Abstract
Somatic mutations in exon 2 of the RNA polymerase II transcriptional Mediator subunit MED12 occur at high frequency in uterine fibroids (UFs) and breast fibroepithelial tumors as well as recurrently, albeit less frequently, in malignant uterine leimyosarcomas, chronic lymphocytic leukemias, and colorectal cancers. Previously, we reported that UF-linked mutations in MED12 disrupt its ability to activate cyclin C (CycC)–dependent kinase 8 (CDK8) in Mediator, implicating impaired Mediator-associated CDK8 activity in the molecular pathogenesis of these clinically significant lesions. Notably, the CDK8 paralog CDK19 is also expressed in myometrium, and both CDK8 and CDK19 assemble into Mediator in a mutually exclusive manner, suggesting that CDK19 activity may also be germane to the pathogenesis of MED12 mutation–induced UFs. However, whether and how UF-linked mutations in MED12 affect CDK19 activation is unknown. Herein, we show that MED12 allosterically activates CDK19 and that UF-linked exon 2 mutations in MED12 disrupt its CDK19 stimulatory activity. Furthermore, we find that within the Mediator kinase module, MED13 directly binds to the MED12 C terminus, thereby suppressing an apparent UF mutation–induced conformational change in MED12 that otherwise disrupts its association with CycC-CDK8/19. Thus, in the presence of MED13, mutant MED12 can bind, but cannot activate, CycC-CDK8/19. These findings indicate that MED12 binding is necessary but not sufficient for CycC-CDK8/19 activation and reveal an additional step in the MED12-dependent activation process, one critically dependent on MED12 residues altered by UF-linked exon 2 mutations. These findings confirm that UF-linked mutations in MED12 disrupt composite Mediator-associated kinase activity and identify CDK8/19 as prospective therapeutic targets in UFs.
Keywords: cyclin-dependent kinase (CDK), gene transcription, RNA polymerase II, tumor, transcription coregulator, cancer, allosteric regulation, CDK19, CDK8, MED12, Mediator, uterine leiomyoma
Introduction
The fate of individual cells during the course of metazoan development and maturation, including the decision to proliferate, differentiate, or die, is critically dependent on controlled expression of enhancer-dependent transcriptional programs enforced by signal-activated transcription factors. The regulatory activity of these mobilized transcription factors is often realized with the assistance of Mediator, a conserved multiprotein interface between enhancer-bound regulators and the RNA polymerase II (Pol II)3 machinery assembled on core promoters (1, 2). In this capacity, Mediator has been proposed to function as a biochemical “bridge” through which the regulatory information conveyed by chromatin-bound transcription factors is converted to transcriptional output via control of Pol II recruitment (3, 4). Nonetheless, Mediator has also been shown to control and coordinate multiple steps in the transcription process following Pol II recruitment, including early initiation events linked to Pol II promoter escape, Pol II pausing and elongation, and co-transcriptional RNA processing (3, 5).
Structurally, Mediator is assembled from a set of core subunits (26 in humans) arranged into 3 distinct modules, termed “head,” “middle,” and “tail,” that bind tightly to Pol II in the so-called “holoenzyme” (6–9). A fourth “kinase” module exists in variable association with core Mediator, and is nominally comprised of 4 subunits designated MED13, MED12, Cyclin C (CycC), and cyclin-dependent kinase 8 (CDK8) (3, 10, 11). Vertebrate paralogs of all kinase module subunits with the exception of CycC have been identified, including MED13L, MED12L, and CDK19 (3, 12). Significantly, prior mass spectrometric-based proteomic analyses indicate that kinase module paralogs assemble into Mediator in a mutually exclusive manner, raising the likelihood that multiple, compositionally distinct Mediator–kinase complexes exist in mammalian cells (13).
Functionally, the kinase module has been implicated in both activation and repression of transcription through mechanisms that are both dependent and independent of its resident CDK activity. CDK-dependent transcription control by Mediator appears to be mechanistically complex and substrate diverse, with targeted phosphorylation events linked to transcription factor turnover, Pol II activity, and chromatin chemistry and functional status (3, 5). Notably, the Mediator kinase module represents a major conduit and variable resistor of signal transduction through Mediator, and mounting evidence suggests that MED12-dependent CDK8 activity is a critical determinant of signal strength and transmission. First, Mediator kinase activity has been implicated as a positive or negative modulator of key developmental and oncogenic signaling pathways with which MED12 itself has been linked biochemically and/or genetically, including the WNT, Sonic hedgehog, Notch, and transforming growth factor β pathways, among others (3, 14–21). Second, MED12 is an established activator of CycC–CDK8 in Mediator (22, 23). In this regard, CDK8 is atypical among cyclin-dependent kinases (CDKs) in its biochemical mode of activation. Thus, although the association of CDK8 with CycC promotes an active conformation through relief of a steric block to the catalytic cleft in accordance with the conserved mechanism of CDK activation, CDK8 does not undergo subsequent phosphorylation within its activation loop, an event commonly required for complete activation of other CDKs (3, 24, 25). Instead, CDK8 is activated through an otherwise obscure allosteric mechanism that nonetheless has been shown to require direct binding of the MED12 N terminus to a phylogenetically conserved surface groove on CycC (22, 23).
The biological significance of MED12-dependent CDK8 activation in Mediator is suggested by the recent discovery that oncogenic mutations in MED12 disrupt its allosteric regulation of CDK8 (23, 26). In this regard, mutations in exon 2 of the Xq13 gene encoding MED12 occur at high frequency (59–80%) in benign stromal tumors of the uterus (uterine leiomyomas) and breast (breast fibroadenomas and phyllodes tumors) (27–30). Furthermore, MED12 exon 2 mutations have been found to occur recurrently, albeit less frequently, in malignant uterine leiomyosarcomas (7–30%), chronic lymphocytic leukemias (5%), and colorectal cancers (0.5%) (3, 31, 32). Among the tumor types in which MED12 exon 2 mutations have been identified, the oncogenic potential of these mutations has been most convincingly demonstrated for uterine leiomyomas.
Uterine leiomyomas (uterine fibroids; UFs) are benign monoclonal neoplasms of the myometrium and represent the most common gynecologic tumor among reproductive age women (33–35). Although benign, UFs nonetheless account for significant morbidity; they are the leading indication for hysterectomy and a major source of gynecologic and reproductive dysfunction, ranging from profuse menstrual bleeding and pelvic pain to infertility, recurrent miscarriage, and pre-term labor (33–35). The majority of UF-linked mutations in MED12 (∼60%) are missense mutations that precipitate substitutions at three highly conserved MED12 amino acids: Leu-36, Gln-43, and Gly-44 (28, 29). The remaining ∼40% of MED12 mutations linked to UFs correspond to missense mutations at other residues or small in-frame insertions and deletions (28, 29). Along with their high-frequency occurrence, two additional lines of genetic evidence strongly support the notion that MED12 mutations are drivers of UF formation. First, monoallelic expression of mutant MED12 from the X chromosome has been observed in all tumors duly examined, indicating the requirement for a functionally altered allele during tumorigenesis (3, 28, 36). Second, conditional expression of a UF-linked MED12 mutant transgene in the mouse uterus elicits tumor formation, providing direct genetic proof of disease causality (37). However, the impact of these mutations on MED12 function and the molecular basis for their tumorigenic activity remain to be fully established. In this regard, it is notably that all UF-linked exon 2 mutations in MED12 thus far identified reside exclusively within its CycC–CDK8 binding and activation domain (aa 1–100), suggesting that Mediator kinase disruption is the principal biochemical defect arising from these genetic alterations (23, 28, 29). Recently, we confirmed this prediction by showing that UF-linked exon 2 mutations in MED12 disrupt its ability to bind to CycC and activate CDK8 (23). Our discovery that UF-linked exon 2 mutations in MED12 disrupt its CycC–CDK8 activation function represents the first description of a molecular defect arising from these mutations and further implicates aberrant CDK8 activity in UF pathogenesis. Nonetheless, in addition to CDK8, its paralog CDK19 is also expressed in myometrium and UF tissues (23). Because these paralogous subunits assemble into the Mediator kinase module in a mutually exclusive manner, the extent to which the tumorigenic potential of MED12 mutations derives from disruption of Mediator-associated CDK8 versus CDK19 activity is therefore presently unclear.
CDK19 (also known as CDK11, CDK8L, and CDC2L6) was originally identified as a CDK8 paralog through a bioinformatics-based homology screen of existing DNA sequence databases and thereafter linked directly to Mediator via mass spectrometric-based analysis of Mediator complexes immunopurified from mammalian cells (38–41). Despite extensive sequence homology between CDK8 and CDK19 (∼76 and ∼83% sequence identity and similarity, respectively), the two paralogs nonetheless differ in certain biological and structural aspects. Thus, whereas CDK8 is ubiquitously expressed, CDK19 is restricted in its tissue distribution, with reported expression limited to salivary gland, thymus, testis, prostate, and uterus (23, 42). Furthermore, whereas the two paralogs share extensive sequence homology within their CycC-binding and catalytic kinase domains (80 and 97%, respectively), they nonetheless exhibit considerable sequence divergence in their corresponding C termini (12, 39). These differences may in part underlie the observation that CDK8 and CDK19 confer distinct transcriptional regulatory properties on Mediator. For example, biochemical studies have revealed that CDK8–Mediator supports, whereas CDK19–Mediator suppresses, herpes simplex virus VP16 transactivation domain function (41). Furthermore, comparative genome-wide expression profiling analyses revealed distinct gene sets uniquely regulated by CDK8 or CDK19, further highlighting their independent roles in transcription (42).
Notwithstanding the divergent transcriptional regulatory properties of CDK8 and CDK19, which may reflect differences in their respective tissue distribution patterns and/or substrate selectivity, the high degree of sequence homology within their corresponding CycC-binding and catalytic kinase domains implies a common biochemical mode of action, including possible allosteric regulation by MED12. However, whether and how MED12 activates CDK19, and the impact of UF-linked MED12 mutations on CDK19 activation, is presently unknown. Herein, we show for the first time that MED12 is an allosteric activator of CDK19 and, furthermore, that UF-linked mutations in MED12 disrupt its CDK19 stimulatory activity. We also identify dynamic subunit interactions within the Mediator kinase module that offer new insight concerning allosteric regulation of CDK8/19 by MED12. In this regard, we show that MED13 binds directly to the C terminus of MED12, thereby suppressing a tumorigenic mutation–induced conformational change in MED12 that otherwise disrupts its association with CycC–CDK8/19. Thus, in the presence of MED13, mutant MED12 can bind to, but not activate, CycC–CDK8/19. Together, these findings indicate that MED12 binding is necessary but not sufficient for CycC–CDK8/19 activation, and identify an additional step in the MED12-dependent activation process, one that is critically dependent on MED12 residues altered by oncogenic exon 2 mutations. We discuss the implications of these novel findings for the regulation of Mediator kinase activity and the molecular basis of MED12 in UF pathogenesis.
Results
MED12 activates CycC–CDK19
Previously, we and others found that MED12 activates CycC–CDK8 within the Mediator kinase module (22, 23). Mechanistically, we showed that this occurs through a direct interaction between the MED12 N terminus and a surface groove on CycC that lies spatially distant (∼22 Å) from the CycC–CDK8 binding interface within the CycC–CDK8 crystal structure (23, 24). In this regard, the CDK8 interaction surface on CycC comprises α-helices within the first of two canonical repeats of the CycC box fold along with N- and C-terminal helices, whereas the CycC surface groove to which MED12 binds is located between the two CycC repeats (24). Thus, CycC can accommodate simultaneous association with both MED12 and CDK8 through distinct binding surfaces, leading to the formation of a ternary MED12–CycC–CDK8 complex that is required for full CDK8 activation. Notably, CDK8 and CDK19 share extensive sequence homology within their corresponding CycC-binding and catalytic kinase domains, suggesting that CDK19, like CDK8, may be similarly activated by MED12.
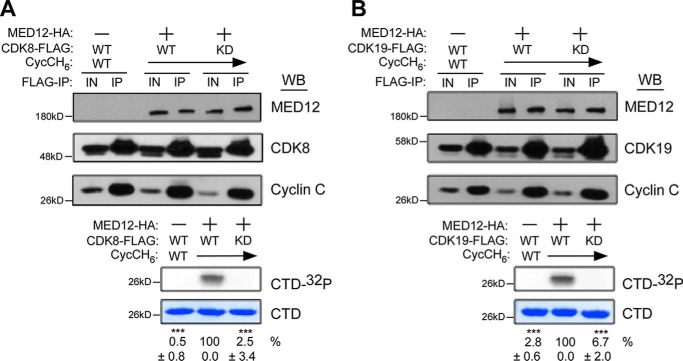
To test this, we examined the impact of MED12 on CycC–CDK19 kinase activity in vitro using recombinant baculovirus-expressed human kinase module subunits. To this end, FLAG-specific immunoprecipitates from insect High Five cells co-expressing FLAG epitope-tagged CDK19 (CDK19–FLAG) and CycC without or with MED12 were monitored for the level of CDK19 kinase activity in the presence of [γ-32P]ATP and a recombinant peptide substrate corresponding to 3 tandem copies of the consensus RNA Pol II C-terminal domain (CTD) heptapeptide repeat sequence (23). Parallel reactions reconstituted with CDK8-FLAG were included as a positive control for the CDK stimulatory activity of MED12. Additionally, reactions reconstituted with kinase-dead derivatives of CDK8/19 were included as controls for the functional selectivity of MED12 in these experiments. Consistent with our previous findings (23), MED12 potently stimulated CycC–CDK8 kinase activity, which was otherwise negligible in its absence (Fig. 1A). Notably, MED12 also stimulated otherwise abeyant CycC–CDK19 kinase activity comparably to CycC–CDK8 (Fig. 1B). Importantly, no kinase activity was detected in reactions reconstituted with kinase-dead CDK8/19 derivatives, confirming that the stimulatory effect of MED12 in these experiments is selective for wildtype CDK8/19 (Fig. 1, A and B). These findings reveal that MED12 is indeed an activator of CDK19. We further confirmed that MED12 activates CDK19, like CDK8, in a manner requiring its N-terminal 100 amino acids (Figs. S1 and S2), corresponding to a region on MED12 that binds directly to a phylogenetically conserved surface groove on CycC (23).
Figure 1.
MED12 stimulates CycC-dependent CDK19 activity. A and B, lysates from baculovirus-infected insect cells co-expressing the indicated combinations of Mediator kinase module subunits MED12–HA, CycCH6, and CDK8–FLAG (A) or CDK19–FLAG (B) (WT or kinase-dead (KD) mutant D173A) were subjected to IP with FLAG epitope-specific antibodies. FLAG-specific IPs were processed by Western blot (WB) using the indicated antibodies (top panels) or incubated with [γ-32P]ATP and purified GST–CTD prior to resolution by SDS-PAGE and PhosphorImager analyses (bottom panels). Input (IN) corresponds to 10% of cell lysate used in IP reactions. Horizontal arrows indicate the presence of specified Mediator subunits in parallel reactions. The level of 32P-labeled GST–CTD (CTD-32P) in each IP–kinase reaction was quantified and expressed relative to the level in the CDK8 WT CycC–MED12 IP (A) or the CDK19 WT–CycC–MED12 IP (B). Data represent the average ± S.E. of 3 independent experiments. Asterisks denote statistically significant differences versus the CDK8 or CDK19 WT–CycC–MED12 IP (Student's t test, ***, p < 0.001).
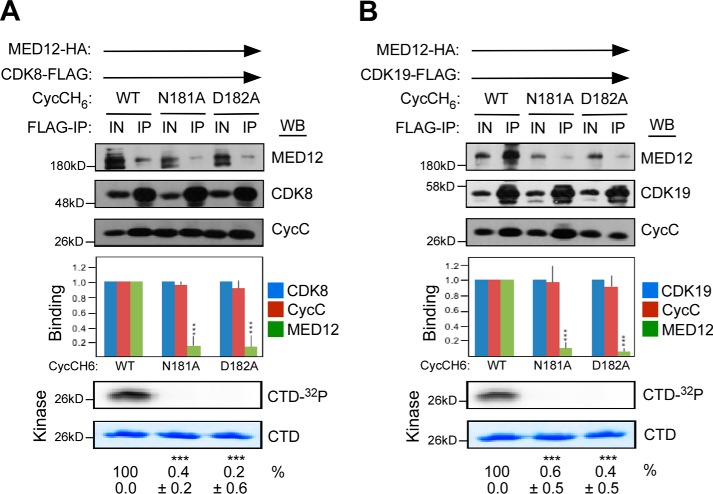
To investigate whether MED12 activates CDK19, like CDK8, through an allosteric mechanism that involves direct interaction of its N terminus with the CycC surface groove, we monitored the impact of CycC surface groove mutations N181A and D182A on the ability of MED12 to bind and activate CycC–CDK19. Previously, we showed that these mutations disrupt the ability of MED12 to bind and activate CycC–CDK8 (23). To this end, FLAG-specific immunoprecipitates from insect cells co-expressing MED12, CDK19–FLAG, and WT or mutant CycC derivatives were monitored for both the presence of MED12 and the level of CDK19 kinase activity. Control reactions reconstituted with CDK8–FLAG were included as a control for the deleterious impact of CycC surface groove mutations on MED12-dependent CDK activation. These analyses confirmed our prior observations that mutagenic disruption of the CycC surface groove severely impaired MED12 binding and consequent activation of CDK8 (Fig. 2A). Importantly, disruption of the MED12-binding interface on CycC also drastically compromised CDK19 activity (Fig. 2B), indicating that MED12 stimulates both CDK8 and CDK19 through a similar allosteric mechanism involving direct interaction of MED12 with CycC.
Figure 2.
MED12 is an allosteric activator of CycC–CDK19. A and B, lysates from insect cells co-expressing baculovirus-produced MED12–HA, CycCH6 (WT or mutant as indicated), and either CDK8–FLAG (A) or CDK19–FLAG (B) were subjected to IP with FLAG-specific antibodies. FLAG-specific IPs were processed by Western blot (WB) using the indicated antibodies (top panels) or incubated with [γ-32P]ATP and purified GST–CTD prior to resolution by SDS-PAGE and PhosphorImager analyses (bottom panels). Input (IN) corresponds to 10% of cell lysate used in IP reactions. Horizontal arrows indicate the presence of specified Mediator subunits in parallel reactions. WBs were quantified and levels of MED12 and CycC in each IP were normalized to those of CDK8 (A) or CDK19 (B) and expressed relative to their corresponding normalized levels in the CDK8 (or CDK19)–CycC WT–MED12 IP (middle panel). 32P-Labeled GST–CTD levels in each IP–kinase reaction were quantified and expressed relative to the level in the CDK8 (or CDK19)–CycC WT–MED12 IP. Data represent the average ± S.E. of 3 independent experiments. Asterisks denote statistically significant differences versus WT (Student's t test, ***, p < 0.001).
UF-linked mutations in MED12 disrupt its CycC–CDK19 stimulatory activity
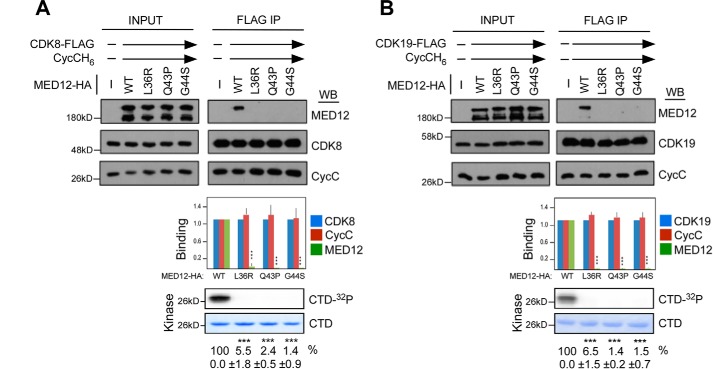
We previously showed that UF-linked mutations in MED12 disrupt its CycC-binding interface, leading to reduced interaction with and activation of CycC–CDK8 (23). Our observation that MED12 also activates CycC–CDK19, coupled with the fact that CDK19, like CDK8, is expressed in both myometrium and UF tissues (Fig. S3) led us to ask whether UF-linked mutations in MED12 similarly disrupt its CycC–CDK19 binding and activation functions. To test this, we monitored the subunit composition and activity of recombinant baculovirus-expressed kinase module variants reconstituted with WT MED12 or UF mutants L36R, Q43P, or G44S. Accordingly, FLAG-specific immunoprecipitates from insect cells co-expressing FLAG-tagged CDK19, CycC, and either WT or mutant MED12 derivatives were monitored for the presence of MED12 and the level of CDK19 kinase activity. As a control for the deleterious impact of UF-linked MED12 mutations on CDK activity, we performed a parallel set of reactions reconstituted with FLAG-tagged CDK8. Consistent with our prior observations (23), all three of the UF-linked MED12 mutants were severely compromised, compared with WT MED12, for their respective CycC–CDK8 binding and activation functions (Fig. 3A). Notably, all three of the UF-linked MED12 mutant derivatives were similarly severely defective for CycC–CDK19 binding and activation (Fig. 3B). These findings extend the immediate collateral defects associated with UF-linked mutations in MED12 beyond CDK8 to include its paralog CDK19, suggesting that disruption of composite Mediator-associated CDK activity is a likely biochemical defect arising from these tumorigenic MED12 mutations.
Figure 3.
UF-linked mutations in MED12 disrupt its CycC–CDK19 binding and stimulatory activities. A and B, lysates from insect cells co-expressing baculovirus-produced CycCH6, MED12–HA (WT or mutant as indicated), and either CDK8-FLAG (A) or CDK19–FLAG (B) were subjected to IP with FLAG epitope-specific antibodies. FLAG-specific IPs were processed by Western blot (WB) using the indicated antibodies (top panels) or subjected to in vitro kinase assay prior to resolution by SDS-PAGE and PhosphorImager analyses (bottom panels). Input corresponds to 10% of cell lysate used in IP reactions. Horizontal arrows indicate the presence of specified Mediator subunits in parallel reactions. WB were quantified and levels of MED12 and CycC in each IP were normalized to those of CDK8 (A) or CDK19 (B) and expressed relative to their corresponding normalized levels in the CDK8 (or CDK19)–CycC–MED12 WT IP (middle panel). 32P-Labeled GST–CTD levels in each IP–kinase reaction were quantified and expressed relative to the level in the CDK8 (or CDK19)–CycC–MED12 WT IP. Data represent the average ± S.E. of 3 independent experiments. Asterisks denote statistically significant differences versus WT (Student's t test, ***, p < 0.001).
MED12 binding is necessary but not sufficient for activation of CycC–CDK8/19
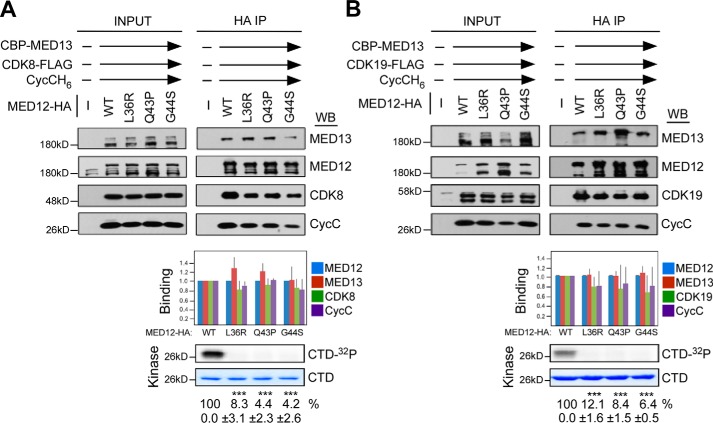
Our observation that mutagenic disruption of the MED12–CycC interface severely impairs MED12-dependent CDK8/19 activation supports the notion that MED12 binding is necessary for CycC–CDK8/19 activation. Nonetheless, whether MED12 binding is also sufficient for CycC–CDK8/19 activation is indeterminable based on our initial experimental design and therefore remains to be established. Notably, our initial in vitro binding and kinase assays were reconstituted in the absence MED13 and therefore included only three of four Mediator kinase module subunits: MED12 (WT or mutant), CycC, and CDK8 or CDK19. These conditions were employed based on our prior observations that MED13 does not appreciably impact the integrity or activity of a trimeric MED12 (WT)–CycC–CDK8 submodule assembly (23). Nonetheless, as naturally occurring Mediator complexes in vivo bear intact four-subunit kinase modules, we sought to investigate the impact of MED13 on the association of UF-linked mutant MED12 derivatives with CycC–CDK8/19. To this end, we repeated in vitro binding and kinase assays using WT or mutant MED12 kinase modules reconstituted from all four subunits, including MED13, MED12, CycC, and CDK8/19–FLAG. Notably, all of the mutant MED12 derivatives were co-precipitated from insect cell lysates by FLAG-specific antibodies comparably to WT MED12, indicating that UF-linked mutations in MED12 do not aberrantly affect its stable incorporation into a four-subunit kinase module (Fig. 4, A and B). Strikingly, the addition of MED13 suppressed the deleterious impact (otherwise observed in its absence) of UF-linked mutations in MED12 on its interaction with CycC–CDK8/19 without rescuing activation. Thus, whereas CycC and CDK8/19 were comparably present in WT and mutant MED12-reconstituted four-subunit kinase modules, CDK activity was nonetheless severely compromised in the latter (Fig. 4, A and B). Note that these reactions were carried out under conditions in which MED13 was saturating for quaternary (MED13–MED12–CycC–CDK) complex formation, ensuring that diminished kinase activity associated with mutant MED12-containing complexes was not attributable to reduced association of CycC–CDK8/19 from residual ternary (MED12–CycC–CDK) complexes (Fig. S4). Together, these results indicate that MED12 binding is necessary but not sufficient for CycC–CDK8/19 activation, and imply an additional step in the activation process, one dependent on MED12 residues (Leu-36, Gln-43, and Gly-44) frequently altered by oncogenic mutations. Because MED13 binds directly to MED12, but not to CycC or CDK8/19 (23), we conclude that MED13 likely renders mutant MED12 derivatives capable of binding CycC–CDK8/19 through a conformational constraint(s) within MED12 induced by direct protein–protein interactions between MED13 and MED12.
Figure 4.
MED13 suppresses the deleterious impact of UF-linked mutations in MED12 on its CycC–CDK8/19 binding, but not its CycC–CDK8/19 stimulatory, activity. A and B, lysates from insect cells co-expressing baculovirus-produced CBP–MED13, CycCH6, MED12–HA (WT or mutant as indicated), and either CDK8–FLAG (A) or CDK19–FLAG (B) were subjected to IP with HA epitope-specific antibodies. HA-specific IPs were processed by Western blot (WB) using the indicated antibodies (top panels) or subjected to in vitro kinase assay prior to resolution by SDS-PAGE and PhosphorImager analyses (bottom panels). Input corresponds to 10% of cell lysate used in IP reactions. Horizontal arrows indicate the presence of specified Mediator subunits in parallel reactions. WBs were quantified and levels of MED13, CycC, and CDK8 (A) or CDK19 (B) in each IP were normalized to those of MED12 and expressed relative to their corresponding normalized levels in the CDK8 (or CDK19)–CycC–MED12 WT–MED13 IP (middle panel). 32P-Labeled GST–CTD levels in each IP–kinase reaction were quantified and expressed relative to the level in the CDK8 (or CDK19)–CycC–MED12–MED13 WT IP. Data represent the average ± S.E. of 3 independent experiments. Asterisks denote statistically significant differences versus WT (Student's t test, ***, p < 0.001).
MED13 binds to the MED12 C terminus that otherwise disrupts the interaction of UF-linked MED12 mutant derivatives with CycC–CDK8/19
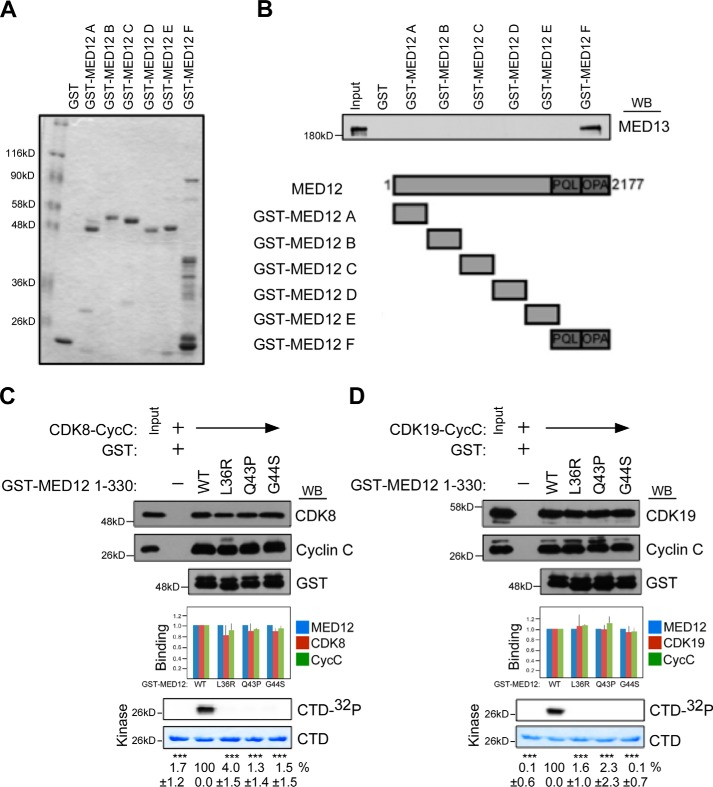
Within the Mediator kinase module, we and others have previously shown that MED12 occupies a central architectural role as a link between MED13 and CycC–CDK8 (11, 23). Thus, MED12 binds directly to CycC, which in turn binds to CDK8. MED12 also binds directly to MED13, which does not bind to either CycC or CDK8. To understand how MED13 suppresses the deleterious impact of UF-linked mutations in MED12 on its interaction with CycC–CDK8/19, we mapped the MED13-binding domain on MED12. To this end, individual GST–MED12 protein fragments spanning the entire length of the MED12 protein were tested for their respective abilities to bind recombinant baculovirus-expressed full-length MED13. This analysis identified the principal MED13-binding domain on MED12 to encompass the C terminus of MED12 (aa 1616–2177) (Fig. 5, A and B).
Figure 5.
MED12 binding is necessary but not sufficient for CycC–CDK8/19 activation. A and B, glutathione-Sepharose-immobilized GST-MED12 fragments A–F (spanning the full-length MED12 protein), as indicated, were incubated with whole cell lysates from insect cells expressing baculovirus-produced MED13. Bound proteins were eluted with Laemmli sample buffer, resolved by SDS-PAGE, and visualized by Coomassie Blue staining (A) or immunoblot analysis using MED13-specific antibody (B, top panel). Schematic diagram of MED12 derivatives used for in vitro binding reactions is also depicted (B, bottom panel). Input corresponds to 10% cell lysate used in binding reactions. C and D, glutathione-Sepharose-immobilized GST–MED12(1–330) derivatives (WT or mutants L36R, Q43P, G44S) as indicated were incubated with whole cell lysates from insect cells co-expressing baculovirus-produced CDK8–FLAG/CycCH6 (C) or CDK19–FLAG/CycCH6 (D). Bound proteins were either eluted with Laemmli sample buffer and processed by Western blot (WB) using the indicated antibodies (top panels) or subjected to in vitro kinase assay prior to resolution by SDS-PAGE and PhosphorImager analyses (bottom panels). Input corresponds to 10% of cell lysate used in binding reactions. Horizontal arrows indicate the presence of specified Mediator subunits in parallel reactions. 32P-Labeled GST–CTD levels were quantified and expressed relative to the level in the CDK8 (or CDK19)–CycC–MED12(1–330) WT reaction. Data represent the average ± S.E. of 3 independent experiments. Asterisks denote statistically significant differences versus WT (Student's t test, ***, p < 0.001).
The observation that MED13 binds to the MED12 C terminus prompted us to investigate whether the MED12 C terminus contributes to the inability of UF-linked MED12 mutant derivatives to bind to CycC–CDK8/19 in the absence of MED13. To do this, we tested the ability of WT and UF-linked MED12 mutant derivatives lacking their C termini to bind and activate CycC–CDK8/19. Thus, WT or mutant GST–MED12 truncation derivatives (comprising MED12 aa 1–330 and thus lacking the entire MED12 C terminus) were incubated with baculovirus-expressed CycC–CDK8/19 prior to processing of protein complexes by immunoblot analysis and in vitro kinase assay. Strikingly, UF-linked MED12 mutants L36R, Q43P, and G44S, none of which bound to CycC–CDK8/19 in the context of full-length MED12 (Fig. 3), nonetheless, bound CycC–CDK8/19 comparably to unmutated MED12 in the context of C-terminally-truncated MED12 (Fig. 5, C and D). Notably, despite their CycC–CDK8/19-binding proficiencies, all three UF-linked MED12 mutant derivatives were severely defective in their CycC–CDK8/19-stimulatory activities (Fig. 5, C and D), once again demonstrating that MED12 binding is necessary but not sufficient for CDK8/19 activation. Importantly, the CycC–CDK8/19-binding and stimulatory activities of all three UF-linked MED12 mutants examined in the context of C-terminally-truncated MED12 recapitulate those observed in the context of full-length MED12 in the presence of MED13. Because MED13 binds directly to the MED12 C terminus, we speculate that MED13 suppresses a UF mutation–induced conformational change in MED12, one involving its C terminus, which otherwise disrupts its association with CycC–CDK8/19. These findings provide new insight regarding subunit interaction dynamics within the Mediator kinase module and suggest that UF-linked mutations in MED12 disrupt Mediator kinase activity without uncoupling CycC–CDK8/19 from MED12 in Mediator.
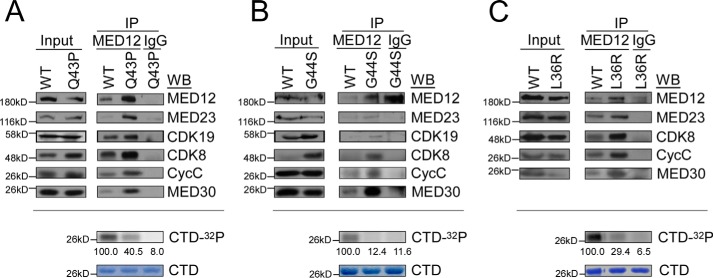
Mediator kinase activity is selectively disrupted in MED12 mutant UFs
To validate these findings in a clinically relevant setting, we comparatively assessed MED12-specific immunoprecipitates from patient-derived MED12 WT and mutant UF tumors for Mediator kinase module integrity and activity. For these experiments, matched tumor samples from patients harboring WT and L36R, Q43P, or G44S mutant MED12 tumors were used for analyses. Notably, all of the mutant MED12 derivatives were expressed and co-precipitated Mediator subunits comparably to WT MED12, indicating that UF-linked mutations in MED12 do not aberrantly affect its stable expression or incorporation into Mediator (Fig. 6, A–C, top panels). Strikingly, whereas CycC, CDK8, and CDK19 were comparably present in WT and mutant MED12–Mediator complexes, Mediator-associated CDK activity was nonetheless significantly impaired in the latter (Fig. 6, A–C, bottom panels). These findings reveal for the first time that Mediator kinase activity is uniquely disrupted in MED12-mutant UFs, and thus provide further evidence to implicate altered CDK8/19 activity in the pathogenesis of MED12 mutation-positive UFs.
Figure 6.
Mediator kinase activity is selectively disrupted in MED12–mutant UF tumors. A–C, whole tissue lysates from three different patient-matched uterine fibroid tumor sets, including MED12 WT and MED12 Q43P (A), G44S (B), or L36R (C) were subjected to IP with MED12-specific antibodies. MED12-specific IPs were resolved by SDS-10% PAGE and processed by Western blot (WB) analysis using the indicated Mediator subunit-specific antibodies (top panels) or subjected to in vitro kinase assay prior to resolution by SDS-PAGE and PhosphorImager analyses (bottom panels). Input corresponds to 10% of tissue lysates used in IPs. 32P-Labeled GST–CTD levels were quantified and expressed relative those obtained in kinase reactions with WT MED12/Mediator IPs. Note that insufficient sample material in C precluded analysis of CDK19 levels by WB.
Discussion
Genetic and biochemical studies have established the Mediator kinase module as a major conduit of developmental and oncogenic signaling through Mediator, and much of its function and pathological dysfunction in signal-dependent gene regulation has been attributed to the MED12-dependent CDK8 activity. Comparably less is known regarding the function and regulation of the CDK8 paralog CDK19 within Mediator, including whether and how WT or oncogenic mutant forms of MED12 impact CDK19 activity. Nonetheless, because CDK19 is coexpressed with CDK8 in human myometrium and the two paralogs incorporate into Mediator in a mutually exclusive manner, these questions are particularly relevant to pathogenesis of MED12-mutant UFs, in which MED12 driver mutations account for ∼70% of tumors.
Canonical CDKs are typically activated sequentially, first through cyclin-induced structural alterations leading to partial activation, and subsequently, by phosphorylation-triggered T-loop reorientation that culminates in complete activation (43–45). Notably, CDK8 is an atypical CDK in several respects, and these unique features underlie its apparent distinct mode of activation. First, CDK8 is unique among CDKs in its mode of interaction with CycC. For example, the cyclin-associated element, present in all CDKs (PFTAIRE or PCTAIRE), is significantly divergent in CDK8 (SMSACRE) (25). Notably, CDK19 also harbors this unique cyclin-associated element, indicating that CDK19 may interact similarly with CycC (25). Second, CDK8 activation occurs in the absence of T-loop phosphorylation, and also relies on an alternative MED12-dependent mechanism through which requisite conformational change in its ATP-binding site is apparently achieved. The fact that CDK8 and CDK19 share extensive sequence homology within their corresponding CycC-binding and catalytic kinase domains suggested that CDK19, like CDK8, might also be activated by MED12. Herein, we confirmed this prediction and demonstrate for the first time that MED12 activates both CDK8 and CDK19 through a similar, if not identical, mechanism that involves direct binding of MED12 exon 2-encoded sequences with a negatively charged surface groove on CycC. We found that disruption of the MED12–CycC interface by reciprocal mutations on either face impaired CDK8/19 activity, thus invoking allostery as a mechanistic basis for MED12-dependent CDK8/19 activation. Our observation that CDK8 and CDK19 are similarly activated by MED12 suggests that differences in gene expression programs controlled by the two paralogs likely reflect differences in their tissue-specific expression patterns and/or respective substrate selectivities as opposed to inherent differences in their biochemical mode of regulation in Mediator. Alternatively, or additionally, it is possible that differences in their respective gene regulatory properties could reflect unique kinase-independent functions for CDK8 and CDK19 in Mediator (46).
Previously, we showed that UF-linked driver mutations in MED12 disrupt its ability to bind and activate CycC–CDK8 in Mediator (23). Herein, we demonstrate that the same tumorigenic mutations in MED12 also disrupt its CycC–CDK19 binding and activation function, suggesting that loss of composite Mediator-associated kinase activity is likely a principal biochemical defect arising from these pathogenic mutations. We validated these findings in patient-derived UF tumors samples, revealing for the first time that Mediator kinase activity is selectively impaired in MED12 mutant tumors compared with MED12 WT tumors. Notably, our biochemical studies indicate that tumorigenic MED12 exon 2 mutations are loss of function mutations, a conclusion at apparent odds with a recent report based on a genetic mouse model that mutant MED12 drives UF formation through an apparent gain of function mechanism (37). In this genetic model, somatic expression of a mutant MED12 transgene was sufficient to drive UF formation on an X chromosome (endogenous) MED12 null or WT background (37). However, the observation that a mutant MED12 transgene drives UF formation in mice need not necessarily be construed as evidence that MED12 mutations are gain of function in nature, as MED12 is an integral subunit of the multiprotein Mediator. In this genetic model, MED12 in Mediator likely undergoes replacement and/or reciprocal exchange with ectopic mutant MED12, resulting in overall impaired Mediator kinase activity. Thus, mutational loss of MED12 function could be a tumorigenic trigger, implicating MED12 as possible tumor suppressor in the myometrium.
Interestingly, we observed quantitative differences among individual UF-linked MED12 exon 2 mutations with regard to their disruptive impact on Mediator-associated kinase activity, indicating that these mutations are not biochemically equivalent and may therefore span a range of tumorigenic potential in vivo. Thus, among Mediator complexes recovered from UF tissues, those harboring MED12 mutant G44S bore significantly less kinase activity than those harboring MED12 mutants L36R or Q43P. Whether these observations reflect a functional hierarchy among MED12 residues Gly-44, Leu-36, and Gln-43 within its overall mechanism of CDK8/19 activation will require further biochemical analyses using recombinant WT and mutant MED12 derivatives. Nonetheless our findings implicate altered CDK8/19 kinase activity in the pathogenesis of MED12-mutant UFs and thus identify prospective druggable targets for therapeutic intervention in this specific genetic setting.
The underlying basis by which impaired Mediator kinase activity contributes to UF pathogenesis is presently unclear, but likely involves dysregulation of signal-dependent gene expression programs that control myometrial stem cell fate. In this regard, the prevailing model for UF pathogenesis invokes the genetic transformation of a single myometrial stem cell into a tumor-initiating cell (UF stem cell) that seeds and sustains clonal tumor growth, characterized by cell proliferation and abundant extracellular matrix production, under the influence of endocrine, autocrine, and paracrine growth factor and hormone receptor signaling (47). Given the critical role of the Mediator kinase module as an integrative hub for cell signaling in the nucleus, disruption of Mediator kinase activity through oncogenic mutations in MED12 could thus trigger widespread transcriptional reprogramming that drives myometrial stem cell transformation. To identify such programs, we previously comparatively profiled the transcriptomes MED12 WT and MED12 mutant UF tumors (36, 48). These analyses revealed unique gene expression signatures characteristic of each genetic tumor type, and further permitted stratification of UFs into different molecular subtypes on the basis of distinct recurrent genetic alterations (i.e. driver mutations) and their corresponding unique gene expression profiles, indicative of separate pathways to tumorigenesis. Future studies comparing gene expression changes specifically in the UF stem cell compartment associated with tumorigenic mutations in MED12 as well as those incurred through independent disruption of CDK8/19 function should clarify the relative contribution of the latter to mutant MED12-driven tumorigenic pathways.
Our analyses of UF-linked mutations in MED12 and their impact on its CycC–CDK8/19 binding and activation functions has further revealed new insight concerning allosteric regulation of CDK8/19 by MED12 as well as hitherto unknown dynamic subunit interactions with the Mediator kinase module. In this regard, analysis of WT and mutant MED12-containing kinase modules reconstituted without and with MED13 revealed that MED13 binds to the MED12 C terminus, thereby suppressing a mutation-induced conformational change in MED12 that otherwise disrupts its association of CycC–CDK8/19. Thus, as validated in patient-derived tumors, UF-linked MED12 mutations disrupt Mediator kinase activity without uncoupling CycC–CDK8/19 from core Mediator (Fig. S5). These findings reveal that MED12 binding is necessary but not sufficient for CycC–CDK8/19 kinase activation and identify an additional step in the activation process that is essentially dependent on MED12 residues mutated in UFs and other tumor types. The biochemical nature of this additional step in the CycC–CDK8/19 activation process is presently unclear. We consider it likely that MED12 residues mutated in UFs and other tumors, including Leu-36, Gln-43, and Gly-44, normally trigger a structural reconfiguration in the CDK8 activation loop (T-loop) sufficient to promote an open activation segment, rearrangements typically driven in other CDKs by T-loop phosphorylation that is absent in CDK8. These structural alterations could impact ATP and/or substrate binding. Further studies, including X-ray crystallographic analyses of WT and mutant MED12 derivatives in complex with CycC–CDK8/19 should help to resolve these biochemical details.
Finally, our finding that UF-linked MED12 mutations disrupt Mediator kinase activity without uncoupling CycC–CDK8/19 from core Mediator, validated in clinical tumor samples, has possible implications for medical treatment of UFs. Current treatment options for UFs are primarily surgical or radiological and range from hysterectomy or myomectomy to minimally invasive options, including uterine artery embolization and magnetic resonance-guided focused ultrasound (MRgFUS) (49–51). However, the deleterious impact of these procedures on reproductive function is clear (hysterectomy) or controversial (uterine artery embolization, magnetic resonance-guided focused ultrasound), rendering these options unsuitable for women who wish to retain future fertility (49–51). Likewise, hormonal therapies designed to blunt the growth-stimulatory effects of estrogen or progesterone on UF growth are contraindicated in women actively pursuing a pregnancy, and are otherwise approved only for short-term use due to long-term safety concerns (34, 49, 52, 53). Accordingly, there is a critical need to develop effective, safe, long-term, and fertility-compatible non-surgical treatment options for UF management. Mechanistically, our finding that catalytically impaired CDK8/19 are nonetheless physically retained in Mediator suggests a possible avenue to rescue abeyant Mediator-associated kinase activity, as for example, with small molecule activators. This strategy, if effective, would ensure reactivation of CDK8/19 in its appropriate physiological context as an integral component of chromatin-bound Mediator. Future studies should clarify whether CDK8/19 represent viable therapeutic targets for the clinical management and/or treatment of UF patients.
Experimental procedures
Antibodies
Antibodies used for immunoprecipitation and immunoblotting assays were as follows: anti-MED12, Bethyl A300–774A; anti-MED13, Bethyl A301-278A; anti-CDK8, Santa Cruz sc-1521; anti-CDK19, Sigma HPA007053; anti-CycC, BD Pharmingen 558903; anti-GST, GE Healthcare 27-457701. Rabbit polyclonal antibodies specific for MED30 and MED4 antibodies are generated as previously described (19).
Cloning and mutagenesis
MED12 cDNA sequence encoding amino acids 1–330 were PCR amplified and cloned into pGEX6P-1. Site-directed mutagenesis of MED12 to generate L36R, Q43P, and G44S mutants was performed using a MED12 fragment spanning amino acids in 1–330 in pGEX6P-1. The baculovirus transfer vector pFastBac1-MED12-HA carrying a C-terminal hemagglutinin (HA) epitope tag appended to full-length MED12 has been described previously (21). A CDK19 derivative carrying a C-terminal FLAG epitope tag (CDK19-FLAG) was generated by PCR amplification and cloned into pFastBac1. Additional pFastBac1 transfer vectors carrying CDK8–FLAG (C-terminally FLAG-tagged CDK8), CycCH6 (C-terminally His6-tagged CycC), and CBP-MED13 (MED13 N-terminally tagged with calmodulin-binding peptide) were generated as previously described (21).
Baculovirus protein expression
pFASTBAC1 donor plasmids encoding MED12-HA, CDK8–FLAG, CDK19–FLAG, CycCH6, and CBP–MED13 were transformed into DH10Bac competent cells (Invitrogen). Following transformation and transposition of MED12–HA, CDK8–FLAG, CDK19–FLAG, CycCH6, and CBP–MED13 DNA open reading frames into bacmid DNAs, colonies containing recombinant bacmids were identified by disruption of the lacZ gene inside the bacmid DNA. The isolated recombinant bacmid DNAs from white LacZ− colonies, which were confirmed by PCR, were used for transfection of Sf9 insect cells. After three rounds of viral amplification, high-titer baculoviruses were used for infection of insect High Five cells (Invitrogen). 48 h postinfection, cells were resuspended and lysed with 0.5 m D buffer (500 mm NaCl, 0.1% Nonidet P-40, 0.1 mm EDTA, pH 8.0, 10% glycerol, 20 mm Hepes pH 7.9) using a Dounce homogenizer at 4 °C. Lysates were clarified by centrifugation at 15,000 rpm for 30 min.
Immunoprecipitation/kinase assays from insect cells
Whole cell lysates from insect cells expressing MED12–HA, CycCH6, and either CDK8–FLAG or CDK19–FLAG were subjected to FLAG-specific immunoprecipitation (IP) for 2 h at 4 °C in 0.2 m NaCl, D buffer and either eluted in Laemmli sample buffer followed by SDS-10% PAGE for immunoblot analysis using the indicated antibodies, or incubated with kinase reaction buffer (25 mm Tris, pH 7.5, 20 mm MgCl2), 2.5 mCi of [γ-32P]ATP, and 2 μg of purified GST or GST-3xCTD substrate. Reactions were incubated for 30 min at 30 °C, eluted in Laemmli sample buffer, processed by SDS-12% PAGE, stained with Coomassie Blue stain, and visualized by PhosphorImager analysis. 32P-Labeled GST–CTD was quantified using ImageQuant software, and the level of phosphorylation in each IP/kinase reaction was expressed relative to that obtained in IP/kinase reactions reconstituted with CycC/CDK8/MED12 WT or CycC/CDK19/MED12 WT.
Whole cell lysates from High Five cells expressing CBP–MED13, MED12–HA–WT or MED12–HA–Mutants (L36R, Q43P, G44S), CycCH6, and CDK8–FLAG or CDK19–FLAG were subjected to HA-specific IPs. IPs were performed for 2 h at 4 °C in 0.2 m NaCl, D buffer (200 mm NaCl, 0.1% Nonidet P-40, 0.1 mm EDTA, pH 8.0, 10% glycerol, 20 mm Hepes, pH 7.9). Immune complexes were washed in 0.2 m NaCl, D buffer and either eluted in Laemmli sample buffer followed by SDS-10% PAGE for immunoblot analysis, or subjected to kinase assay, which contained 25 mm Tris, pH 7.5, 20 mm MgCl2, 2.5 mCi of [γ-32P]ATP, and 2 μg of purified GST or GST-3xCTD substrate. 32P-Labeled GST-3xCTD was resolved by SDS-12% PAGE, stained with Coomassie stain and visualized by PhosphoImager analysis. The 32P-labeled GST-3xCTD was quantitated using ImageQuant software, and levels of phosphorylation obtained in IP/kinase reactions reconstituted with MED12 mutant derivatives were relatively compared with levels obtained in reactions reconstituted with MED12 WT.
GST pulldown and kinase assays
Lysates from Escherichia coli BL21 cells expressing GST–MED12(1–330) WT or Mutant (L36R, Q43P, G44S) derivatives were incubated with glutathione-Sepharose 4B for 1 h at 4 °C. Beads were washed 4 times with Lysis250 (50 mm Tris, pH 7.5, 250 mm NaCl, 5 mm EDTA) and immobilized GST–MED12(1–330) derivatives were then incubated with whole cell lysates from insect High Five cells expressing CycCH6/CDK8–FLAG or CycCH6/CDK19–FLAG for 1 h at 4 °C. Complexes were washed 4 times in Lysis250 and either eluted in Laemmli sample buffer followed by SDS-10% PAGE for immunoblot analysis, or incubated with kinase reaction buffer (25 mm Tris, pH 7.5, 20 mm MgCl2), 2.5 mCi of [γ-32P]ATP and 2 μg of purified GST or GST-3xCTD substrate. Reactions were incubated for 30 min at 30 °C, eluted in Laemmli sample buffer, processed by SDS-12% PAGE, and stained with Coomassie stain and visualized by PhosphorImager analysis. 32P-Labeled GST–CTD was quantified using ImageQuant software, and the level of phosphorylation in each IP/kinase reaction is expressed relative to that obtained in reactions reconstituted with CycC–CDK8–MED12(1–330) WT or CycC–CDK19–MED12(1–330) WT complexes.
Immunoprecipitation from human tissue samples
Written informed consent was obtained from patients and these studies were approved by the appropriate ethics review board of the Helsinki University Central Hospital, Finland, and Institutional Review Board of the University of Texas Health Science Center at San Antonio. Fresh frozen tissues, including myometrium and UFs (MED12 mutation negative and MED12 mutations positive) from women undergoing hysterectomy or myomectomy were used for study purposes. All samples were screened for MED12 exons 1 and 2 mutations using Sanger sequencing as reported previously (28, 36).
Tissues were homogenized at 4 °C in protein lysis buffer (40 mm Tris, pH 7.4, 500 mm NaCl, 0.5% sodium deoxycholic acid, 1% Triton X-100, and 1 mm EDTA). Tissue homogenates were precleared by incubation with protein A-agarose. Anti-MED12 antibody, covalently coupled to protein A-agarose, was mixed with each sample. As a negative control, tissue homogenates were incubated with normal rabbit IgG–agarose conjugate (Santa Cruz Biotechnology, Inc.). Immunoprecipitation reactions were carried out for 3 h at 4 °C on a rocking platform. The beads were washed three times with wash buffer (40 mm Tris, pH 7.4, 500 mm NaCl, 1 mm EDTA). Immunoprecipitates were either eluted in Laemmli sample buffer prior to SDS-10% PAGE for immunoblot analysis, or subjected to an in vitro kinase assay in the presence of 25 mm Tris, pH 7.5, 20 mm MgCl2, 2.5 mCi of [γ-32P]ATP and 2 μg of purified GST or GST-3xCTD substrate. The 32P-labeled GST-3xCTD was quantitated using ImageQuant software, and phosphorylation levels obtained in IP/kinase reactions from MED12-mutant tumor lysates were relatively compared with those in patient-matched MED12 WT tumor lysates.
Author contributions
M. J. P., H. S., J. M. S., S. E. B., A. A.-H., R. S. S., L. A. A., and T. G. B. conceptualization; M. J. P. data curation; M. J. P., H. S., J. M. S., S. E. B., A. A.-H., R. S. S., and T. G. B. formal analysis; M. J. P., H. S., J. M. S., J. H. T., C. F., J. F. K., J. M., S. K. H., and Q. Y. investigation; M. J. P., H. S., J. M. S., J. H. T., C. F., J. F. K., J. M., S. K. H., and Q. Y. methodology; M. J. P., S. E. B., A. A.-H., R. S. S., L. A. A., and T. G. B. writing-original draft; M. J. P., S. E. B., A. A.-H., R. S. S., L. A. A., and T. G. B. writing-review and editing; A. A.-H., R. S. S., L. A. A., and T. G. B. supervision; T. G. B. funding acquisition; T. G. B. project administration.
Supplementary Material
Acknowledgments
We thank members of the Boyer laboratory and P. Renee Yew for helpful discussions.
This work was supported by U.S. Dept. of Health and Human Services, National Institute of Health Grant R01HD087417 (to T. G. B.) and the National Center for Advancing Translational Sciences, National Institutes of Health, through the Clinical and Translational Science Award (CTSA) UL1 TR001120. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5.
- Pol II
- RNA polymerase II
- UFs
- uterine fibroids
- CTD
- C-terminal domain
- IP
- immunoprecipitation
- HA
- hemagglutinin
- CBP
- calmodulin-binding peptide
- GST
- glutathione S-transferase
- CDK
- cyclin-dependent kinase
- CycC
- cyclin C
- aa
- amino acid(s).
References
- 1. Kornberg R. D. (2005) Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235–239 10.1016/j.tibs.2005.03.011 [DOI] [PubMed] [Google Scholar]
- 2. Malik S., and Roeder R. G. (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 10.1038/nrg2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clark A. D., Oldenbroek M., and Boyer T. G. (2015) Mediator kinase module and human tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 50, 393–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eychenne T., Werner M., and Soutourina J. (2017) Toward understanding of the mechanisms of Mediator function in vivo: focus on the preinitiation complex assembly. Transcription 8, 328–342 10.1080/21541264.2017.1329000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allen B. L., and Taatjes D. J. (2015) The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 10.1038/nrm3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cevher M. A., Shi Y., Li D., Chait B. T., Malik S., and Roeder R. G. (2014) Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 21, 1028–1034 10.1038/nsmb.2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nozawa K., Schneider T. R., and Cramer P. (2017) Core Mediator structure at 3.4-Å extends model of transcription initiation complex. Nature 545, 248–251 10.1038/nature22328 [DOI] [PubMed] [Google Scholar]
- 8. Tsai K. L., Tomomori-Sato C., Sato S., Conaway R. C., Conaway J. W., and Asturias F. J. (2014) Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 158, 463 10.1016/j.cell.2014.06.036 [DOI] [PubMed] [Google Scholar]
- 9. Tsai K. L., Yu X., Gopalan S., Chao T. C., Zhang Y., Florens L., Washburn M. P., Murakami K., Conaway R. C., Conaway J. W., and Asturias F. J. (2017) Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201 10.1038/nature21393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harper T. M., and Taatjes D. J. (2017) The complex structure and function of Mediator. J. Biol. Chem. September 14 10.1074/jbc.R117.794438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsai K. L., Sato S., Tomomori-Sato C., Conaway R. C., Conaway J. W., and Asturias F. J. (2013) A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 20, 611–619 10.1038/nsmb.2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bourbon H. M. (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 10.1093/nar/gkn349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daniels D. L., Ford M., Schwinn M. K., Benink H., Galbreath M. D., Amunugama R., Jones R., Allen D., Okazaki N., Yamakawa H., Miki F., Nagase T., Espinosa J. M., and Urh M. (2013) Mutual exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs revealed with the CDK-Mediator kinase module. J. Proteomics Bioinform. S2, 004 10.4172/jpb.S2-004 [DOI] [Google Scholar]
- 14. Al-Hendy A., Laknaur A., Diamond M. P., Ismail N., Boyer T. G., and Halder S. K. (2017) Silencing Med12 gene reduces proliferation of human leiomyoma cells mediated via Wnt/β-catenin signaling pathway. Endocrinology 158, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carrera I., Janody F., Leeds N., Duveau F., and Treisman J. E. (2008) Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc. Natl. Acad. Sci. U.S.A. 105, 6644–6649 10.1073/pnas.0709749105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang S., Hölzel M., Knijnenburg T., Schlicker A., Roepman P., McDermott U., Garnett M., Grernrum W., Sun C., Prahallad A., Groenendijk F. H., Mittempergher L., Nijkamp W., Neefjes J., Salazar R., et al. (2012) MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signaling. Cell 151, 937–950 10.1016/j.cell.2012.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Janody F., Martirosyan Z., Benlali A., and Treisman J. E. (2003) Two subunits of the Drosophila mediator complex act together to control cell affinity. Development 130, 3691–3701 10.1242/dev.00607 [DOI] [PubMed] [Google Scholar]
- 18. Janody F., and Treisman J. E. (2011) Requirements for mediator complex subunits distinguish three classes of notch target genes at the Drosophila wing margin. Dev. Dyn. 240, 2051–2059 10.1002/dvdy.22705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S., Xu X., Hecht A., and Boyer T. G. (2006) Mediator is a transducer of Wnt/β-catenin signaling. J. Biol. Chem. 281, 14066–14075 10.1074/jbc.M602696200 [DOI] [PubMed] [Google Scholar]
- 20. Zhou H., Kim S., Ishii S., and Boyer T. G. (2006) Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol. Cell. Biol. 26, 8667–8682 10.1128/MCB.00443-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou H., Spaeth J. M., Kim N. H., Xu X., Friez M. J., Schwartz C. E., and Boyer T. G. (2012) MED12 mutations link intellectual disability syndromes with dysregulated GLI3-dependent Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 19763–19768 10.1073/pnas.1121120109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knuesel M. T., Meyer K. D., Donner A. J., Espinosa J. M., and Taatjes D. J. (2009) The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 29, 650–661 10.1128/MCB.00993-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Turunen M., Spaeth J. M., Keskitalo S., Park M. J., Kivioja T., Clark A. D., Mäkinen N., Gao F., Palin K., Nurkkala H., Väahärautio A., Aavikko M., Kämpjärvi K., Vahteristo P., Kim C. A., et al. (2014) Uterine leiomyoma-linked MED12 mutations disrupt Mediator-associated CDK activity. Cell Rep. 7, 654–660 10.1016/j.celrep.2014.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider E. V., Böttcher J., Blaesse M., Neumann L., Huber R., and Maskos K. (2011) The structure of CDK8/CycC implicates specificity in the CDK/cyclin family and reveals interaction with a deep pocket binder. J. Mol. Biol. 412, 251–266 10.1016/j.jmb.2011.07.020 [DOI] [PubMed] [Google Scholar]
- 25. Xu W., and Ji J. Y. (2011) Dysregulation of CDK8 and Cyclin C in tumorigenesis. J. Genet. Genomics 38, 439–452 10.1016/j.jgg.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alderton G. K. (2014) Transcription: mediating tumorigenesis. Nat. Rev. Cancer 14, 382 10.1038/nrc3755 [DOI] [PubMed] [Google Scholar]
- 27. Lim W. K., Ong C. K., Tan J., Thike A. A., Ng C. C., Rajasegaran V., Myint S. S., Nagarajan S., Nasir N. D., McPherson J. R., Cutcutache I., Poore G., Tay S. T., Ooi W. S., Tan V. K., et al. (2014) Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat. Genet. 46, 877–880 10.1038/ng.3037 [DOI] [PubMed] [Google Scholar]
- 28. Mäkinen N., Mehine M., Tolvanen J., Kaasinen E., Li Y., Lehtonen H. J., Gentile M., Yan J., Enge M., Taipale M., Aavikko M., Katainen R., Virolainen E., Böhling T., Koski T. A., et al. (2011) MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 334, 252–255 10.1126/science.1208930 [DOI] [PubMed] [Google Scholar]
- 29. Mehine M., Mäkinen N., Heinonen H. R., Aaltonen L. A., and Vahteristo P. (2014) Genomics of uterine leiomyomas: insights from high-throughput sequencing. Fertil. Steril. 102, 621–629 10.1016/j.fertnstert.2014.06.050 [DOI] [PubMed] [Google Scholar]
- 30. Tan J., Ong C. K., Lim W. K., Ng C. C., Thike A. A., Ng L. M., Rajasegaran V., Myint S. S., Nagarajan S., Thangaraju S., Dey S., Nasir N. D., Wijaya G. C., Lim J. Q., Huang D., et al. (2015) Genomic landscapes of breast fibroepithelial tumors. Nat. Genet. 47, 1341–1345 10.1038/ng.3409 [DOI] [PubMed] [Google Scholar]
- 31. Kämpjärvi K., Järvinen T. M., Heikkinen T., Ruppert A. S., Senter L., Hoag K. W., Dufva O., Kontro M., Rassenti L., Hertlein E., Kipps T. J., Porkka K., Byrd J. C., de la Chapelle A., and Vahteristo P. (2015) Somatic MED12 mutations are associated with poor prognosis markers in chronic lymphocytic leukemia. Oncotarget 6, 1884–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kämpjarvi K., Mäkinen N., Kilpivaara O., Arola J., Heinonen H. R., Böhm J., Abdel-Wahab O., Lehtonen H. J., Pelttari L. M., Mehine M., Schrewe H., Nevanlinna H., Levine R. L., Hokland P., Böhling T., et al. (2012) Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br. J. Cancer 107, 1761–1765 10.1038/bjc.2012.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bulun S. E. (2013) Uterine fibroids. N. Engl. J. Med. 369, 1344–1355 10.1056/NEJMra1209993 [DOI] [PubMed] [Google Scholar]
- 34. Sabry M., and Al-Hendy A. (2012) Medical treatment of uterine leiomyoma. Reprod. Sci. 19, 339–353 10.1177/1933719111432867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stewart E. A. (2015) Clinical practice: UTerine fibroids. N. Engl. J. Med. 372, 1646–1655 10.1056/NEJMcp1411029 [DOI] [PubMed] [Google Scholar]
- 36. Kämpjärvi K., Park M. J., Mehine M., Kim N. H., Clark A. D., Bützow R., Böhling T., Böhm J., Mecklin J. P., Järvinen H., Tomlinson I. P., van der Spuy Z. M., Sjöberg J., Boyer T. G., and Vahteristo P. (2014) Mutations in Exon 1 highlight the role of MED12 in uterine leiomyomas. Hum. Mutat. 35, 1136–1141 10.1002/humu.22612 [DOI] [PubMed] [Google Scholar]
- 37. Mittal P., Shin Y. H., Yatsenko S. A., Castro C. A., Surti U., and Rajkovic A. (2015) Med12 gain-of-function mutation causes leiomyomas and genomic instability. J. Clin. Invest. 125, 3280–3284 10.1172/JCI81534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malumbres M., Harlow E., Hunt T., Hunter T., Lahti J. M., Manning G., Morgan D. O., Tsai L. H., and Wolgemuth D. J. (2009) Cyclin-dependent kinases: a family portrait. Nat. Cell Biol. 11, 1275–1276 10.1038/ncb1109-1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 40. Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A., Jin J., Cai Y., Washburn M. P., Conaway J. W., and Conaway R. C. (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 10.1016/j.molcel.2004.05.006 [DOI] [PubMed] [Google Scholar]
- 41. Tsutsui T., Umemura H., Tanaka A., Mizuki F., Hirose Y., and Ohkuma Y. (2008) Human mediator kinase subunit CDK11 plays a negative role in viral activator VP16-dependent transcriptional regulation. Genes Cells 13, 817–826 10.1111/j.1365-2443.2008.01208.x [DOI] [PubMed] [Google Scholar]
- 42. Tsutsui T., Fukasawa R., Tanaka A., Hirose Y., and Ohkuma Y. (2011) Identification of target genes for the CDK subunits of the Mediator complex. Genes Cells 16, 1208–1218 10.1111/j.1365-2443.2011.01565.x [DOI] [PubMed] [Google Scholar]
- 43. Huse M., and Kuriyan J. (2002) The conformational plasticity of protein kinases. Cell 109, 275–282 10.1016/S0092-8674(02)00741-9 [DOI] [PubMed] [Google Scholar]
- 44. Endicott J. A., Noble M. E., and Johnson L. N. (2012) The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81, 587–613 10.1146/annurev-biochem-052410-090317 [DOI] [PubMed] [Google Scholar]
- 45. Nolen B., Taylor S., and Ghosh G. (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 10.1016/j.molcel.2004.08.024 [DOI] [PubMed] [Google Scholar]
- 46. Audetat K. A., Galbraith M. D., Odell A. T., Lee T., Pandey A., Espinosa J. M., Dowell R. D., and Taatjes D. J. (2017) A kinase-independent role for cyclin-dependent kinase 19 in p53 response. Mol. Cell. Biol. 37, e00626–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moravek M. B., and Bulun S. E. (2015) Endocrinology of uterine fibroids: steroid hormones, stem cells, and genetic contribution. Curr. Opin. Obstet. Gynecol. 27, 276–283 10.1097/GCO.0000000000000185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mehine M., Kaasinen E., Heinonen H. R., Mäkinen N., Kämpjärvi K., Sarvilinna N., Aavikko M., Vähärautio A., Pasanen A., Bützow R., Heikinheimo O., Sjöberg J., Pitkanen E., Vahteristo P., and Aaltonen L. A. (2016) Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc. Natl. Acad. Sci. U.S.A. 113, 1315–1320 10.1073/pnas.1518752113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Levy G., Hill M. J., Beall S., Zarek S. M., Segars J. H., and Catherino W. H. (2012) Leiomyoma: genetics, assisted reproduction, pregnancy and therapeutic advances. J. Assist. Reprod. Genet. 29, 703–712 10.1007/s10815-012-9784-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martin J., Bhanot K., and Athreya S. (2013) Complications and reinterventions in uterine artery embolization for symptomatic uterine fibroids: a literature review and meta analysis. Cardiovasc. Intervent. Radiol. 36, 395–402 10.1007/s00270-012-0505-y [DOI] [PubMed] [Google Scholar]
- 51. van der Kooij S. M., Ankum W. M., and Hehenkamp W. J. (2012) Review of nonsurgical/minimally invasive treatments for uterine fibroids. Curr. Opin. Obstet. Gynecol. 24, 368–375 10.1097/GCO.0b013e328359f10a [DOI] [PubMed] [Google Scholar]
- 52. Islam M. S., Protic O., Giannubilo S. R., Toti P., Tranquilli A. L., Petraglia F., Castellucci M., and Ciarmela P. (2013) Uterine leiomyoma: available medical treatments and new possible therapeutic options. J. Clin. Endocrinol. Metab. 98, 921–934 10.1210/jc.2012-3237 [DOI] [PubMed] [Google Scholar]
- 53. Parsanezhad M. E., Azmoon M., Alborzi S., Rajaeefard A., Zarei A., Kazerooni T., Frank V., and Schmidt E. H. (2010) A randomized, controlled clinical trial comparing the effects of aromatase inhibitor (letrozole) and gonadotropin-releasing hormone agonist (triptorelin) on uterine leiomyoma volume and hormonal status. Fertil. Steril. 93, 192–198 10.1016/j.fertnstert.2008.09.064 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.