Abstract
Heterotrimeric G protein complexes are molecular switches relaying extracellular signals sensed by G protein–coupled receptors (GPCRs) to downstream targets in the cytoplasm, which effect cellular responses. In the plant heterotrimeric GTPase cycle, GTP hydrolysis, rather than nucleotide exchange, is the rate-limiting reaction and is accelerated by a receptor-like regulator of G signaling (RGS) protein. We hypothesized that posttranslational modification of the Gα subunit in the G protein complex regulates the RGS-dependent GTPase cycle. Our structural analyses identified an invariant phosphorylated tyrosine residue (Tyr166 in the Arabidopsis Gα subunit AtGPA1) located in the intramolecular domain interface where nucleotide binding and hydrolysis occur. We also identified a receptor-like kinase that phosphorylates AtGPA1 in a Tyr166-dependent manner. Discrete molecular dynamics simulations predicted that phosphorylated Tyr166 forms a salt bridge in this interface and potentially affects the RGS protein–accelerated GTPase cycle. Using a Tyr166 phosphomimetic substitution, we found that the cognate RGS protein binds more tightly to the GDP-bound Gα substrate, consequently reducing its ability to accelerate GTPase activity. In conclusion, we propose that phosphorylation of Tyr166 in AtGPA1 changes the binding pattern with AtRGS1 and thereby attenuates the steady-state rate of the GTPase cycle. We coin this newly identified mechanism “substrate phosphoswitching.”
Keywords: heterotrimeric G protein, regulator of G protein signaling (RGS), GTPase, GTPase-activating protein (GAP), phosphotyrosine signaling, guanine-nucleotide exchange factor, protein phosphorylation, substrate phosphoswitching, GTP hydrolysis, G protein-coupled receptor (GPCR)
Introduction
The heterotrimeric G protein complex functions as a molecular switch that couples extracellular signals sensed by cell-surface G protein–coupled receptors (GPCRs)2 to downstream targets in the cytoplasm that affect cellular behavior. The G protein complex contains Gα, Gβ, and Gγ subunits; the Gα subunit binds guanine nucleotides. When Gα binds GTP, the complex becomes active by separating into a conformationally different Gα subunit and a released Gβγ dimer, each able to initiate downstream signaling through interaction with cellular targets called effectors. After the bound GTP is hydrolyzed to GDP by the intrinsic GTPase activity of Gα, the heterotrimeric complex reforms its inactive GDP-bound complex (resting state). This process is termed the GTPase cycle. The GTPase cycle is modulated by three known classes of proteins: GTPase-accelerating proteins (GAPs; e.g. regulator of G signaling (RGS) proteins (1, 2)), guanine-nucleotide exchange factors (GEFs; e.g. GPCRs), and the guanine nucleotide dissociation inhibitor (GDI). In animals, but not plants and protists, nucleotide exchange is the rate-limiting step of the GTPase cycle (1).
Unlike Gα homologs in metazoans and yeast, plant and protist Gα subunits spontaneously exchange guanine nucleotide; therefore, they self-activate without the need of a GPCR or other GEF (3, 4). Consequently, the rate-limiting step is the GTP hydrolysis reaction. In the genetic model Arabidopsis thaliana, acceleration of GTP hydrolysis by the canonical Gα subunit, AtGPA1, is catalyzed by the prototype seven-transmembrane RGS protein, AtRGS1 (4–7). In the current model, AtRGS1 maintains G protein in the resting state by its GAP activity (8, 9). Although sustained activation does occur through decoupling AtRGS1 from AtGPA1 by phosphorylation-dependent AtRGS1 endocytosis (10, 11), this cannot be the main mechanism because AtRGS1 endocytosis itself requires prior G protein activation (10). Therefore, this leaves three possible mechanisms by which AtRGS1 keeps the GTPase cycle in the resting state: 1) accelerated hydrolysis at a rate faster than guanine-nucleotide exchange alone shifts AtGPA1GDP to the predominant state in the GTPase cycle; 2) the GTPase cycle lingers or stops at the AtGPA1GDP state; or 3) a combination of both hydrolysis and cycling control. In animals, phosphorylation of the Gα subunit affects its activity. Tyrosine phosphorylation by pp60c-src or epidermal growth factor receptor increases Gαs GTPγS binding and GTPase activity (12–14). Serine phosphorylation by p21-activated protein kinase (PAK)-1 or PKC decreases the affinity of Gαz with its Gβγ subunit and confers Gαz resistance to GAP proteins (15–17). Although none of these reported phosphorylation sites are conserved in plant Gα subunits, such as AtGPA1, reversible phosphorylation as a regulatory mechanism is not excluded. This prospect is explored here.
Based on acceptor specificity, plant kinases are classified into serine/threonine and serine/tyrosine dual-specificity kinases. Despite the Arabidopsis genome lacking canonical tyrosine kinases (18), there are: 1) canonical phosphotyrosine phosphatases (19–21), 2) phosphotyrosine PTMs (22–24), and 3) Ser/Thr receptor kinases that have dual specificity to tyrosine (25, 26). Specifically, some members of the leucine-rich repeat receptor-like kinase (LRR RLK) family phosphorylate tyrosine residues (27). The Arabidopsis genome encodes at least 223 LRR RLKs (28), and several of these LRR RLKs participate in the regulation of the same plant growth, defense, and stress pathways (29–34) as do the G protein components (35, 36). Among the LRR RLK family members, BRI1 and BRI1-associated receptor kinase 1 (BAK1) have dual specificity, which initiates brassinosteroid signaling by auto- and trans-phosphorylation (34). BAK1 also phosphorylates AtRGS1 and regulates G protein signaling in response to stimulation by the pathogen-associated molecular pattern flagellin peptide 22 (flg22) (10).
AtGPA1 phosphorylation at Tyr166 is induced by several plant hormones (37). We show that the corresponding Tyr166 phosphorylation mimetic alters how AtRGS1 acts upon its substrate AtGPA1. Tyr166 phosphorylation significantly slows the rate of the GTPase cycle. We coined this newly described substrate–enzyme relationship “substrate phosphoswitching.”
Results
SAPH-ire predicts that AtGPA1 Tyr166 is an important phosphorylation site in one of three major clusters in the Gα protein family
SAPH-ire (structural analysis of PTM hot spots) is a quantitative informatics method to prioritize the function potential of co-aligning PTMs based on protein sequence and structural features. In brief, multiple sequence and structural features, including PTM frequency across all eukaryotes, PTM residue conservation, protein interface residence, solvent-accessible surface area, and nearest neighbor features, are integrated through a neural network model trained on modified alignment positions (MAPs) with known biological function. As a result, each MAP receives a probability score that can be used to rank-order the MAP from low to high function potential (low to high probability score, respectively). Previous results showed that SAPH-ire is highly effective for identification of functional PTM hot spots and was validated both computationally and empirically (38, 39).
Nearly 160 PTMs, comprising 70 clusters designated MAPs, were experimentally observed in the protein family (Fig. S1) represented specifically by Gα subunits (IPR001019), the vast majority of which are due to phosphorylation (38, 39). Tyr166 of AtGPA1 occurs within MAP IPR001019-1010, which exhibits several features consistent with functional PTM hot spots, including multiple observations of phosphorylation, moderate solvent accessibility, a high degree of sequence identity between family members, and the presence of neighboring MAPs (38). Consequently, SAPH-ire ranks the function potential of IPR001019–1010 as the 22nd out of 70 MAPs (approximately the 30th percentile) in the Gα protein family, above which ∼60% of all MAPs with known regulatory function are ranked (Fig. 1A).
Figure 1.
SAPH-ire analyses of AtGPA1 Tyr166. A, log plot of experimentally observed PTM hot spots in the Gα protein family IPR001019. PTM hot spots generated by SAPH-ire were plotted in rank order with respect to calculated function potential probability score (SAPH-ire NN Score). Hot spots known to be involved in regulating protein function were color- and size-coded by the number of literature sources (known function source count, KFSC) that provide evidence of the biological function. Hot spot IPR001019-1010, containing AtGPA1 pTyr166 was colored green for ease of viewing. B, PTM hot spot cluster density from N to C terminus of the Gα protein family (Alignment Position (AP)). The native residue position of AtGPA1 (a member of the Gα family) is shown within the nested x axis for clarity (Native Position (NP)). Circle size is proportional to the number of PTMs found within the ±2 residue cluster centered on each hot spot. Circle color indicates whether the hot spot has a known function (red), is known by proximity to a hot spot with known function (yellow), or is uncharacterized (gray).
Surveying from N to C terminus, the density of MAPs along the length of a protein family reveals regions of local structure that exceed the average density of modification for the family, a characteristic that is common for functional MAPs (38). Therefore, we compared the MAP cluster density for every MAP between the N and C terminus of the Gα family, reporting the number of total MAPs contained within ±2 residues of each modified position (Fig. 1B). We observed three distinct MAP clusters in the family: the base of the N-terminal α-helix (αN) that interfaces with receptors (alignment position (AP)682–AP685), the P-loop that is a well-conserved structure that is necessary for nucleotide/phosphate coordination (AP761–AP771), and the αE helix that exhibits an uncharacterized phosphorylation cluster that includes AtGPA1 Tyr166 (AP1009–AP1019) (Fig. 1B). In addition to Tyr166, the αE helix is phosphorylated in several different positions across mouse and human Gα subunits, including mGαS2 (pTyr177), mGα14 (pTyr156), hGα11 (pThr162), and mGαo (pSer159) (40–43).
The functional relevance of phosphorylation sites in the αE helix has not yet been determined. MAP IPR001019-1010 exhibits 100% identity across all family members, indicating that substitution at this site is evolutionarily constrained (Fig. 2A, middle). In addition to AtGPA1 Tyr166, phosphorylation of mouse Gαs2 Tyr177 (equivalent to Arabidopsis Tyr167) was also observed experimentally in more than 10 independent high-throughput experiments across humans, mice, and rats (43). Despite having high sequence conservation, high PTM observation frequency, or measured occurrence in diverse organisms, neither Tyr166 nor homologous sites in mammalian Gα subunits were functionally characterized at the onset of this study. Given that Tyr166 is remote from switch II and the Gβ subunit binding interface (Fig. 2B, left), we presumed that it is unlikely that its phosphorylation directly affects AtGPA1 binding to the Gβγ subunit. Rather, Tyr166 is located in the intramolecular domain interface for GTP hydrolysis (Fig. 2B (right) and Fig. 3A), suggesting that phosphorylation at this site may have an effect on GTPase activity, either intrinsic or accelerated by AtRGS1. Indeed, the corresponding residue of AtGPA1 Tyr166, bovine Gαt Tyr150, participates in guanine ring binding (44). Although such a structural important residue might belong to a rigid local structure, nonetheless phosphorylation occurs at or near this positon in the αE helix in other G subunits (40–43), indicating that the αE helix is a prominent, yet uncharacterized, phosphorylation hot spot occurring in plants and animals. We hypothesized that phosphorylation of Tyr166 within this region plays a regulatory role in the function of the Gα subunit.
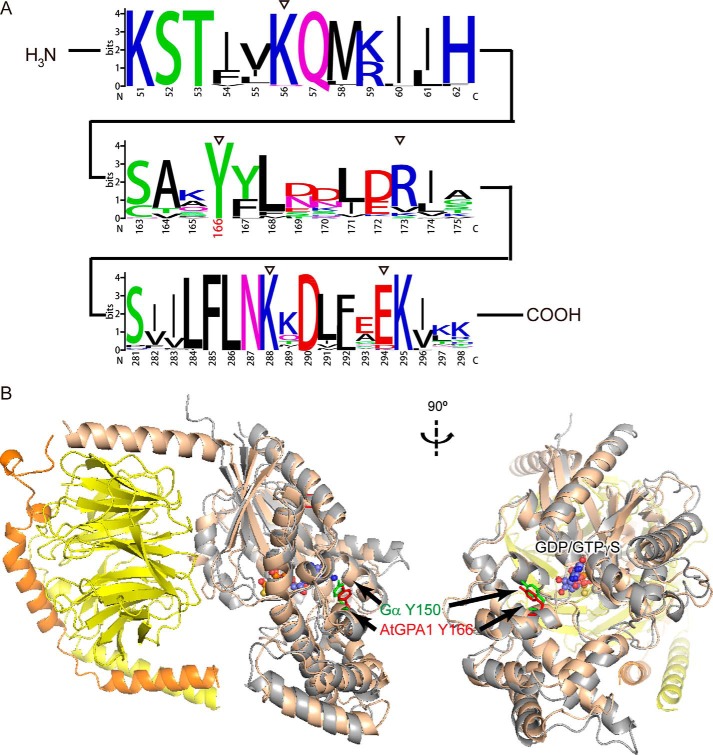
Figure 2.
Conservation analysis of AtGPA1 Tyr166. A, conservation of α1 helix (top, amino acid residues 51–62), αE helix (middle, amino acid residues 163–175), and β5 brand + αG helix (bottom, amino acid residues 281–298) were aligned and calculated by the online WebLogo program (73). The x axis represents the corresponding position on AtGPA1, and the y axis represents the bit score. Empty triangles denote the amino acid residues Lys56, Tyr166, Lys173, Lys288, and Lys294, respectively. B, crystal structure of AtGPA1 (gray, PDB code 2XTZ (3)) aligned with a heterotrimeric G protein (tan for Bos hybrid Gα subunit, yellow for Gβ, and orange for Gγ; PDB code 1GOT (74)). Arrows point to Tyr166 on AtGPA1 and its corresponding residue Tyr150 on Bos hybrid Gα subunit.
Figure 3.
DMD simulations for unphosphorylated, phosphorylated, and phosphomimetic AtGPA1. A, positions of Lys56, Tyr166, Arg173, Lys288, and Lys294 around the intramolecular domain interface are shown. B–Q, distance histograms between the phenolic oxygen of unphosphorylated (B–E), phosphorylated Tyr166 (F–I), phosphomimetic Y166E (J–M), or phosphomimetic Y166D (N–Q) and the amino nitrogen or guanidinium carbon of nearby positive charge residues (Lys56, Arg173, Lys288, and Lys294). Peaks in the histograms in the range indicating salt bridge formation (<6 Å) are highlighted (*).
Discrete molecular dynamics (DMD) simulations predict pTyr166 salt bridging at the intramolecular domain interface
Phosphorylation changes protein structure and consequently affects protein stability or catalytic activity. We hypothesized that phosphorylation of Tyr166 changes AtGPA1 conformation. The local effect of phosphorylation is to change a neutral amino acid to a negatively charged amino acid. We expected that the major effects of phosphorylation would be evident in the formation of new salt bridges. To test our hypothesis in silico, we performed DMD simulations of phosphorylated and nonphosphorylated AtGPA1. We computed distance distributions between the phenolic oxygen of Tyr166 and positively charged groups in nearby amino acids (Fig. 3A) and compared the results from simulations of phosphorylated and unphosphorylated Tyr166, using a distance of ≤6 Å as a metric for salt bridge formation. Positive charges at residue positions 56 and 288 (AtGPA1 numbering) are 100% conserved across eukaryotic G subunits, mostly conserved at position 173 and poorly conserved at position 294, although a lysine at position 295 is 100% conserved (Fig. 2A). Predicted salt bridges formed between Arg173 and pTyr166 and between Arg173 and the phosphomimetic mutation Tyr166 → Glu (Fig. 3, G and K), but not unphosphorylated Tyr166 or the Tyr166 → Asp mutant (Fig. 3, C and O). The small population of short distances between Tyr166 and Arg173 (Fig. 3C) may be the result of hydrogen bonding between Tyr166 and Arg173; however, the effect was noticeably less pronounced than for either pTyr166 or Y166E. Both phosphorylated and unphosphorylated Tyr166 interact with Lys288 (Fig. 3, D and H). However, whereas the Y166E mutation promoted interaction with Lys288, the Y166D mutation disrupted interaction with Lys288 (Fig. 3, L and P). Moreover, Lys288 forms a hydrophobic interaction with the nucleotide, and therefore its rotation away to form the salt bridge would be expected to alter the intrinsic exchange rate (45, 46). As will be shown later, a phosphomimetic did not alter the intrinsic exchange rate; therefore, this potential pTyr166–Lys288 salt bridge is given low probability. A salt bridge formed between Lys294 and pTyr166 or Y166E (Fig. 3, I and M), but not unphosphorylated Tyr166 or Y166D (Fig. 3, E and Q). Finally, a salt bridge formed between Lys56 and pTyr166 (Fig. 3F) but not between Lys56 and unphosphorylated Tyr166, Y166E, or the Y166D mutant (Fig. 3, B, J, and N). On the basis of these simulation results, we predict that AtGPA1 Y166E mimics pTyr166, whereas the Y166D substitution is insufficient to form salt bridges. All implicated residues except Lys294 are conserved (Fig. 2A), emphasizing the importance of the potential function elicited by formation of these salt bridges. This predicted distinction between the two mimetic forms is tested below.
Amino acid residues that potentially form salt bridges with pTyr166 face toward the interface between the helical and Ras domains (Fig. 3A), prompting the hypothesis that phosphorylation changes the local but not the global conformation of AtGPA1. To test this hypothesis, CD spectroscopy was employed to measure the secondary structure formation (Fig. 4A). The two minimum peaks appeared near 208 and 222 nm, indicating that all of the mutant proteins contained the characteristic wildtype helical structure. The mutations also showed thermal stability comparable with that of wildtype AtGPA1, excluding aggregation caused by mutagenesis (Fig. 4B). Taken together, we conclude that the Y166E mutation conserved the global structure of AtGPA1. GTP binding and hydrolysis rates that are at wildtype levels, which are described below, further support the likelihood that the point mutation does not affect global structure.
Figure 4.
AtGPA1 mutants retain global wildtype structure. A, CD spectra of His-tagged AtGPA1 wildtype (red) and its mutations Y166D (blue) and Y166E (green) on far-UV spectra (185–260 nm) in 0.5-nm scan steps at 20 °C. The protein solutions were all present at 100 nm, and the cell path length was 0.5 cm. The spectra results were analyzed with Chirascan software. B, fast quantitative cysteine reactivity unfolding curves for His-tagged AtGPA1 wildtype (red circle) and its mutations Y166D (blue square) and Y166E (green triangle).
Identification of 18 AtGPA1 kinases, one of which requires Tyr166 for AtGPA1 phosphorylation
To identify kinase(s) that phosphorylate a tyrosine residue on AtGPA1, we purified 70 recombinant LRR RLKs, containing the intracellular juxtamembrane, the catalytic kinase, and the C-terminal domains (47). We biochemically screened these kinases for AtGPA1 substrate specificity (Fig. 5A and Fig. S2). No signal was found in [γ-32P]ATP-containing buffer (lane 1) or the “GST only” reaction (lane 2), indicating that no contaminating kinases were present. Not all of the LRR RLKs showed autophosphorylation, suggesting that 1) some plant RLKs do not autophosphorylate, 2) the conditions for the in vitro kinase assay were not optimal for every kinase to autophosphorylate, or 3) some LRR RLKs lost phosphorylation activity during purification. Among the 70 kinase domains, 18 transphosphorylated AtGPA1 under the conditions tested, and not all of these kinases displayed autophosphorylation (Table S1). As shown in Fig. 5B, these candidates were predicted to be involved in many biological processes and functional categorization (48), including protein metabolism; developmental processes; response to stress, signal transduction, and abiotic or biotic stimulus; cell organization; and biogenesis. The DAVID bioinformatics resource (49, 50) was used to analyze pathways shared by these LRR RLKs. Among them, LRR RLK family protein (PSY1R, AT1G72300, lane 19), LRR RLK family protein (AT5G37450, lane 66), LLR transmembrane protein kinase (GSO1, AT4G20140, lane 31), and PEP1 receptor 1 (PEPR1, AT1G73080, lane 20) are involved in protein-tyrosine kinase signaling pathways; however, none of the four were previously reported to have tyrosine-protein kinase activity either in vivo or in vitro; BAK1 (AT4G33430, lane 12), LRR RLK family protein (BRI1, AT4G39400, lane 35), and somatic embryogenesis receptor-like kinase 1 (SERK1, AT1G71830, lane 18) are involved in the brassinosteroid-mediated signaling pathway and are known dual-specificity kinases (27, 51, 52).
Figure 5.
Phosphorylation of AtGPA1 by leucine-rich repeat receptor-like kinases. A, in vitro kinase assays were performed to screen kinases for AtGPA1. Naive reactions ([γ-32P]ATP-containing buffer only (lane 1) and GST (lane 2)) were performed as negative controls. Seventy purified LRR RLKs (47) (the migration range on SDS-PAGE is denoted by the vertical line, lanes 3–72) were incubated with GST-AtGPA1 (∼70 kDa, empty triangle) in kinase assay buffer containing [γ-32P]ATP as described under “Experimental procedures.” Purified RGS + Ct (∼28 kDa, solid triangle) was employed as a positive control for the kinase activity of LRR RLKs. Apparent molecular masses were indicated on the left in kDa. The TAIR locus number of each LRR RLK is indicated at the top of the corresponding lane, and the lane number is at the bottom. Autophosphorylation and transphosphorylation were detected by autoradiography. See Table S1. B, functional categorization by annotation for AtGPA1 kinase candidates based on gene ontology biological process. The number indicates the percentage of annotations to terms in each gene ontology slim category with regard to the total annotations to terms in ontology. C, detection of phosphotyrosine residues on wildtype AtGPA1. Triangles denote the position of GST-AtGPA1, circles denote the position of His-BAK1, and squares denote the position of GST. Apparent molecular masses (kDa) are indicated on the right. Tyrosine phosphorylation on AtGPA1 or BAK1 was detected by a phosphotyrosine-specific antibody. Polyhistidine-tagged BAK1 was detected by His tag antibody. Total GST-GPA1 and GST were detected by GST tag antibody. D, the phosphorylation of AtGPA1 wildtype, Y166D, and Y166E was detected by phosphotyrosine-specific antibody. Total GST-GPA1 was detected by GST tag antibody. The intensity of each band was quantified with ImageJ. The ratio of phosphorylated/total AtGPA1 was calibrated to wildtype. The quantitative results were expressed as the means ± S.D. (error bars) of three experiments. Statistical significance was determined by an analysis of variance (ANOVA). **, differences with p values of <0.01. IB, immunoblotting.
We chose BAK1 (AT4G33430, lane 12) to characterize not only because it is a known dual-specificity kinase (27, 53), but also because it operates in G protein–coupled signaling (10, 54, 55). The stoichiometry of BAK1 phosphorylation of AtGPA1 (i.e. serine, threonine, and tyrosine) was 5.5 mol of Pi to 1.0 mol of AtGPA1, as described under “Experimental procedures,” indicating multiple phosphorylation sites on AtGPA1. The phosphotyrosine specificity of BAK1 on AtGPA1 was further confirmed using a phosphotyrosine antibody. BAK1 phosphorylated GST-tagged AtGPA1 (Fig. 5C, open triangle). There was no band at the corresponding position of the lanes with either GST-GPA1 only or BAK1 only, despite detectable BAK1 autophosphorylation (Fig. 5C, open circle), indicating an active BAK1 kinase in the reaction. The total amount of BAK1 was determined immunochemically using an antibody to the BAK1 polyhistidine tag (Fig. 5C, solid circle). GST and GST-AtGPA1 were detected by anti-GST tag antibody (Fig. 5C, solid square and solid triangle).
A requirement for Tyr166 for BAK1-mediated phosphorylation was confirmed by site-directed mutagenesis in combination with immunodetection using a phosphotyrosine-specific antibody. As shown in Fig. 5D, Tyr166 → Asp/Glu mutagenesis reduced phosphorylation 2-fold, confirming that Tyr166 is required for BAK1 phosphorylation of AtGPA1. SAPH-ire indicated that phosphorylation at Tyr166 has the largest functional potential. Nonetheless, we did not detect a pTyr166 tryptic peptide by tandem MS. This indicates that either 1) Tyr166 is not directly phosphorylated by BAK1 or 2) the pTyr166 tryptic peptide is not detected by tandem mass spectrometry. Because TiO2-enriched tryptic peptides containing pTyr166 have been repeatedly detected (37), the latter is less plausible, leaving the possibility that BAK1 does not directly phosphorylate Tyr166. Because we have shown that Tyr166 is necessary for some BAK1 phosphorylation of AtGPA1 (Fig. 5D), we do not rule out the possibility that pTyr166 is required for other BAK1 phosphorylation sites.
The AtGPA1 phosphorylation mimetic mutation affects the RGS-dependent GTPase cycle
Based on the predicted importance of pTyr166, Tyr166 → Glu mutagenesis was used as a phosphorylation mimetic protein in further experiments, whereas Tyr166 → Asp mutagenesis was used as a negative control for reasons described above (Fig. 3). Nonetheless, we do not exclude the possibility that other phosphorylated AtGPA1 residues play functional roles, although the lack of conservation lowers the priority for investigation at this time.
The intrinsic single turnover and steady-state rates of GTP hydrolysis were unaffected by the Tyr166 → Asp/Glu mutations (Fig. 6, A and C, respectively). GTP hydrolysis accelerated by AtRGS1 was slightly reduced by the Tyr166 → Glu mutation (Fig. 6B). At a low concentration of AtRGS1 (∼1:1 ratio AtRGS1/AtGPA1), GTP hydrolysis was about half for the phosphomimetic mutant compared with wildtype. At a high concentration of AtRGS1, the reaction approached saturation.
Figure 6.
AtGPA1 Y166E impairs AtRGS1-accelerated GTPase cycle. The GTPase hydrolysis rate was measured by a single-turnover GTP hydrolysis assay in the absence (A) or presence (B) of AtRGS1. Wildtype AtGPA1 or equivalent mutants were incubated with [γ-32P]GTP at 20 °C in the presence of GTPγS. The reactions were quenched at the indicated time points (A) or at 10 min (B), and the amount of 32PO4 in solution was measured by scintillation counting. The GTPase cycle was measured by a steady-state GTP hydrolysis assay in the absence (C) or presence of AtRGS1 (D). Wildtype AtGPA1 or equivalent mutants were incubated with [γ-32P]GTP at 20 °C. The reactions were quenched at the indicated time points (C) or at 10 min (D), and the amount of 32PO4 in solution was measured by scintillation counting. E, GTPγS-binding ability was measured by a GTPγS-binding assay in the absence or presence of 1 μm AtRGS1. Wildtype AtGPA1 or equivalent mutants were incubated with [γ-35S]GTPγS on ice. The reactions were stopped at the indicated time points. The amount of AtGPA1-bound [γ-35S]GTPγS was filtered on nitrocellulose membrane and measured by scintillation counting. The quantitative results were expressed as the means ± S.D. (error bars) of at least three experiments and fitted to exponential one-phase association functions (A and E), linear regression (C), or log (agonist) versus response (D) using GraphPad Prism version 5.0. Statistical significance was determined by ANOVA. ***, differences with p values <0.001. Supporting material for D is shown in Table 1.
Most informative is the effect of AtRGS1 on GTP hydrolysis at steady state. The AtRGS1-accelerated steady-state GTP hydrolysis was reduced in the Tyr166 → Glu mutant (Fig. 6D and Table 1). As predicted (see Fig. 3), the Tyr166 → D mutant behaved like wildtype AtGPA1 (Fig. 6D).
Table 1.
Reaction rate for steady-state GTPase assay in the presence of AtRGS1
The reaction rates for the steady-state GTPase assay in the presence of AtRGS1 were calculated based on the results in Fig. 6D. The quantitative results were fitted to linear regression using GraphPad Prism version 5.0.
| AtRGS1 | AtGPA1 wildtype | AtGPA1 Y166D | AtGPA1 Y166E |
|---|---|---|---|
| nm | min−1 | min−1 | min−1 |
| 0 | 0.034 ± 0.004 | 0.041 ± 0.009 | 0.027 ± 0.004 |
| 30 | 0.046 ± 0.005 | 0.051 ± 0.009 | 0.041 ± 0.008 |
| 100 | 0.069 ± 0.007 | 0.101 ± 0.012 | 0.035 ± 0.005 |
| 300 | 0.086 ± 0.01 | 0.146 ± 0.017 | 0.051 ± 0.006 |
| 1000 | 0.19 ± 0.02 | 0.222 ± 0.02 | 0.078 ± 0.01 |
| 3000 | 0.36 ± 0.007 | 0.396 ± 0.05 | 0.149 ± 0.02 |
To understand the basis for this difference in steady-state cycling, we examined the nucleotide exchange rate using a GTPγS-binding assay. In the absence of AtRGS1, GTPγS binding by the mutants was statistically indistinguishable from that of wildtype (Fig. 6E). AtRGS1 (1 μm) slightly reduced the GTPγS binding rate nearly equally by both the wildtype and mutant (Fig. 6E). Thus, the strong reduction in AtRGS1-accelerated GTPase cycle by the phosphomimetic mutation was not caused by an altered nucleotide exchange rate.
Substrate phosphoswitching
As a modulator, AtRGS1 acts as a GAP to accelerate GTP hydrolysis by AtGPA1; therefore, it reduces the free energy of the transition state for the GTPase reaction (4, 6). RGS proteins selectively bind Gα subunits in their transition state conformation, which is mimicked by Gα·GDP aluminum tetrafluoride (Gα-GDP AlF4−) (56). As shown previously (e.g. see Refs. 6 and 57), AtRGS1 preferentially binds the transition state of the wildtype AtGPA1 (Fig. 7A, lane 3 versus lanes 1 and 2). However, AtRGS1 strongly bound both the transition state and GDP-bound form of the phosphomimetic Y166E mutant AtGPA1 (Fig. 7, A (lanes 4 and 6) and B).
Figure 7.
AtGPA1 Y166E changes its binding specificity with AtRGS1. A, direct interaction between GST-tagged AtRGS1 (RGS + Ct) and His-tagged AtGPA1 wildtype or Y166E mutant in the presence of GDP, GTPγS, or GDP-AlF4− was detected by an in vitro pulldown assay. Protein complexes were purified by glutathione-Sepharose, separated by SDS-PAGE, and detected by anti-GST or anti-His antibodies. B, the intensities of His-AtGPA1 wildtype and Y166E mutant were quantitated with ImageJ and normalized as relative values to each interaction in the presence of GDP-AlF4−. The quantitative results were expressed as the means ± S.D. (error bars) of four experiments. Statistical significance was determined by ANOVA. *, differences with p values of <0.05. **, differences with p values of <0.01. The binding affinity constants of AtRGS1 and AtGPA1 wildtype or Y166E mutant were measured by surface plasmon resonance with a ProteOn XPR36 instrument. The GST-tagged AtRGS1 (RGS + Ct) was immobilized on the surface of a GLC sensor chip as ligand, and the His-tagged AtGPA1 wildtype or Y166E in the GDP-bound, GTPγS-bound, or GDP-AlF4−-bound state were diluted into dosage concentrations as analyte. The affinity constants were calculated by kinetic analysis (C–H) or equilibrium analysis (I-K) and are shown in Table 2. IP, immunoprecipitation; IB, immunoblotting.
To confirm and quantitate these different binding affinities by an independent method, we used surface plasmon resonance with AtRGS1 immobilized on the chip and the wildtype and the Y166E mutant AtGPA1 as the analytes (Fig. 7 (C–K) and Table 2). There was no specific binding between AtRGS1 and GDP- or GTPγS-bound AtGPA1 (Fig. 7, C and D), whereas AtRGS1 bound to wildtype AtGPA1 in its transition state with a binding affinity of 2.51 nm from kinetic analysis (Fig. 7E) and 4.83 nm from equilibrium analysis (Fig. 7I). Although there was no specific binding with the AtGPA1 Y166E mutant in its GTPγS-bound state (Fig. 7G), AtRGS1 bound to the GDP-bound state with a binding affinity of 5.99 nm from kinetic analysis (Fig. 7F) and 23.4 nm from equilibrium analysis (Fig. 7J). This was similar to its binding affinity in the transition state, which was 5.39 nm from kinetic analysis (Fig. 7H) and 11.1 nm from equilibrium analysis (Fig. 7K). The affinities calculated from kinetic and equilibrium analyses were comparable, and the χ2 values were small compared with the Rmax, indicating that these affinity constants were reliable. These results were perfectly consistent with the binding pattern shown by co-immunoprecipitation (Fig. 7, A and B).
Table 2.
Summary of affinity constants between AtRGS1 and Gα subunits
Binding affinity constants were measured using a ProteOn XPR36 surface plasmon resonance instrument. For the kinetic analysis, the original curves were fit with a 1:1 Langmuir binding model. ka, association rate constant; kd, dissociation rate constant; KD, calculated by ka/kd; Rmax, maximum response; χ2, the average of squared residuals. For the equilibrium analysis, KD was calculated by response units at steady state. RU, response units; NA, no specific binding was detected with the GDP-bound state of wildtype AtGPA1.
| Kinetic analysis |
Equilibrium analysis |
|||||||
|---|---|---|---|---|---|---|---|---|
| ka | kd | kD | Rmax | χ2 | KD | Rmax | χ2 | |
| m−1 s−1 | s−1 | nm | RU | RU | nm | RU | RU | |
| AtGPA1 wildtype | ||||||||
| GDP | NA | NA | NA | NA | NA | NA | NA | NA |
| GDP-AlF4− | 1.24E+06 | 3.11E−03 | 2.51 | 27.12 | 2.81 | 4.83 | 30.01 | 0.148 |
| AtGPA1 Y166E | ||||||||
| GDP | 1.22E+05 | 7.29E−04 | 5.99 | 32.81 | 8.25 | 23.4 | 33.92 | 1.36 |
| GDP-AlF4− | 6.56E+05 | 3.54E−03 | 5.39 | 15.19 | 1.90 | 11.1 | 17.48 | 0.121 |
To test the effect of Tyr166 phosphorylation on AtGPA1-AtRGS1 interaction in vivo, FRET experiments were conducted using a donor AtGPA1 with and without the Tyr166 phosphomimetic mutation tagged with a C-terminal CFP and coexpressed with acceptor AtRGS1 protein (wildtype and phosphorylation and GTPase mutants, 3SA and E320K, respectively) with a C-terminal YFP tag in Nicotiana benthamiana. As shown in Fig. 5A (left), there was no difference in FRET efficiency (percentage) between the wildtype AtGPA1 donor and the wildtype AtRGS1 versus phosphorylation-dead mutant (3SA) FRET acceptors. As expected, mutations that disrupt the AtRGS1-AtGPA1 interactions brought FRET efficiency to its baseline value (∼3.3-fold lower, p < 0.039).
As shown in Fig. 5A (right), as compared with wildtype AtGPA1, the Tyr166 phosphomimetic mutation had a lower FRET efficiency, indicating either a greater distance between the FRET pairs or a different orientation between AtGPA1 and wildtype AtRGS1 (Fig. 8A, ∼1.5 fold, p < 0.047). The 3SA AtRGS1 mutation abrogated this decrease, indicating that the phosphorylation state of AtRGS1 is also important for its interaction orientation with the AtGPA1 isoforms.
Figure 8.
Tyr166 modulates AtGPA1 interaction with AtRGS1 in vivo. A, FRET efficiency between transiently expressed C-terminal YFP-tagged AtRGS1 wildtype, phosphorylation mutant (3SA), or GAP mutant (E320K) and C-terminal CFP-tagged AtGPA1 wildtype or Y166E mutant in N. benthamiana. Values are based on Equation 1 (see “Experimental procedures”). B, FRET changes in response to 3-min 1 μm flg22 treatment between AtRGS1 and AtGPA1 wildtype or Y166E mutant in N. benthamiana. The quantitative results were expressed as the means ± S.D. (error bars) of regions of interest (n = 6–37). Statistical significance was determined by t test. *, differences with p values of <0.05.
AtRGS1 phosphorylation by BAK1 is important for AtRGS1 endocytosis, G protein dynamics, and G protein activation in response to flg22 (10). AtGPA1 Tyr166 phosphorylation shown here fits into this mechanism, whereby increasing the AtRGS1-AtGPA1 transient complex consequently increases freed Gβγ dimer to recruit kinases that phosphorylate AtRGS1, necessary for its endocytosis and thus sustained activation.
In response to flg22, the AtGPA1- AtRGS1 alters orientation within 3 min (Fig. 8, right) (p < 0.032); however, the donor AtGPA1 Y166E-AtRGS1 complex appeared to be already in this altered orientation in the absence of flg22.
Conclusions
We propose that phosphorylation on Tyr166 of the substrate AtGPA1 changes the binding pattern with its enzyme, AtRGS1. As a consequence, the steady-state rate of the GTPase cycle attenuates. Phosphorylation of Tyr166 increases the binding affinity of the GDP state of AtGPA1 to AtRGS1. Typically, RGS1 proteins only bind the GDP + Pi transition state of their cognate Gα subunit.
This unprecedented change in AtRGS1-AtGPA1 binding pattern prompts an obvious question as to whether or not AtRGS1 theoretically behaves as a GDI to the phosphorylated Gα subunit. Typically, GDIs impair the dissociation rate of GDP from the Gα subunit, thus preventing guanine-nucleotide exchange and consequently attenuating the GTPase cycle (1). A statistical difference between wildtype and the AtGPA1 Tyr166 mutant was not observed here (Fig. 6E), as determined by GTPγS binding.
In animals, RGS proteins typically bind the switch region on their corresponding Gα proteins (58–62). The reported GAP-resistant phosphorylation sites on Gαz were all at the N terminus near the switch region (15–17). To our surprise, Tyr166 is located in the intramolecular domain interface for GTP hydrolysis, spatially remote from the switch region (3). Thus, this leaves two possible mechanisms by which phosphomimetic mutagenesis on Tyr166 affects the interaction: 1) AtRGS1 binds to a different region on AtGPA1 where it can be affected by Tyr166, or 2) the Tyr166 → Glu mutant changes the conformation of the switch region. Although we cannot exclude the possibility that a novel RGS-binding region exists on Arabidopsis Gα protein, given that both AtGPA1 and the RGS domain of AtRGS1 are conserved among the respective eukaryotic homologs (3, 6), the first hypothesis is less likely. Therefore, according to the molecular dynamics prediction (Fig. 3), we propose that phosphorylation at Tyr166 forms a salt bridge with positive residues near the guanine-nucleotide–binding pocket to serve as a sensor of the state of nucleotide binding and consequently affects AtRGS1 binding.
Definitive tests involve reconstitution of a full-length seven-transmembrane AtRGS1 protein with the other G protein complex subunits and await a method to produce the full-length AtRGS1 protein. Although we developed a method by which the full-length protein can be expressed and purified from a cell-free system (63), it does not yet produce sufficient quantity to move forward at this time.
Experimental procedures
Protein purification
Briefly, His-tagged LRR RLK cDNA encoding the complete cytoplasmic domain was transformed into BL21 (DE3) pLysS cells (47) as described previously (10).
AtGPA1 mutants were made by the QuikChange Lightning Site-Directed MutagenesisTM kit (Agilent Technologies, catalog no. 210518) and cloned into the pDEST15 or pDEST17 for GST-tagged or His-tagged recombinant proteins. Wildtype and mutant proteins were expressed and purified as described previously (10, 63). The purification procedure for GST-tagged AtGPA1 was similar to that for His-tagged AtGPA1 purification with minor modifications. To determine the specific activity of the protein, the concentrations of purified AtGPA1 wildtype and its mutants were compared with total [γ-35S]GTPγS binding.
His-tagged and GST-tagged RGS + Ct (AtRGS1, residues 284–459) were cloned into pDEST17 or pDEST15 destination vector as described previously (11). The procedure for expression and purification was similar to His-tagged and GST-tagged AtGPA1 purification except without GDP. Detailed information is provided in the supporting Experimental procedures.
SAPH-ire analyses
SAPH-ire is described in detail (38, 39) and was utilized here with only minor adjustments. Briefly, neural network models for function potential probability scores were generated using maximum observed solvent-accessible surface area for each hot spot. PTM hot spot cluster analysis was accomplished by determining the count of neighboring hot spots within ±2 residues and including the hot spot in question. All PTM and structural data used for SAPH-ire correspond to experimental values and strictly exclude predicted or putative information.
DMD simulations
The starting structure for all simulations was the previously published crystal structure for AtGPA1 (PDB code 2XTZ). Simulations were performed using the Medusa force field (64). Phosphorylated tyrosine is not a natural amino acid included within the Medusa force field. For consistency, side chains for both phosphorylated and unphosphorylated Tyr166 in AtGPA1 were modeled as ligands. The Y166A mutant was modeled using the protein design platform Eris (65, 66). Eris was used to perform 100 independent Monte Carlo simulations optimizing side positions within 10 Å of the mutation site. The lowest energy structure was selected. The remaining portions of the Tyr166 and pTyr166 side chains were modeled and optimized using Gaussian 09 at the B3LYP 6-31+G* level of theory (67). The energy-optimized groups were aligned to the position of Tyr166 in the original crystal structure and restrained to Cβ using the same distance, angle, and dihedral potentials used for the native tyrosine residue in the Medusa force field. AtGPA1 Y166D and AtGPA1 Y166E mutations were modeled using Eris to perform 200 independent Monte Carlo side-chain optimizations for residues within 10 Å of the mutation site.
Note that we did not consider the formation of a cation-π interaction. The reason is 2-fold. First the distance, while not unreasonable for cation-π interaction, is more consistent with hydrogen bonding. The peak in question is centered at ∼4 Å. The expected distance between the measured groups for hydrogen bonding is also ∼4 Å, whereas for a cation-π interaction, the distance is ∼5 Å. The second reason is that cation-π and aromatic stacking interactions are not typically well-captured by MD simulations.
For each system, energy minimization was performed according to the protocol implemented in Chiron (68). Systems were then equilibrated by linearly increasing the simulation temperature from 0.1 to 0.45 kcal mol−1 kB−1 over 100,000 steps of discrete molecular dynamics (DMD) (69, 70). Five replicate DMD simulations were performed for each system at a constant temperature of 0.45 kcal mol−1 kB−1. Each simulation was performed for 2 million steps (∼100 ns).
CD spectroscopy
The CD spectra of purified AtGPA1 and its mutations were collected with a Chirascan plus CD spectrometer at 20 °C from 185 to 260 nm in 0.5-nm scan steps in CD spectroscopy buffer (100 nm in 4.3 mm Na2HPO4, 1.4 mm NaH2PO4, 50 mm Na2SO4, 50 μm GDP). The results were analyzed with Chirascan software.
Protein thermostability
Thermostability of AtGPA1 was analyzed by the method of Isom et al. (71). Briefly, 100 ng/μl AtGPA1 wildtype or the mutants were incubated with 1 mm 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole (TCI America; A5597) in Tris buffer (20 mm Tris-HCl, pH 7.6, 50 mm NaCl, 1 mm MgCl2, 1 mm phenylmethylsulfonyl fluoride, 50 μm GDP). 20-μl aliquots were distributed into thin-walled 0.5-ml microcentrifuge tubes and then placed in a gradient thermocycler (Biometra TProfessional Thermocycler), incubated for 3 min at the indicated temperatures. Then the samples were aliquoted into a 384-well plate, and the fluorescence values were measured by a BMG Labtech PHERAstar plate reader with excitation and emission bandpass filters of 400 and 500 nm. The quantitative results were expressed as individual curves.
In vitro kinase assays
To screen the potential kinase for AtGPA1, purified LRR RLKs was mixed with 1.25 μg of RGS + Ct and 2.5 μg of GST-AtGPA1 in 25 μl of kinase buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 10 mm MnCl2, 1 mm DTT, 1 μg/ml leupeptin, 0.1 μm calyculin A, and 1 μm [γ-32P]ATP (PerkinElmer Life Sciences, NEG004Z250UC)) at ambient temperature. Previously, a set of 70 LRR RLKs was screened for AtRGS1 phosphorylation (10). As described therein, we used purified His-tagged RGS + Ct as the positive control for the activity of each kinase. The reactions were quenched by 8 μl of 4× strength Laemmli buffer (62.5 mm Tris-HCl, pH 6.8, 10% glycerol, 1% SDS, 0.005% bromphenol blue) after 8 h of incubation and separated by SDS-PAGE. The phosphorylated proteins were detected by autoradiography.
To further confirm the transphosphorylation by BAK1, GST-AtGPA1 (8 μg) or GST was incubated with 1 μg of His-BAK1 in 30 μl of kinase buffer (50 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 50 μm ATP, and 1 mm DTT) for 2 h at 25 °C. The reactions were quenched by 4× Laemmli buffer and separated by SDS-PAGE. The transphosphorylation and autophosphorylation on AtGPA1 and BAK1 were detected by anti-phosphotyrosine (Santa Cruz Biotechnology, Inc., PY99, sc-7020). GST-tagged or His-tagged protein was detected by polyclonal rabbit anti-GST antibody (Life Technologies, Inc., A5800) or mouse anti-His tag monoclonal antibody (Roche Applied Science, clone BMG-His-1, reference no. 11 922 416 001).
AtGPA1 GTP hydrolysis rate
For measuring the single-turnover GTP hydrolysis rate, purified His-AtGPA1 (wildtype or mutant) was preloaded with [γ-32P]GTP (PerkinElmer Life Sciences, NEG004Z250UC) in single-turnover GTPase buffer (50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 1 mm DTT, and 0.05% Thesit) for 5 min on ice. The hydrolysis reaction was then started by adding an equal volume of GTPγS-containing buffer (50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 1 mm DTT, 0.05% Thesit, and 400 μm GTPγS) with or without His-tagged RGS cytoplasmic domain and moved to a 20 °C water bath. At the indicated times, duplicate samples were denatured by mixing with 1 ml of quench buffer (5% (w/v) activated charcoal, 50 mm phosphoric acid, pH 2.0) to remove non-hydrolyzed [γ-32P]GTP and proteins. The charcoal-treated samples were centrifuged, and the amount of γ-32P in solution was measured by scintillation counting.
The steady-state hydrolysis rate of AtGPA1 was measured as described previously with some modification (4, 8). Briefly, purified His-AtGPA1 (wildtype or mutant) was preloaded with [γ-32P]GTP (PerkinElmer Life Sciences, NEG004Z250UC) in steady-state GTPase buffer (50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 1 mm DTT, and 0.05% Thesit) at 20 °C for 5 min. Then the reaction was started by adding an equal volume of 20 mm MgCl2-containing buffer with or without His-tagged RGS + Ct. At the indicated time points, 1 ml of ice-cold quench buffer (5% (w/v) activated charcoal, 50 mm phosphoric acid, pH 2.0) was added to remove non-hydrolyzed [γ-32P]GTP and proteins. The charcoal-treated samples were centrifuged, and the amount of γ-32P in solution was measured by scintillation counting.
[γ-35S]GTPγS binding
The rate of GTPγS binding was measured by [γ-35S]GTP (PerkinElmer Life Sciences, NEG030H250UC) (9). The entire procedure was performed on ice. Purified His-AtGPA1 (wildtype or mutant) was mixed with GTPγS-binding buffer (50 mm Tris-HCl, pH 7.0, 5 mm MgCl2, 1 mm EDTA, and 5 μm GTPγS (∼5,000 cpm/pmol)) with or without His-tagged RGS + Ct. The reaction was quenched at the indicated time points by quench buffer (20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 25 mm MgCl2, and 50 μm GTP) and vacuum-filtered onto 0.45-μm nitrocellulose membrane (Bio-Rad, 1620115) by a 1225 sampling manifold (Millipore, XX2702550). The filter was washed three times with 3 ml of quench buffer and dried thoroughly. The γ-35S bound to the filter was quantified by scintillation counting.
In vitro pulldown assays
3 μg of His-tagged AtGPA1 (wildtype or Y166E mutant) was incubated with 3 μg of GST-tagged RGS + Ct in 500 μl of pulldown buffer (25 mm Tris-HCl (pH 8.0), 50 mm NaCl, 1 mm DTT, 10 mm MgCl2), in the presence of either 50 μm GDP, 50 μm GTPγS, or AlF4− (50 μm GDP, 10 mm NaF, 100 μm AlCl3). GST-tagged RGS + Ct were precipitated by glutathione-Sepharose 4B (GE Healthcare, product code 17075601) and washed three times with pulldown buffer. The precipitated proteins were eluted by Laemmli sample buffer, separated by SDS-PAGE, and detected by polyclonal rabbit anti-GST antibody (Life Technologies, A5800) or mouse anti-His tag monoclonal antibody (Roche Applied Science, clone BMG-His-1, reference no. 11 922 416 001).
Surface plasmon resonance
Binding affinity constants were measured by the ProteOn XPR36 protein interaction array system and a ProteOnTM GLC Sensor Chip (Bio-Rad, catalog no. 1765011). GST-tagged RGS + Ct (AtRGS1, residues 284–459) was immobilized as ligand. His-tagged AtGPA1 wildtype or Y166E mutant was dosage-diluted by running buffer (10 mm HEPES, pH 8.0, 150 mm NaCl, 10 mm MgCl2, 3 mm EDTA, 10 μm GDP, and 0.01% Tween 20) in the absence or presence of 10 μm GTPγS or AlF4 (50 μm GDP, 10 mm NaF, and 100 μm AlCl3) as analyte. Both association and dissociation were performed for 300 s at a flow rate of 50 μl/min at 25 °C. Kinetic analysis was performed by fitting the original curves with a 1:1 Langmuir binding model. The affinity constants were also calculated by equilibrium analysis.
FRET analyses
Transient expression in N. benthamiana for FRET was performed as described by Tunc-Ozdemir and Jones (72). Briefly, Agrobacterium carrying a binary plasmid encoding either AtRGS1-YFP, AtRGS13SA-YFP, AtRGS1E320K AtGPA1-CFP, AtGPA1 Y166E-CFP, or RNA-silencing suppressor P19 was infiltrated into N. benthamiana leaves' abaxial side with a needleless syringe. Here, the AtGPA1-CFP coding cDNA was carried by pEG100. The coding region of the CFP was inserted between amino acids 97 and 98 of AtGPA1. For acceptor photobleaching FRET, 514- and 458-nm argon lasers were used to excite YFP (acceptor) and CFP (donor), respectively. Acceptor and donor channels' emissions were detected within the range of 516–596 and 460–517 nm, respectively. Regions of interest were scanned five times (each for 50 iterations) using a 514-nm argon laser line at 100% intensity with a pinhole diameter set to 1.00 Airy units. Acceptor was photobleached until it reached ∼20–30% of its initial value. FRET efficiency was then estimated via Zen software (Carl Zeiss). Regions of interests with similar intensity levels to start with were selected, and samples with decreased donor fluorescence intensity after bleaching were excluded in the calculations,
| (1) |
where FDpost represents fluorescence intensity of donor after photobleaching, Fbckgrnd is fluorescence intensity of a non-fluorescence background area, and FDpre is fluorescence intensity of donor before photobleaching.
Author contributions
B. L., M. T. -O., D. U., H. J., E. G. W., D. D. M., M. P. T., and A. M. J. conceptualization; B. L., M. T. -O., E. G. W., D. D. M., and M. P. T. data curation; B. L., M. T. -O., H. J., E. G. W., D. D. M., and M. P. T. formal analysis; B. L., M. T. -O., D. U., H. J., D. D. M., M. P. T., and A. M. J. investigation; B. L., M. T. -O., H. J., D. D. M., and M. P. T. methodology; B. L., D. D. M., M. P. T., and A. M. J. writing-original draft; B. L., M. T. -O., D. U., M. P. T., and A. M. J. writing-review and editing; M. T. -O. visualization; L. M. H., M. P. T., and A. M. J. funding acquisition; L. M. H., N. V. D., M. P. T., and A. M. J. project administration; N. V. D. supervision.
Supplementary Material
Acknowledgments
We are grateful to Dr. Stephen R. Sprang (University of Montana) for insightful comments; Dr. Henrik Dohlman (University of North Carolina, Chapel Hill, NC) for sharing equipment; and Kevin Knight, Jing Yang, and Catherine Jones for valuable technical assistance. This research is based in part upon work conducted using the UNC Michael Hooker Proteomics Center, which is supported in part by NCI, National Institutes of Health, Grant CA016086 (to the Lineberger Comprehensive Cancer Center).
This work was supported by NIGMS, National Institutes of Health (NIH), Grant R01GM065989 and National Science Foundation (NSF) Grant MCB-1713880 (to A. M. J.); NSF Grant MCB-1552522 (to L. M. H); NIGMS, NIH, Grants R01GM080742, R01GM11401, R01GM064803, and 1R01GM123247 (to N. V. D.); and NIH Grant R01 GM117400 (to M. P. T.). The plant-based experimental part of this project was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy through Grant DE-FG02-05er15671 (to A. M. J.). This work was also supported by the NCI, NIH, Grant P30CA016086. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1, Figs. S1 and S2, and supporting Experimental procedures.
- GPCR
- G protein–coupled receptor
- GAP
- GTPase-accelerating/activating protein
- GEF
- guanine-nucleotide exchange factor
- GDI
- guanine-nucleotide dissociation inhibitor
- PAK-1
- p21-activated protein kinase
- PTM
- post-translational modification
- LRR
- leucine-rich repeat
- RLK
- receptor-like kinase
- SAPH-ire
- structural analysis of PTM hot spots
- MAP
- modified alignment position
- DMD
- discrete molecular dynamics
- BAK1
- BRI1-associated receptor kinase 1
- RGS
- regulator of G signaling
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- flg22
- flagellin peptide 22
- YFP
- yellow fluorescent protein
- CFP
- cyan fluorescent protein
- PDB
- Protein Data Bank.
References
- 1. Siderovski D. P., and Willard F. S. (2005) The GAPs, GEFs, and GDIs of heterotrimeric G-protein α subunits. Int. J. Biol. Sci. 1, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neubig R. R., and Siderovski D. P. (2002) Regulators of G-protein signalling as new central nervous system drug targets. Nat. Rev. Drug Discov. 1, 187–197 10.1038/nrd747 [DOI] [PubMed] [Google Scholar]
- 3. Jones J. C., Duffy J. W., Machius M., Temple B. R. S., Dohlman H. G., and Jones A. M. (2011) The crystal structure of a self-activating G protein α subunit reveals its distinct mechanism of signal initiation. Sci. Signal. 4, ra8 10.1126/scisignal.2001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnston C. A., Taylor J. P., Gao Y., Kimple A. J., Grigston J. C., Chen J.-G., Siderovski D. P., Jones A. M., and Willard F. S. (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 17317–17322 10.1073/pnas.0704751104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen J.-G., and Jones A. M. (2004) AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 389, 338–350 10.1016/S0076-6879(04)89020-7 [DOI] [PubMed] [Google Scholar]
- 6. Chen J.-G., Willard F. S., Huang J., Liang J., Chasse S. A., Jones A. M., and Siderovski D. P. (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301, 1728–1731 10.1126/science.1087790 [DOI] [PubMed] [Google Scholar]
- 7. Sprang S. R. (2016) Invited review: activation of G proteins by GTP and the mechanism of Gα-catalyzed GTP hydrolysis. Biopolymers 105, 449–462 10.1002/bip.22836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones J. C., Temple B. R. S., Jones A. M., and Dohlman H. G. (2011) Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana. J. Biol. Chem. 286, 13143–13150 10.1074/jbc.M110.190355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Urano D., Jones J. C., Wang H., Matthews M., Bradford W., Bennetzen J. L., and Jones A. M. (2012) G protein activation without a GEF in the plant kingdom. PLoS Genet. 8, e1002756 10.1371/journal.pgen.1002756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tunc-Ozdemir M., Urano D., Jaiswal D. K., Clouse S. D., and Jones A. M. (2016) Direct modulation of heterotrimeric G protein-coupled signaling by a receptor kinase complex. J. Biol. Chem. 291, 13918–13925 10.1074/jbc.C116.736702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Urano D., Phan N., Jones J. C., Yang J., Huang J., Grigston J., Taylor J. P., and Jones A. M. (2012) Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat. Cell Biol. 14, 1079–1088 10.1038/ncb2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poppleton H., Sun H., Fulgham D., Bertics P., and Patel T. B. (1996) Activation of Gsα by the epidermal growth factor receptor involves phosphorylation. J. Biol. Chem. 271, 6947–6951 10.1074/jbc.271.12.6947 [DOI] [PubMed] [Google Scholar]
- 13. Moyers J. S., Linder M. E., Shannon J. D., and Parsons S. J. (1995) Identification of the in vitro phosphorylation sites on Gsα mediated by pp60c-src. Biochem. J. 305, 411–417 10.1042/bj3050411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hausdorff W. P., Pitcher J. A., Luttrell D. K., Linder M. E., Kurose H., Parsons S. J., Caron M. G., and Lefkowitz R. J. (1992) Tyrosine phosphorylation of G protein α subunits by pp60c-src. Proc. Natl. Acad. Sci. U.S.A. 89, 5720–5724 10.1073/pnas.89.13.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glick J. L., Meigs T. E., Miron A., and Casey P. J. (1998) RGSZ1, a G(z)-selective regulator of G protein signaling whose action is sensitive to the phosphorylation state of G(z)α. J. Biol. Chem. 273, 26008–26013 10.1074/jbc.273.40.26008 [DOI] [PubMed] [Google Scholar]
- 16. Wang J., Ducret A., Tu Y., Kozasa T., Aebersold R., and Ross E. M. (1998) RGSZ1, a Gz-selective RGS protein in brain: structure, membrane association, regulation by Gαz phosphorylation, and relationship to a Gz GTPase-activating protein subfamily. J. Biol. Chem. 273, 26014–26025 10.1074/jbc.273.40.26014 [DOI] [PubMed] [Google Scholar]
- 17. Wang J., Frost J. A., Cobb M. H., and Ross E. M. (1999) Reciprocal signaling between heterotrimeric G proteins and the p21-stimulated protein kinase. J. Biol. Chem. 274, 31641–31647 10.1074/jbc.274.44.31641 [DOI] [PubMed] [Google Scholar]
- 18. Rudrabhatla P., Reddy M. M., and Rajasekharan R. (2006) Genome-wide analysis and experimentation of plant serine/threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 60, 293–319 10.1007/s11103-005-4109-7 [DOI] [PubMed] [Google Scholar]
- 19. Luan S. (2003) Protein phosphatases in plants. Annu. Rev. Plant Biol. 54, 63–92 10.1146/annurev.arplant.54.031902.134743 [DOI] [PubMed] [Google Scholar]
- 20. Xu Q., Fu H. H., Gupta R., and Luan S. (1998) Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene. Plant Cell 10, 849–857 10.1105/tpc.10.5.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yeh Y.-H., Panzeri D., Kadota Y., Huang Y.-C., Huang P.-Y., Tao C.-N., Roux M., Chien H.-C., Chin T.-C., Chu P.-W., Zipfel C., and Zimmerli L. (2016) The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28, 1701–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang S., and Klessig D. F. (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9, 809–824 10.1105/tpc.9.5.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suzuki K., and Shinshi H. (1995) Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with a fungal elicitor. Plant Cell 7, 639–647 10.1105/tpc.7.5.639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stratmann J. W., and Ryan C. A. (1997) Myelin basic protein kinase activity in tomato leaves is induced systemically by wounding and increases in response to systemin and oligosaccharide elicitors. Proc. Natl. Acad. Sci. U.S.A. 94, 11085–11089 10.1073/pnas.94.20.11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oh M. H., Wu X., Kim H. S., Harper J. F., Zielinski R. E., Clouse S. D., and Huber S. C. (2012) CDPKs are dual-specificity protein kinases and tyrosine autophosphorylation attenuates kinase activity. FEBS Lett. 586, 4070–4075 10.1016/j.febslet.2012.09.040 [DOI] [PubMed] [Google Scholar]
- 26. Kim D., Ntui V. O., Zhang N., and Xiong L. (2015) Arabidopsis Yak1 protein (AtYak1) is a dual specificity protein kinase. FEBS Lett. 589, 3321–3327 10.1016/j.febslet.2015.09.025 [DOI] [PubMed] [Google Scholar]
- 27. Oh M.-H., Wang X., Kota U., Goshe M. B., Clouse S. D., and Huber S. C. (2009) Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 658–663 10.1073/pnas.0810249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shiu S.-H. H., and Bleecker A. B. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768 10.1073/pnas.181141598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tör M., Lotze M. T., and Holton N. (2009) Receptor-mediated signalling in plants: molecular patterns and programmes. J. Exp. Bot. 60, 3645–3654 10.1093/jxb/erp233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Lorenzo L., Merchan F., Laporte P., Thompson R., Clarke J., Sousa C., and Crespi M. (2009) A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell 21, 668–680 10.1105/tpc.108.059576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Becraft P. W. (2002) Receptor kinase signaling in plant development. Annu. Rev. Cell Dev. Biol. 18, 163–192 10.1146/annurev.cellbio.18.012502.083431 [DOI] [PubMed] [Google Scholar]
- 32. Greeff C., Roux M., Mundy J., and Petersen M. (2012) Receptor-like kinase complexes in plant innate immunity. Front. Plant Sci. 3, 209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stahl Y., and Simon R. (2012) Receptor kinases in plant meristem development. Signal. Commun. Plants 13, 23–39 10.1007/978-3-642-23044-8_2 [DOI] [Google Scholar]
- 34. Kim T.-W., and Wang Z.-Y. (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61, 681–704 10.1146/annurev.arplant.043008.092057 [DOI] [PubMed] [Google Scholar]
- 35. Urano D., Miura K., Wu Q., Iwasaki Y., Jackson D., and Jones A. M. (2016) Plant morphology of heterotrimeric G protein mutants. Plant Cell Physiol. 57, 437–445 10.1093/pcp/pcw002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Urano D., Chen J.-G., Botella J. R., and Jones A. M. (2013) Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 3, 120186 10.1098/rsob.120186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Y., Hoehenwarter W., and Weckwerth W. (2010) Comparative analysis of phytohormone-responsive phosphoproteins in Arabidopsis thaliana using TiO2-phosphopeptide enrichment and mass accuracy precursor alignment. Plant J. 63, 1–17 [DOI] [PubMed] [Google Scholar]
- 38. Torres M. P., Dewhurst H., and Sundararaman N. (2016) Proteome-wide structural analysis of PTM hotspots reveals regulatory elements predicted to impact biological function and disease. Mol. Cell Proteomics 15, 3513–3528 10.1074/mcp.M116.062331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dewhurst H. M., Choudhury S., and Torres M. P. (2015) Structural analysis of PTM hotspots (SAPH-ire): a quantitative informatics method enabling the discovery of novel regulatory elements in protein families. Mol. Cell Proteomics 14, 2285–2297 10.1074/mcp.M115.051177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., and Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 [DOI] [PubMed] [Google Scholar]
- 42. Tweedie-Cullen R. Y., Reck J. M., and Mansuy I. M. (2009) Comprehensive mapping of post-translational modifications on synaptic, nuclear, and histone proteins in the adult mouse brain. J. Proteome Res. 8, 4966–4982 10.1021/pr9003739 [DOI] [PubMed] [Google Scholar]
- 43. Wiśniewski J. R., Nagaraj N., Zougman A., Gnad F., and Mann M. (2010) Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J. Proteome Res. 9, 3280–3289 10.1021/pr1002214 [DOI] [PubMed] [Google Scholar]
- 44. Noel J. P., Hamm H. E., and Sigler P. B. (1993) The 2.2 Å crystal structure of transducin-α complexed with GTPγS. Nature 366, 654–663 10.1038/366654a0 [DOI] [PubMed] [Google Scholar]
- 45. Sunahara R. K., Tesmer J. J., Gilman A. G., and Sprang S. R. (1997) Crystal structure of the adenylyl cyclase activator Gsα. Science 278, 1943–1947 10.1126/science.278.5345.1943 [DOI] [PubMed] [Google Scholar]
- 46. Coleman D. E., and Sprang S. R. (1998) Crystal structures of the G protein Giα1 complexed with GDP and Mg2+: a crystallographic titration experiment. Biochemistry 37, 14376–14385 10.1021/bi9810306 [DOI] [PubMed] [Google Scholar]
- 47. Mitra S. K., Chen R., Dhandaydham M., Wang X., Blackburn R. K., Kota U., Goshe M. B., Schwartz D., Huber S. C., and Clouse S. D. (2015) An autophosphorylation site database for leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. Plant J. 82, 1042–1060 10.1111/tpj.12863 [DOI] [PubMed] [Google Scholar]
- 48. Berardini T. Z., Mundodi S., Reiser L., Huala E., Garcia-Hernandez M., Zhang P., Mueller L. A., Yoon J., Doyle A., Lander G., Moseyko N., Yoo D., Xu I., Zoeckler B., Montoya M., Miller N., Weems D., and Rhee S. Y. (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135, 745–755 10.1104/pp.104.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang D. W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 51. Shah K., Vervoort J., and de Vries S. C. (2001) Role of threonines in the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J. Biol. Chem. 276, 41263–41269 10.1074/jbc.M102381200 [DOI] [PubMed] [Google Scholar]
- 52. Lin W., Li B., Lu D., Chen S., Zhu N., He P., and Shan L. (2014) Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. U.S.A. 111, 3632–3637 10.1073/pnas.1318817111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., and Walker J. C. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- 54. Aranda-Sicilia M. N., Trusov Y., Maruta N., Chakravorty D., Zhang Y., and Botella J. R. (2015) Heterotrimeric G proteins interact with defense-related receptor-like kinases in Arabidopsis. J. Plant Physiol. 188, 44–48 10.1016/j.jplph.2015.09.005 [DOI] [PubMed] [Google Scholar]
- 55. Liu J., Ding P., Sun T., Nitta Y., Dong O., Huang X., Yang W., Li X., Botella J. R., and Zhang Y. (2013) Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol. 161, 2146–2158 10.1104/pp.112.212431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Berman D. M., Kozasa T., and Gilman A. G. (1996) The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J. Biol. Chem. 271, 27209–27212 10.1074/jbc.271.44.27209 [DOI] [PubMed] [Google Scholar]
- 57. Urano D., Maruta N., Trusov Y., Stoian R., Wu Q., Liang Y., Jaiswal D. K., Thung L., Jackson D., Botella J. R., and Jones A. M. (2016) Saltational evolution of the heterotrimeric G protein signaling mechanisms in the plant kingdom. Sci. Signal. 9, ra93 10.1126/scisignal.aaf9558 [DOI] [PubMed] [Google Scholar]
- 58. Chen Z., Singer W. D., Danesh S. M., Sternweis P. C., and Sprang S. R. (2008) Recognition of the activated states of Gα13 by the rgRGS domain of PDZRhoGEF. Structure 16, 1532–1543 10.1016/j.str.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Slep K. C., Kercher M. A., Wieland T., Chen C.-K., Simon M. I., and Sigler P. B. (2008) Molecular architecture of Gαo and the structural basis for RGS16-mediated deactivation. Proc. Natl. Acad. Sci. U.S.A. 105, 6243–6248 10.1073/pnas.0801569105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nance M. R., Kreutz B., Tesmer V. M., Sterne-Marr R., Kozasa T., and Tesmer J. J. G. (2013) Structural and functional analysis of the regulator of G protein signaling 2-Gαq complex. Structure 21, 438–448 10.1016/j.str.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Taylor V. G., Bommarito P. A., and Tesmer J. J. G. (2016) Structure of the regulator of G protein signaling 8 (RGS8)-Gαq complex: molecular basis for Gα selectivity. J. Biol. Chem. 291, 5138–5145 10.1074/jbc.M115.712075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Z., Singer W. D., Sternweis P. C., and Sprang S. R. (2005) Structure of the p115RhoGEF rgRGS domain–Gα13/i1 chimera complex suggests convergent evolution of a GTPase activator. Nat. Struct. Mol. Biol. 12, 191–197 10.1038/nsmb888 [DOI] [PubMed] [Google Scholar]
- 63. Li B., Makino S. I., Beebe E. T., Urano D., Aceti D. J., Misenheimer T. M., Peters J., Fox B. G., and Jones A. M. (2016) Cell-free translation and purification of Arabidopsis thaliana regulator of G signaling 1 protein. Protein Expr. Purif. 126, 33–41 10.1016/j.pep.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ding F., and Dokholyan N. V. (2006) Emergence of protein fold families through rational design. PLoS Comput. Biol. 2, e85 10.1371/journal.pcbi.0020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yin S., Ding F., and Dokholyan N. V. (2007) Eris: an automated estimator of protein stability. Nat. Methods 4, 466–467 10.1038/nmeth0607-466 [DOI] [PubMed] [Google Scholar]
- 66. Yin S., Ding F., and Dokholyan N. V. (2010) Computational evaluation of protein stability change upon mutations. Methods Mol. Biol. 634, 189–201 10.1007/978-1-60761-652-8_14 [DOI] [PubMed] [Google Scholar]
- 67. Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Petersson G. A., Nakatsuji H., Li X., Caricato M., Marenich A., Bloino J., Janesko B. G., et al. (2016) Gaussian 09, Revision A.02., Gaussian, Inc., Wallingford CT [Google Scholar]
- 68. Ramachandran S., Kota P., Ding F., and Dokholyan N. V. (2011) Automated minimization of steric clashes in protein structures. Proteins 79, 261–270 10.1002/prot.22879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shirvanyants D., Ding F., Tsao D., Ramachandran S., and Dokholyan N. V. (2012) Discrete molecular dynamics: an efficient and versatile simulation method for fine protein characterization. J. Phys. Chem. B 116, 8375–8382 10.1021/jp2114576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ding F., Tsao D., Nie H., and Dokholyan N. V. (2008) Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure 16, 1010–1018 10.1016/j.str.2008.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Isom D. G., Marguet P. R., Oas T. G., and Hellinga H. W. (2011) A miniaturized technique for assessing protein thermodynamics and function using fast determination of quantitative cysteine reactivity. Proteins 79, 1034–1047 10.1002/prot.22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tunc-Ozdemir M., and Jones A. M. (2017) Ligand-induced dynamics of heterotrimeric G protein-coupled receptor-like kinase complexes. PLoS One 12, e0171854 10.1371/journal.pone.0171854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Crooks G. E., Hon G., Chandonia J.-M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lambright D. G., Sondek J., Bohm A., Skiba N. P., Hamm H. E., and Sigler P. B. (1996) The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379, 311–319 10.1038/379311a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.