidlich S., and Sapkota G. P. (2013) OTUB1 enhances TGFβ signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat. Commun. 4, 2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goncharov T., Niessen K., de Almagro M. C., Izrael-Tomasevic A., Fedorova A. V., Varfolomeev E., Arnott D., Deshayes K., Kirkpatrick D. S., and Vucic D. (2013) OTUB1 modulates c-IAP1 stability to regulate signalling pathways. EMBO J. 32, 1103–1114 10.1038/emboj.2013.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li S., Zheng H., Mao A. P., Zhong B., Li Y., Liu Y., Gao Y., Ran Y., Tien P., and Shu H. B. (2010) Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 285, 4291–4297 10.1074/jbc.M109.074971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Millour J., de Olano N., Horimoto Y., Monteiro L. J., Langer J. K., Aligue R., Hajji N., and Lam E. W. (2011) ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol. Cancer Ther. 10, 1046–1058 10.1158/1535-7163.MCT-11-0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun X. X., Challagundla K. B., and Dai M. S. (2012) Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 31, 576–592 10.1038/emboj.2011.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Millour J., Constantinidou D., Stavropoulou A. V., Wilson M. S., Myatt S. S., Kwok J. M., Sivanandan K., Coombes R. C., Medema R. H., Hartman J., Lykkesfeldt A. E., and Lam E. W. (2010) FOXM1 is a transcriptional target of ERα and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene 29, 2983–2995 10.1038/onc.2010.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stanisić V., Malovannaya A., Qin J., Lonard D. M., and O'Malley B. W. (2009) OTU Domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) α and affects ERα transcriptional activity. J. Biol. Chem. 284, 16135–16145 10.1074/jbc.M109.007484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Zhou X., Xu M., Weng W., Zhang Q., Yang Y., Wei P., and Du X. (2016) OTUB1-catalyzed deubiquitination of FOXM1 facilitates tumor progression and predicts a poor prognosis in ovarian cancer. Oncotarget 7, 36681–36697 10.18632/oncotarget.9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xue J., Lin X., Chiu W. T., Chen Y. H., Yu G., Liu M., Feng X. H., Sawaya R., Medema R. H., Hung M. C., and Huang S. (2014) Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer metastasis. J. Clin. Investig. 124, 564–579 10.1172/JCI71104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Iglesias-Gato D., Chuan Y. C., Jiang N., Svensson C., Bao J., Paul I., Egevad L., Kessler B. M., Wikström P., Niu Y., and Flores-Morales A. (2015) OTUB1 de-ubiquitinating enzyme promotes prostate cancer cell invasion in vitro and tumorigenesis in vivo. Mol. Cancer 14, 8 10.1186/s12943-014-0280-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou Y., Wu J., Fu X., Du W., Zhou L., Meng X., Yu H., Lin J., Ye W., Liu J., Peng H., Liu R. Y., Pan C., and Huang W. (2014) OTUB1 promotes metastasis and serves as a marker of poor prognosis in colorectal cancer. Mol. Cancer 13, 258 10.1186/1476-4598-13-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuan L., Yuan P., Yuan H., Wang Z., Run Z., Chen G., Zhao P., and Xu B. (2017) miR-542-3p inhibits colorectal cancer cell proliferation, migration and invasion by targeting OTUB1. Am. J. Cancer Res. 7, 159–172 [PMC free article] [PubMed] [Google Scholar]
- 44. Jin G., Lee S. W., Zhang X., Cai Z., Gao Y., Chou P. C., Rezaeian A. H., Han F., Wang C. Y., Yao J. C., Gong Z., Chan C. H., Huang C. Y., Tsai F. J., Tsai C. H., et al. (2015) Skp2-mediated RagA ubiquitination elicits a negative feedback to prevent amino-acid-dependent mTORC1 hyperactivation by recruiting GATOR1. Mol. Cell 58, 989–1000 10.1016/j.molcel.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang T., Yin L., Cooper E. M., Lai M. Y., Dickey S., Pickart C. M., Fushman D., Wilkinson K. D., Cohen R. E., and Wolberger C. (2009) Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J. Mol. Biol. 386, 1011–1023 10.1016/j.jmb.2008.12.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deng L., Jiang C., Chen L., Jin J., Wei J., Zhao L., Chen M., Pan W., Xu Y., Chu H., Wang X., Ge X., Li D., Liao L., Liu M., et al. (2015) The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol. Cell 58, 804–818 10.1016/j.molcel.2015.03.033 [DOI] [PubMed] [Google Scholar]
- 47. Song W. J., Shi X., Zhang J., Chen L., Fu S. X., and Ding Y. L. (2018) Akt-mTOR signaling mediates abnormalities in the proliferation and apoptosis of ovarian granulosa cells in patients with polycystic ovary syndrome. Gynecol. Obstet. Invest. 10.1159/000464351 10.1159/000464351 [DOI] [PubMed] [Google Scholar]
- 48. Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y. C., O'Donnell L., Kumakubo A., Munro M., Sicheri F., Gingras A. C., Natsume T., Suda T., and Durocher D. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 10.1038/nature09297 [DOI] [PubMed] [Google Scholar]
- 49. Chen L., Liu T., Tu Y., Rong D., and Cao Y. (2016) Cul1 promotes melanoma cell proliferation by promoting DEPTOR degradation and enhancing cap-dependent translation. Oncol. Rep. 35, 1049–1056 10.3892/or.2015.4442 [DOI] [PubMed] [Google Scholar]