Abstract
As a master regulator of endothelial cell function, vascular endothelial growth factor receptor-2 (VEGFR2) activates multiple downstream signaling pathways that are critical for vascular development and normal vessel function. VEGFR2 trafficking through various endosomal compartments modulates its signaling output. Accordingly, proteins that regulate the speed and direction by which VEGFR2 traffics through endosomes have been demonstrated to be particularly important for arteriogenesis. However, little is known about how these proteins control VEGFR2 trafficking and about the implications of this control for endothelial cell function. Here, we show that Rab GTPase–binding effector protein 2 (RABEP2), a Rab-effector protein implicated in arteriogenesis, modulates VEGFR2 trafficking. By employing high-resolution microscopy and biochemical assays, we demonstrate that RABEP2 interacts with the small GTPase Rab4 and regulates VEGFR2 endosomal trafficking to maintain cell-surface expression of VEGFR2 and VEGF signaling. Lack of RABEP2 also led to prolonged retention of VEGFR2 in Rab5-positive sorting endosomes, which increased VEGFR2's exposure to phosphotyrosine phosphatase 1b (PTP1b), causing diminished VEGFR2 signaling. Finally, the loss of RABEP2 increased VEGFR2 degradation by diverting VEGFR2 to Rab7-positive endosomes destined for the lysosome. These results implicate RABEP2 as a key modulator of VEGFR2 endosomal trafficking, and demonstrate the importance of RABEP2 and Rab4 for VEGFR2 signaling in endothelial cells.
Keywords: angiogenesis, endothelial cell, Rab, receptor endocytosis, vascular endothelial growth factor (VEGF)
Introduction
The vascular endothelial growth factor receptor-2 (VEGFR2)2 signaling pathway plays a central role in vascular development. Deletion of VEGFR2 causes embryonic lethality while haploinsufficiency of its major ligand, VEGFA, is incompatible with life (1–3). VEGFR2 exerts its influential role in the formation of a functional cardiovascular system by mediating multiple vascular processes, including vasculogenesis, angiogenesis, and arteriogenesis (4). The role of VEGF in regulating vasculogenesis, the formation of the primitive vascular plexus, and angiogenesis, the sprouting of new blood vessels from pre-existing vasculature, has been extensively studied (5–7). However, we are only just beginning to understand VEGFR2-mediated signaling mechanisms that govern the formation of arteries through the process of arteriogenesis.
VEGFR2 mediates multiple cellular events involved in arteriogenesis, including endothelial cell proliferation, migration, survival, and lumen expansion through a complex series of downstream signaling pathways that include the phosphatidylinositol 3′-kinase (PI3K)/Akt and mitogen-activated protein kinase (MAPK)/extracellular response kinase (ERK) signaling cascades (8–11). Activation of ERK signaling has been demonstrated to be of particular importance for arteriogenesis, with decreased activation of ERK leading to impaired artery formation during both development and disease (12–14).
Not surprisingly, VEGF signaling is tightly controlled at multiple levels (15). Recently, much attention has been given to VEGF receptors endosomal trafficking in endothelial cells (16–18). The speed and direction by which VEGFR2 traffics through different endosomal compartments add additional layers to VEGFR2 signal modulation and output (19, 20). Upon ligand binding, VEGFR2 dimerization induces receptor autophosphorylation and internalization via clathrin-coated pits, after which VEGFR2 traffics through different endosomal compartments by a process mediated by Rab GTPases (Rab5, Rab4, Rab11, and Rab7) and Rab effector proteins (21–24). VEGFR2 first enters Rab5-positive (Rab5+) sorting endosomes to either be returned to the cell surface by Rab4+ (fast) or Rab11+ (slow) endosomal recycling pathways for additional rounds of signaling, or to the lysosome for degradation via Rab7+ endosomes (25–27). In contrast to receptor tyrosine kinase dogma that stipulates receptor tyrosine kinases signal exclusively at the plasma membrane, VEGFR2 continues to signal following internalization as it moves through these various endosomal compartments (13, 28). Therefore, endothelial mutations that alter VEGFR2 trafficking can consequentially impair VEGFR2 signaling, and thus endothelial cell function (13, 29, 30). For example, genetic disruption of retrograde endocytic vesicle transport slowed VEGFR2 trafficking through the Rab5 endosomal compartment and caused reduced VEGFR2 phosphorylation, reduced ERK activation, and consequently, impaired arteriogenesis (13).
In the absence of ligand, VEGFR2 is also internalized and trafficked through the cell. Quiescent VEGFR2 has been documented to be constitutively internalized, first entering the Rab5+ endosomal compartment and then returned to the plasma membrane by a fast-loop recycling process mediated by Rab4+ endosomes (26, 27). Little is known about constitutive Rab4-dependent trafficking of VEGFR2 and its implications for endothelial cell function.
Rab-GTPase effector protein-2 (RABEP2), a Rab effector protein with putative Rab4- and Rab5-binding sites documented to regulate early endosomal fusion, has recently been implicated in arteriogenesis (31). Analyses of congenic mouse strains that display varying degrees of arteriogenesis revealed an association between mutations in Rabep2 and impaired artery formation (32). Moreover, mice deficient for Rapeb2 displayed significant arteriogenic defects (33). Here we examined the role of RABEP2 in regulating VEGFR2 signaling in endothelial cells. We demonstrate that RABEP2 is required to maintain VEGFR2 protein expression and signaling. In the absence of RABEP2, VEGFR2 fails to recycle to the plasma membrane and is instead retained and de-activated in the Rab5+ compartment to then be trafficked to lysosomes for degradation. We show that RABEP2 mediates Rab4-dependent trafficking of VEGFR2. Thus, through its interaction with RAB4, RABEP2 plays a critical role in regulating VEGFR2 signaling events required for arteriogenesis.
Results
RABEP2 regulates VEGFR2 expression and signaling
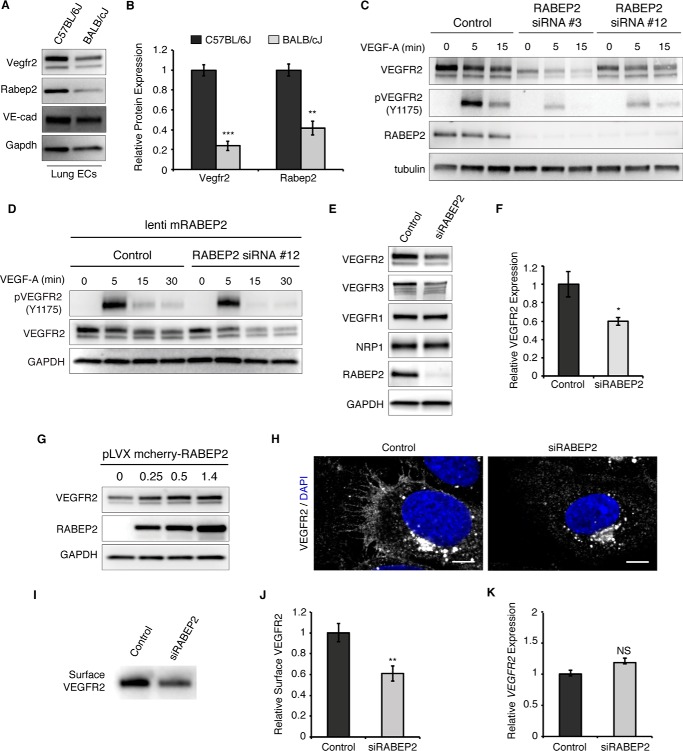
Activation of VEGFR2 signaling is central to new artery formation. To evaluate the link between VEGFR2 expression and arteriogenesis, we compared VEGFR2 expression in endothelial cells derived from mice showing robust (C57Bl/6) and decreased (BALB/c) arteriogenesis (34). In agreement with these phenotypes, VEGFR2 levels were significantly higher in C57Bl/6J endothelial cells (Fig. 1, A and B). A recent publication linked a gene encoding the Rab-effector protein, RABEP2, to the observed reduction in collateral blood vessel number and impaired arteriogenesis displayed by the BALB/c congenic mouse strain (33). In line with this observation, BALB/cJ endothelial cells displayed a 50% reduction in RABEP2 protein expression, as compared with C57Bl/6J (Fig. 1, A and B). These findings suggested that the published in vivo arteriogenesis phenotypes of RABEP2 mutant mice may be due to deficient VEGFR2 activity. We thus hypothesized that RABEP2 regulates VEGFR2 function in endothelial cells and proceeded to assess this hypothesis in vitro.
Figure 1.
RABEP2 regulates VEGFR2 expression. A, protein lysate isolated from either C57Bl/6J or BALB/cJ lung endothelial cells was probed for Vegfr2, Rabep2, and VE-cadherin (VE-cad) to validate endothelial cell identity. Gapdh served as a loading control. B, quantification of Western blot showing relative Vegfr2 and Rabep2 protein expression. Quantification based on 3 independent cell isolations (mean ± S.D., **, p < 0.01; ***, p < 0.01). C, validation of RABEP2 siRNA. HUVEC were exposed to two different siRNA with unique human RABEP2 targeting sequences (RABEP2 siRNA #3 and RABEP2 siRNA #12). Both siRNA constructs depleted RABEP2 expression and caused reduced VEGFR2 expression and activation. RABEP2 siRNA #12 was chosen for all future experiments, as it had the least impact on cell growth. D, specificity of RABEP2 siRNA #12 validated by showing lentiviral infection with mouse RABEP2 construct (mRABEP2) rescues VEGFR2 expression and activation. E, Western blot of protein lysates isolated from HUVEC transfected with scrambled siRNA (control) or siRNA targeting RABEP2 (siRABEP2). F, quantification of the relative total VEGFR2 protein expression based on 3 independent experiments (mean ± S.D., *, p < 0.05). G, Western blot of total VEGFR2 expression following increasing levels of RABEP2 overexpression via lentiviral infection in HUVEC. H, immunocytochemistry of control or siRABEP2 HUVEC stained for VEGFR2 (white) and DAPI (blue) to mark cell nuclei. I, Western blot probed for VEGFR2 of total surface-biotinylated protein isolated from control or siRABEP2 HUVEC. J, quantification of relative surface VEGFR2 levels based on 3 independent surface biotinylation assays (mean ± S.D., **p < 0.01). K, relative VEGFR2 transcript levels in control and siRABEP2 HUVEC (mean ± S.D., NS, not significant).
To determine whether RABEP2 regulates VEGFR2 expression in endothelial cells, we used silencing RNA to target RABEP2 transcripts (siRABEP2) and assessed VEGFR2 expression after validating knockdown efficiency and specificity (Fig. 1, C and D). We chose to use human umbilicus venous endothelial cells (HUVEC) to assess RABEP2 function in endothelial cells, since HUVEC are a firmly established cell model in vascular biology; and despite their name, HUVEC are derived from the lining of umbilical blood vessels that carry arterial blood and express markers of arterial endothelial cells (35). HUVEC with RABEP2 knockdown displayed statistically significant reduced VEGFR2 protein expression, whereas expression of VEGFR1 and the VEGFR2 co-receptor, Neuropilin-1 (NRP1), remained unchanged (Fig. 1, E and F). In agreement with previously published work demonstrating that loss of VEGFR2 caused reduced VEGFR3 expression, we also observed reduced VEGFR3 protein levels in HUVEC deficient for RABEP2 (Fig. 1E) (36, 37). To further support our finding that RABEP2 regulates VEGFR2 protein levels, overexpression of RABEP2 resulted in a dose-dependent increase in total VEGFR2 protein expression (Fig. 1G). Immunocytochemistry and surface biotinylation demonstrated that surface levels of VEGFR2 were reduced in HUVEC following RABEP2 knockdown, as compared with control (Fig. 1, H–J). Interestingly, VEGFR2 transcript levels were unchanged in HUVEC deficient for RABEP2 (Fig. 1K). Taken together, these data suggest that RABEP2 functions post-transcriptionally to regulate VEGFR2 expression in endothelial cells.
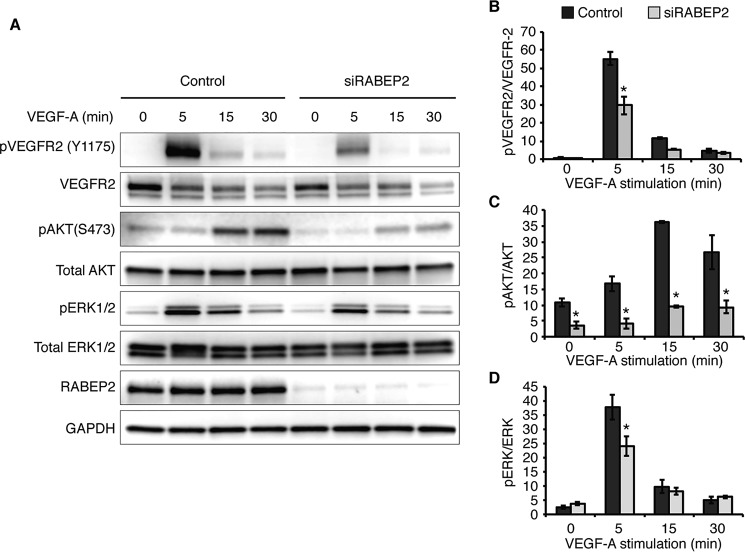
To assess the role of RABEP2 in regulating VEGFR2 signaling, endothelial cells transfected with siRABEP2 were serum-starved for 12 h and then stimulated with VEGF-A165 for 5, 15, and 30 min, after which VEGFR2 phosphorylation and downstream signaling via MAPK/MEK/ERK and PI3K/Akt were assessed by Western blot. After 5 min of VEGF-A stimulation, phosphorylation of VEGFR2 at tyrosine 1175, an event critical for VEGF signaling, was dramatically reduced in siRABEP2-transfected HUVEC, as compared with control (Fig. 2A). Quantification revealed a statistically significant reduction in phosphorylated VEGFR2 in siRABEP2 HUVEC when VEGFR2 phosphorylation levels were normalized to reduced total VEGFR2 (Fig. 2B). Thus, RABEP2 regulates VEGFR2 signaling independent of its role in maintaining total VEGFR2 expression. In line with these findings, we observed reduced activation of downstream signaling pathways associated with VEGFR2, including reduced pAKT and pERK in siRABEP2 HUVEC, as compared with control cells (Fig. 2, C and D).
Figure 2.
RABEP2 regulates VEGFR2 signaling. A, HUVEC were serum-starved for 12 h (0 min) and then stimulated with 50 ng/ml of VEGF-A165 for 5, 15, or 30 minutes (min). Protein lysates were collected and analyzed by Western blot using antibodies against the tyrosine 1175 phosphorylation site of VEGFR2 (pVEGFR2 Y1175), total VEGFR2, the serine 473 phosphorylation site of AKT (pAKT S473), total AKT, phosphorylated ERK1 and -2 (pERK1/2), total ERK1 and 2 (ERK1/2), RABEP2, and GAPDH as a loading control. B, quantification of pVEGFR2 (Y1175) normalized to total VEGFR2 levels. C, quantification of pAkt (S473) normalized to total Akt. D, quantification of pERK1/2 normalized to total ERK1/2. B–D, quantification was based on 4 independent experiments (mean ± S.D., *, p < 0.05).
RABEP2-deficient endothelial cells display abnormal VEGFR2 endosomal trafficking
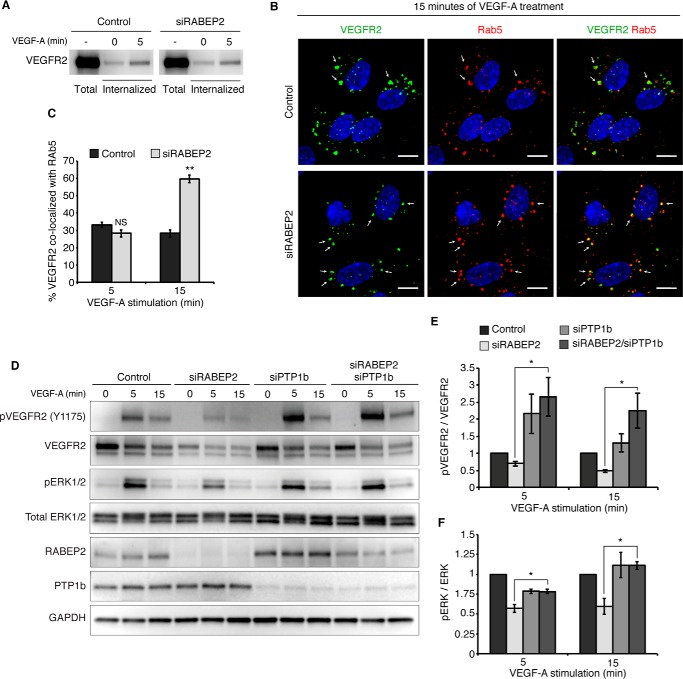
The magnitude of VEGFR2 phosphorylation and its activation of downstream signaling pathways is mediated, in part, by its trafficking through various endosomal compartments (15). Therefore, we next set out to explore if RABEP2 regulates endosomal trafficking of VEGFR2. To rule out a VEGFR2 internalization defect, we performed a protein internalization assay where HUVEC were incubated with biotin to label surface proteins and then stimulated with VEGF-A for 5 min. We then specifically probed the pool of internalized protein for VEGFR2 and observed no difference in VEGFR2 internalization between control and RABEP2-deficient HUVEC (Fig. 3A).
Figure 3.
RABEP2 regulates VEGFR2 endosomal trafficking. A, VEGFR2 internalization assay. Following incubation with biotin, cells were either harvested prior to stimulation to assess total surface VEGFR2 (total) or stimulated with 50 ng/ml of VEGF-A165 for 0 or 5 min (internalized), then stripped of the remaining surface biotin and lysed to assess levels of internalized biotin-labeled protein via Western blot probed for VEGFR2. B, HUVEC co-stained for internalized VEGFR2 (green) and Rab5 (red) after 15 min of stimulation with 50 ng/ml of VEGF-A165. White arrows point to Rab5+ endosomes containing internalized VEGFR2. DAPI marks cell nuclei in blue; scale bar, 10 μm. C, quantification of percent co-localization of internalized VEGFR2 and Rab5 after 5 and 15 min of VEGF-A165 stimulation. Quantification was performed using a co-localization plugin of Image J in at least 10 independent fields (mean ± S.D., NS, not significant; **, p < 0.01). D, HUVEC transfected with scrambled siRNA (control), scrambled siRNA and siRNA targeting RABEP2 (siRABEP2), scrambled siRNA and siRNA targeting PTP1b (siPTP), or siRNA targeting RABEP2 and PTP1b (siRABEP2/siPTP1b), serum starved for 12 h and then stimulated for 5 or 15 min with 50 ng/ml of VEGF-A165. Protein lysates were assessed by Western blots to assess VEGF signaling using antibodies against the tyrosine 1175 phosphorylation site of VEGFR2 (pVEGFR2 Y1175), total VEGFR2, RABEP2, and PTP1b to validate knockdown and GAPDH as a loading control. E, quantification of pVEGFR2 Tyr1175 normalized to total VEGFR2 of 3 independent experiments (mean ± S.D., *, p < 0.05). F, quantification of pERK1/2 normalized to total ERK1/2 of 3 independent experiments (mean ± S.D., *, p < 0.05).
Following internalization, VEGFR2 first traffics through Rab5+ endosomes to be sorted for additional rounds of signaling or for degradation. To observe the movement of internalized VEGFR2 through various endosomal compartments, we employed a previously described method where HUVEC are incubated prior to VEGF stimulation with a monoclonal antibody that specifically binds the extracellular domain of VEGFR2 (13). After VEGF stimulation, we then assessed co-localization of internalized, antibody-labeled surface VEGFR2 with different endosomal compartments by fluorescently labeling internalized VEGFR2 using an appropriate secondary antibody and co-staining with various endosomal markers. After 15 min of VEGF-A stimulation, a significantly higher proportion of internalized VEGFR2 remained co-localized with Rab5+ endosomes (Fig. 3, labeled, in part, by white arrows) in RABEP2-deficient HUVEC, whereas in control endothelial cells VEGFR2 moved on to other endosomal compartments (Fig. 3, B and C). We observed no difference in internalized VEGFR2 and Rab5 co-localization after 5 min of VEGF-A stimulation, demonstrating that loss of RABEP2 does not alter VEGFR2 entry into Rab5+ sorting endosomes (Fig. 3C).
Prolonged localization of VEGFR2 in Rab5+ endosomes increases its exposure to the phosphatase activity of PTP1b, leading to reduced phosphorylation at the Tyr1175 site and impaired arteriogenesis (13, 38). To test if a similar mechanism contributes to the reduced VEGFR2 signaling displayed by RABEP2-deficient endothelial cells, we tested the effect of PTP1b knockdown in RABEP2-deficinent HUVEC. Additional knockdown of PTP1b in siRABEP2-transfected HUVEC restored VEGFR2 Tyr1175 and ERK phosphorylation levels to control levels (Fig. 3, D–F). Collectively, these data suggest that RABEP2 is required for VEGFR2 exit from the Rab5 endosomal compartment. In the absence of RABEP2, VEGFR2 stalls in Rab5+ endosomes, thereby submitting VEGFR2 to prolonged PTP1b-mediated de-phosphorylation resulting in reduced VEGFR2 signaling.
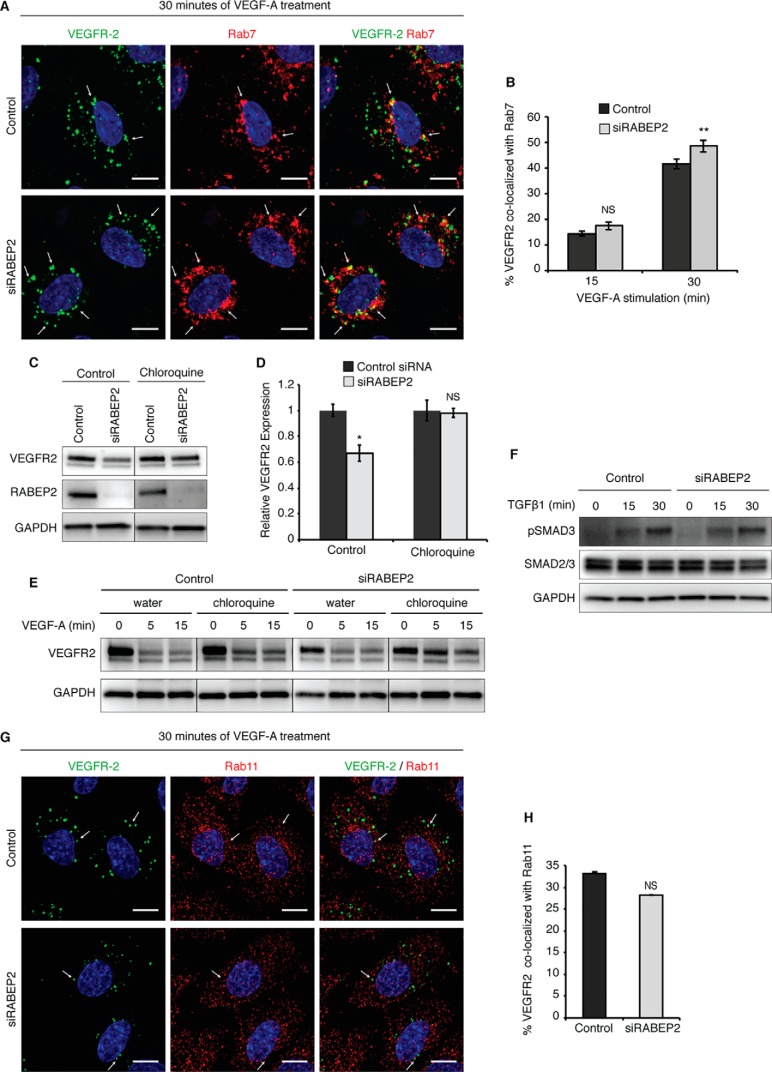
Following its stay in Rab5+ sorting endosomes, movement of VEGFR2 through subsequent endosomal compartments is mediated by Rab4, Rab11, or Rab7. To be recycled back to the plasma membrane for additional rounds of signaling, VEGFR2 enters either Rab4+ or Rab11+ recycling endosomes, whereas VEGFR2 destined for degradation enters Rab7+ endosomes to be trafficked to the lysosome. We assessed VEGFR2 movement through these different endosomal compartments in response to VEGFA stimulation. Although at 15 min post-stimulation there were no differences in internalized VEGFR2 co-localization with Rab7 (white arrows) in RABEP2-deficient versus control cells, at 30 min there was a statistically significant increase in VEGFR2 co-localizing with Rab7+ endosomes in siRABEP2 HUVEC (Fig. 4, A and B). Increased Rab7 co-localization of VEGFR2 suggested that a higher proportion of internalized VEGFR2 is targeted for lysosomal degradation in the absence of RABEP2. To test this possibility, siRABEP2 HUVEC were treated with the lysosomal inhibitor chloroquine. Chloroquine administration restored VEGFR2 to normal protein levels in HUVEC deficient for RABEP2 cultured in complete media (Fig. 4, C and D). Furthermore, chloroquine rescued VEGFR2 protein levels in siRABEP2 HUVEC stimulated with VEGFA, demonstrating that increased degradation causes reduced VEGFR2 protein levels associated with RABEP2 deficiency (Fig. 4E).
Figure 4.
Assessment of VEGFR2 trafficking mediated by Rab7 and Rab11. A, HUVEC co-stained for internalized VEGFR2 (green) and Rab7 (red) after 30 min of stimulation with 50 ng/ml of VEGF-A165. White arrows point to Rab7+ endosomes containing internalized VEGFR2. DAPI marks cell nuclei in blue; scale bar, 10 μm. B, quantification of co-localization of internalized VEGFR2 and Rab7 after 15 and 30 min of 50 ng/ml of VEGF-A165 stimulation. Quantification was performed using a co-localization plugin of ImageJ in at least 10 independent fields (mean ± S.D., NS, not significant, **, p < 0.01). C, Western blot probed for VEGFR2 of lysates isolated from control and RABEP2 knockdown (siRABEP2) HUVEC grown in complete media and treated with either water (control) or 100 μmol/liter of the lysosomal inhibitor chloroquine for 6 h prior to lysate collection. D, quantification of 3 independent experiments of total VEGFR2 levels normalized to GAPDH in siRABEP2 HUVEC treated with water or chloroquine, relative to control siRNA cells (mean ± S.D., *, p < 0.05). E, Western blot probed for VEGFR2 of lysates isolated from control and RABEP2 knockdown (siRABEP2) HUVEC serum starved for 12 h and then treated with either water (control) or 100 μmol/liter of the lysosomal inhibitor chloroquine for 6 h prior to stimulation for 5 or 15 min with 50 ng/ml of VEGF-A165. GAPDH used as a loading control. F, Western blot of lysates isolated from control and siRABEP2 HUVEC serum starved for 12 h and then stimulated with 1 ng/ml of transforming growth factor β1 (TGFβ1) for 0, 15, and 30 min. Western blots were probed for pSMAD3, a mediator of TGFβ downstream signaling, total SMAD2/3, and GAPDH as a loading control. G, HUVEC co-stained for internalized VEGFR2 (green) and Rab11 (red) after 30 min of stimulation with 50 ng/ml of VEGF-A165. White arrows point to Rab11+ endosomes containing internalized VEGFR2. DAPI marks the cell nuclei in blue; scale bar, 10 μm. H, quantification of co-localization of internalized VEGFR2 and Rab11 after 30 min of 50 ng/ml of VEGF-A165 stimulation. Quantification was performed using a co-localization plugin of ImageJ in at least 10 independent fields (mean ± S.D., NS, not significant).
We next addressed whether RABEP2 affects VEGFR2 trafficking through Rab11+ recycling endosomes. Rab11-dependent recycling appeared unchanged, as signaling by TGFRβ, which recycles exclusively by Rab11+ endosomes was normal in siRABEP2-transfected HUVEC (Fig. 4F). Furthermore, we observed normal co-localization of internalized VEGFR2 and Rab11 (white arrows) after 30 min of VEGF-A stimulation in siRABEP2 HUVEC (Fig. 4, G and H). Thus, we conclude that RABEP2 does not regulate VEGFR2 recycling through the Rab11+ endosomal compartment.
RABEP2 interaction with Rab proteins that mediate endosomal trafficking
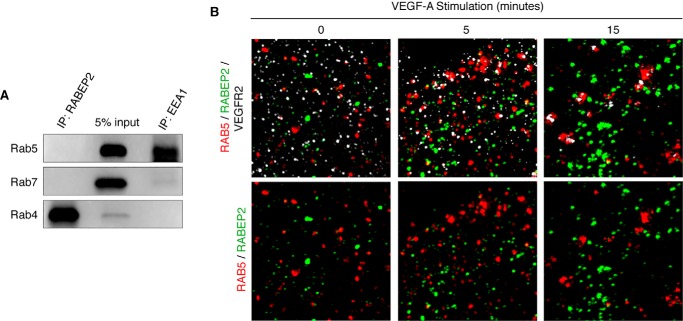
Altered endosomal trafficking of VEGFR2 caused by RABEP2 deficiency suggested that RABEP2 may selectively interact with various Rab proteins involved in this process. To test this, we pulled down endogenous RABEP2 in HUVEC and probed for candidate binding partners. RABEP2 failed to precipitate Rab5, although Rab5 did co-precipitate with its known binding partner, early endosome antigen 1 (EEA1), suggesting that Rab5 and RABEP2 do not co-localize in endothelial cells (Fig. 5A). This data were confirmed by structural illumination microscopy (SIM), which revealed only minimal co-localization of RABEP2 and Rab5 in serum-starved and VEGF-A-stimulated endothelial cells (Fig. 5B). RABEP2 also failed to co-precipitate Rab7 (Fig. 5A). In contrast, we observed a high level of co-precipitation of RABEP2 with the recycling endosome marker Rab4 (Fig. 5A).
Figure 5.
Assessment of RABEP2 and Rab GTPase interaction. A, Western blot of lysates isolated from HUVEC grown in full media following immunoprecipitation (IP) with antibody against RABEP2 (IP: RABEP2) or EEA (IP: EEA1) and then probed for Rab5, Rab7, and Rab4. 5% input is shown in the middle lane. B, HUVEC infected with adenovirus expressing V5-tagged RABEP2, serum starved overnight, and stimulated for 0, 5, or 15 min with 50 ng/ml of VEGF-A165. Following fixation, cells were co-stained for V5 to visualize RABEP2 (green), RAB5 (red), and VEGFR2 (white) and imaged using SIM with an x-y resolution of 130 nm.
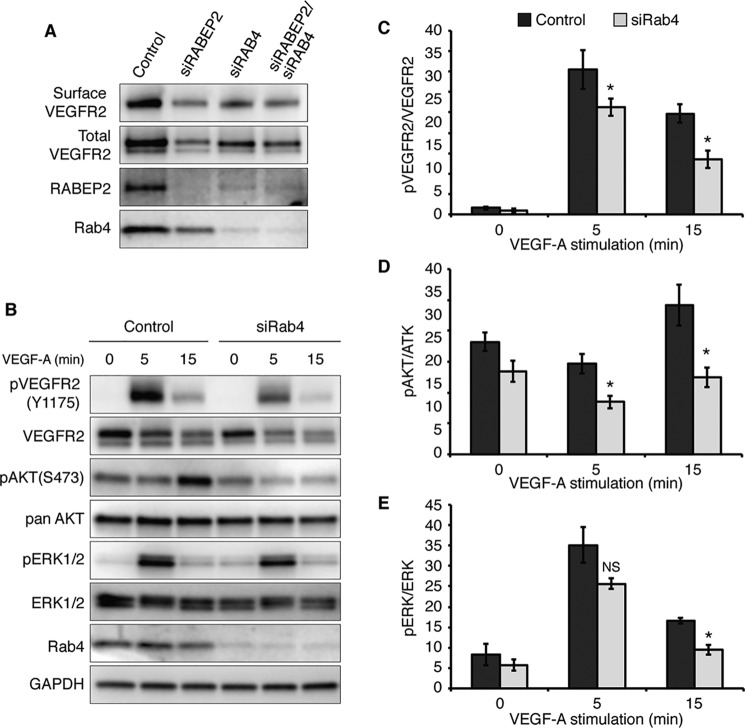
Rab4-positive endosomes mediate fast-loop recycling of VEGFR2, which is not only responsible for the recycling of ligand-activated VEGFR2, but also constitutive maintenance of cell-surface VEGFR2 in the absence of ligand (39). This contrasts with Rab11-positive endosomes, which exclusively recycle activated VEGFR2 via a ligand-dependent slow-loop pathway. We hypothesized that Rab4 and RABEP2 interact to mediate cell-surface VEGFR2 expression. Consistent with our hypothesis, loss of either RABEP2 or RAB4 resulted in reduced surface expression of VEGFR2 in serum-starved endothelial cells, demonstrating a role for both RABEP2 and RAB4 in maintaining constitutive VEGFR2 surface levels (Fig. 6A). Moreover, we observed that knockdown of either Rab4 or RABEP2 decreased the expression levels of the other; suggesting a reciprocal Rab-effector correlation between Rab4 and RABEP2 (Fig. 6A).
Figure 6.
Analysis of RAB4-dependent VEGFR2 trafficking. A, assessment of surface VEGFR2 levels. HUVEC transfected with scrambled siRNA (control), scrambled siRNA and siRNA targeting RABEPS (siRABEP2), scrambled siRNA and siRNA targeting RAB4a and RAB4b (siRAB4), or siRNA targeting Rab4a, Rab4b, and RABEP2 (siRab4/siRABEP2). HUVEC were serum starved for 12 h, incubated with biotin to label surface proteins, and biotin-labeled proteins were probed for VEGFR2 (surface VEGFR2). 5% input was loaded and probed for VEGFR2 (total VEGFR2), RABEP2, and Rab4 to validate knockdown. B, HUVEC transfected with scrambled siRNA (control) and siRNA targeting RAB4a and RAB4b transcripts (siRAB4) were serum starved for 12 h and then stimulated with 50 ng/ml of VEGF-A165 for 0, 5, or 15 min. Lysates were collected and probed using antibodies against the tyrosine 1175 phosphorylation site of VEGFR2 (pVEGFR2 Y1175), total VEGFR2, the serine 473 phosphorylation site of AKT (pAKT S473), total AKT, phosphorylated ERK1 and -2 (pERK1/2), total ERK1 and -2 (ERK1/2), RAB4 to validate knockdown, and GAPDH as a loading control. C, quantification of pVEGFR2 (Tyr1175) normalized to total VEGFR2 levels. D, quantification of pAkt (Ser473) normalized to total Akt. E, quantification of pERK1/2 normalized to total ERK1/2. C–E, quantification based on 4 independent experiments (mean ± S.D., NS, not significant; *, p < 0.05).
Accordingly, when HUVEC transfected with siRNA targeting RAB4 were stimulated with VEGF-A following overnight starvation, we observed reduced phosphorylation of VEGFR2 at Tyr1175 and reduced activation of VEGFR2 downstream signaling pathways, a pattern reminiscent of HUVEC deficient for RABEP2 (Fig. 6, B–E). Taken together, these data suggest that RABEP2 and RAB4 interact to maintain VEGFR2 surface levels in the absence of VEGF-A stimulation. Functionally, our findings demonstrate that loss of RABEP2 and its ability to promote Rab4-dependent VEGFR2 trafficking results in reduced VEGFR2 expression and downstream signaling events required for arteriogenesis.
Discussion
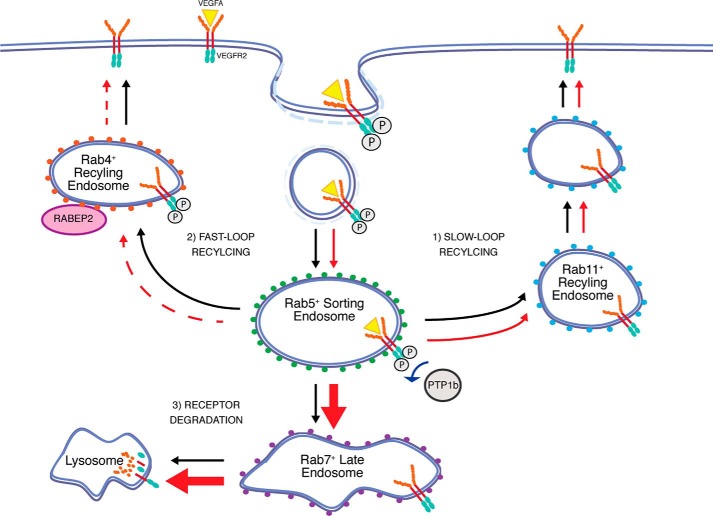
A balance is struck in endothelial cells between VEGFR2 synthesis, recycling, and degradation to ultimately regulate total expression of the receptor and its presence on the plasma membrane. The results of this study identify RABEP2 as a novel regulator of VEGFR2 trafficking involved in maintaining this balance. We show that RABEP2 interacts with Rab4 to maintain constitutive VEGFR2 levels at the plasma membrane. When RABEP2 is depleted (Fig. 7, red arrows), greater amounts of VEGFR2 are localized in Rab5-positive sorting endosomes, where VEGFR2 is partially deactivated by the phosphatase PTP1b. Furthermore, in the absence of RABEP2, a disproportionately high amount of VEGFR2 is trafficked to the lysosome for degradation by Rab7-positive endosomes (Fig. 7, thick red arrows). Thus, knockdown of RABEP2 reduces total and cell-surface VEGFR2 expression, mutes its intracellular signaling, and promotes VEGFR2 degradation.
Figure 7.
Schematic of RABEP2 function in VEGFR2 trafficking. Under normal conditions (black arrows), VEGFR2 is internalized by clathrin-coated pits and then enters Rab5+ sorting endosomes. From the Rab5+ endosome compartment, there are 3 possible routes through which VEGFR2 can traffic: 1) VEGFR2 can be recycled to the plasma membrane by Rab11+ endosomes via a slow-loop recycling route; 2) VEGFR2 can be recycled to the plasma membrane by Rab4+ endosomes via a fast-loop recycling route; or 3) VEGFR2 can traffic through Rab7+ endosomes to the lysosome for degradation. VEGFR2 continues to signal from Rab4+ endosomes. In the absence of RABEP2 (red arrows), Rab4-dependent trafficking of VEGFR2 to the plasma membrane is impaired (dashed red arrow), Rab11-dependent trafficking is normal, and an increased level of VEGFR2 (thick red arrow) is trafficked by Rab7+ endosomes to the lysosome for degradation. Lack of RABEP2 also stalls VEGFR2 trafficking through Rab5+ endosomes, prolonging its exposure to the phosphatase, PTP1b. As a result, loss of RABEP2 causes reduced total VEGFR2 protein levels, reduced surface VEGFR2 expression, and impaired VEGFR2 signaling.
Several lines of evidence support these conclusions. We demonstrated that loss of RABEP2 increased localization of internalized VEGFR2 in Rab7+ endosomes destined for the lysosome and that subsequent inhibition of lysosomal degradation can rescue total VEGFR2 protein levels. We also observed prolonged localization of internalized VEGFR2 in Rab5+ endosomes after 30 min of VEGF-A stimulation and observed rescued VEGFR2 signaling with additional knockdown of PTP1b in HUVEC deficient for RABEP2. Last, we documented that RABEP2 specifically interacts with Rab4 and that knockdown of Rab4 phenocopies knockdown of RABEP2, thus implicating RABEP2 in Rab4-dependent VEGFR2 recycling.
To date, only a handful of proteins, such as Nrp1 and the clathrin-associated proteins, Numb and Numb-like (Numbl), have been connected to VEGFR2 recycling (17, 18). Like RABEP2 deficiency, loss of endothelial Numb expression reduced total VEGFR2 protein levels and impaired VEGFR2 signaling due to increased trafficking through Rab7-positive endosomes and subsequent lysosomal degradation (18). This common phenotype shared by deficiency of two VEGFR2 recycling mediators, RABEP2 and Numb, suggests that the Rab7-positive late endosomal compartment may be a default trafficking route for VEGFR2, and that VEGFR2 recycling serves to maintain VEGFR2 expression by diverting it from lysosomal degradation.
To avoid degradation, internalized VEGFR2 can be recycled to the plasma membrane from the Rab5-positive endosomal compartment by either the Rab4 or Rab11 endosome-associated Rab GTPase (27). Rab4 mediates a fast-loop recycling route, whereas Rab11 controls slow-loop recycling to the plasma membrane. These two recycling loops differ in multiple ways. VEGFR2-phosphorylation status is maintained as it moves through Rab4-positive endosomes, whereas VEGFR2 is first de-phosphorylated before entering the slow-loop Rab11-mediated recycling pathway (26). VEGFR2 can therefore continue to signal as it moves through Rab4, but not Rab11-mediated recycling. Additionally, Rab4-mediated VEGFR2 recycling is unique because it occurs not only following VEGF-A stimulation, but also in the absence of ligand-activation (39). Our work demonstrates that RABEP2 specifically regulates Rab4-dependent VEGFR2 endosomal trafficking. We observed a strong protein interaction between RABEP2 and Rab4, but not Rab5 or Rab7, and we observed no difference in VEGFR2 co-localization with Rab11 in HUVEC deficient for RABEP2.
Impaired Rab4-mediated VEGFR2 trafficking in RABEP2-knockdown cells manifested in several cellular phenotypes. First, deletion of RABEP2 caused reduced VEGFR2 signaling, even when normalized to reduced total VEGFR2 expression. Second, loss of RABEP2 caused increased VEGFR2 degradation. Third, loss of RABEP2 caused reduced surface expression of VEGFR2 both in the presence and absence of ligand. We suggest that these phenotypes arise from a loss of VEGFR2 signaling from the Rab4-positive endosomal compartment due to the impaired capability of Rab4 to constitutively recycle VEGFR2 to the plasma membrane in the absence of RABEP2. Thus, RABEP2 functions in Rab4-mediated VEGFR2 trafficking to enhance VEGFR2 signaling by three mechanisms: (i) RABEP2 diverts VEGFR2 from degradation to maintain total VEGFR2 levels, (ii) RABEP2 maintains VEGFR2 surface levels in the absence of ligand, and (iii) RABEP2 promotes Rab4-dependent VEGFR2 trafficking to prolong VEGFR2 signaling. Overall, by serving these multiple functions in endothelial VEGFR2 trafficking, RABEP2 is a potent supporter of VEGFR2 signaling. Exactly how RABEP2 interacts with Rab4 to regulate VEGFR2 recycling is an important vascular question that remains to be determined.
The critical role played by RABEP2 in endothelial VEGFR2 signaling is apparent from the in vivo phenotypes that accompany its deletion. Mice deficient for RABEP2 displayed significantly impaired arteriogenesis; a process defined by arterial fate specification, endothelial proliferation, and vessel lumenization, and consequently, new artery formation (33, 40). In contrast, the process of angiogenesis governs capillary formation through the sprouting, proliferation, and migration of endothelia cells from pre-existing vessels (41, 42). Both arteriogenesis and angiogenesis are driven by endothelial VEGFR2 signaling and both processes are critical for the formation of a hierarchal vascular network (40). Yet, an unresolved question remains: how can VEGFR2, a single receptor tyrosine kinase, mediate such divergent vascular outcomes?
We propose that modulators of VEGFR2 trafficking, such as RABEP2, allow for the differential endothelial outcomes dictated by VEGFR2 signaling. Data described here and previously, show that endosomal trafficking of VEGFR2 specifically modulates arteriogenesis. Endothelial deletion of the retrograde motor protein, myosin-VI or its binding partner synectin, slowed VEGFR2 endosomal trafficking and resulted in reduced VEGFR2 signaling (13). In mice, this translated to a marked reduction in arteriogenesis, whereas angiogenic responses were conserved (13, 29). Similarly, RABEP2-deficient mice displayed normal angiogenesis, despite significantly impaired artery formation (33). Moreover, RABEP2 deletion in mice caused an enlargement of early endosomes in the brain pial vessels of mice deficient for RABEP2, suggestive of impaired endosomal trafficking. Thus, the speed and route by which VEGFR2 moves through different endosomal compartments appears to be critical for arteriogenesis, yet dispensable for angiogenesis. In vivo findings demonstrating a specific role for RABEP2 in arteriogenesis, combined with our data showing that RABEP2 regulates Rab4-dependent trafficking of VEGFR2 suggest that constitutive, fast-loop VEGFR2 recycling may be specifically required for new artery formation. The specificity of RABEP2 function in arteriogenesis is an exciting opportunity to develop novel therapeutics that can specifically target artery formation whereas avoiding off-target angiogenic effects. Accordingly, continued exploration of how RABEP2 and Rab4 regulate VEGFR2 trafficking may uncover novel strategies for the treatment of diseases characterized by vascular obstruction, such as stroke and coronary artery occlusions.
Experimental procedures
Animals
C57Bl/6J or BALB/cJ mice were obtained from Jackson Laboratories. All animal husbandry and experiments were performed with respect and in compliance with the guidelines established by institutional review boards.
Reagents and antibodies
The following reagents were used: recombinant human VEGF-A165 (R&D Systems, 293-VE-010), recombinant human TGF-β1 (R&D Systems, 240-B-002/CF), fibronectin (Sigma, F0895), and chloroquine diphosphate salt (Sigma, C6628). Cells lysates were collected in 1% Nonidet P-40 Lysis Buffer (Boston BioProducts, BP119) containing protease (Roche Applied Science, Complete Mini, EDTA-free, number 11836170001) and phosphatase inhibitors (Roche Applied Science, Phos STOP Easy, number 04-906-837001). The following antibodies were used for Western blot and/or immunocytochemistry (ICC) (dilutions shown for Western blot, unless otherwise specified): anti-VEGFR2 (Cell Signaling, number 2479, 1:1000), anti-human RABEP2 (Abcam, ab69045, 1:500), anti-mouse RABEP2 (Abcam, ab190003, 1:500), anti-Ve-cadherin (Santa Cruz, sc-6458, 1:200), anti-GAPDH (cell signaling, number 21185, 1:1000), anti-VEGFR3 (R&D Systems, AF349, 1:2000), anti-VEGFR1 (Abcam, ab32152, 1:2000), anti-NRP1 (Cell Signaling number 3725, 1:1000), anti-pVEGFR2 (Tyr1175) (Cell Signaling number 9106, 1:1000), anti-pAKT (Ser473) (Cell Signaling, number 4058, 1:1000), anti-AKT (Cell Signaling, number 9272, 1:1000), anti-pERK1/2 (Thr202/Tyr204) (Cell Signaling #9106, 1:1000), anti-ERK1/2 (Cell Signaling, number 9102, 1:1000), anti-PTP1b (BD Biosciences, catalog number 610139, 1:2000), anti-pSMAD3 (Abcam, ab52903, 1:2000), anti-SMAD2/3 (Cell Signaling, number 8685, 1:1000), anti-Rab5 (Cell Signaling, number 3547, 1:1000, 1:200 for ICC), anti-Rab7 (Cell Signaling, number 9367, 1:1000, 1:200 for ICC), anti-Rab4 (Santa Cruz, sc376243, 1:200), anti-Rab11 (Cell Signaling, number 3539, 1:1000, 1:200 for ICC), and anti-V5 (Abcam, ab27671, 1:1000 for ICC). The following antibodies were used for immunoprecipitation: anti-RABEP2 (Abcam, ab69045, 1:100) and anti-EEA1 (Cell Signaling, number 3288, 1:100). Secondary antibodies conjugated to horseradish peroxidase (Vector Laboratories) were used for Western blots. Alexa Fluor-conjugated secondary antibodies (Invitrogen) were used for immunocytochemistry.
Isolation of primary murine lung endothelial cells
Primary endothelial cells were isolated from mouse lungs using a previously described protocol (43). Briefly, lungs were isolated from either C57Bl/6J or BALB/cJ (Jackson Laboratories) 5-week-old wildtype mice. Lungs from 3 mice were pooled, mechanically disrupted, and digested with 2 mg/ml of type 1 collegenase (Sigma, C-0130) for 45 min at 37 °C. Endothelial cell sorting was performed using sheep anti-rat dynabeads (Invitrogen, 110.35) coated with rat anti-mouse CD31 (BD Pharmingen, 553370) antibody.
Human umbilicus venous endothelial cell culture
Passage (P)1 HUVEC were obtained from the Yale Vascular Biology and Therapeutics Tissue Culture Core Laboratory. HUVEC were maintained in EMG2 Endothelial Cells Growth Media (Lozna, CC3162) on 0.1% gelatin (Millipore, ES-006-B)-coated plates at 5% CO2. All experiments were performed on HUVEC between P2 and P6. Overexpression of RABEP2 was achieved using a lentiviral construct expression mcherry-tagged (N-terminal) C57Bl/6 mouse RABEP2. Lentiviral infection was achieved by co-transfecting 293T cells with the RABEP2 lentiviral construct and appropriate viral packaging constructs. 48 h of transfection, HUVEC were infected with varying quantities of media from transfected 293T and lysates collected 48 h after infection.
siRNA transfection
HUVEC were transfected with 25 pmol/liter of siRNA per well of a 6-well plate using Lipofectamine RNAiMAX (Invitrogen, 13778) according to the manufacturer's instructions. 24 h after transfection, HUVEC were switched to either full media or serum-starvation media containing 0.5% FBS (Invitrogen). The following siRNAs were used: AllStars Neg. Control siRNA (Qiagen, 1027281), HS_RABEP2_12 (Qiagen, S104776520), HS_RAB4A_6 (Qiagen, S102655030), HS_RAB4B_6 (Qiagen, S102662793), and PTPN1 siRNA (Sigma, SAS1_Hs01_00230699). All experiments were performed 48 h after transfection.
Immunocytochemistry
Immunocytochemistry was performed on cells grown on fibronectin-coated 35-mm glass-bottom plates (MatTek, P35-1.5-20C). Cells were first washed with PBS and then fixed and permeabilized by incubation in 4% paraformaldehyde for 10 min, followed by incubation in 2% paraformaldehyde, 0.1% Triton X-100, 0.1% Nonidet P-40 for an additional 10 min. Blocking was performed by incubation in 3% BSA (in PBS) for 1 h at room temperature. Cells were then incubated overnight at 4 °C in 1% BSA (in PBS) containing appropriate primary antibodies. The following day, cells were washed with PBS and incubated in 1% BSA (in PBS) containing appropriate Alexa Fluor-conjugated secondary antibodies used at a dilution of 1:400 for 1 h at room temperature. After washing in PBS, cells were mounted with glass coverslips using Vectashield containing DAPI (Vector, H1200). Imaging was performed using laser-scanning confocal microscopy with ×63 objective (Zeiss LSM 510 Meta) or SIM microscopy as described in detail below.
Surface biotinylation assays
To assess cell-surface expression of VEGFR2, cells were grown to confluence on 0.1% gelatin-coated plates, placed on ice, washed with cold PBS, and then incubated in 0.25 mg/ml of EZ-Link Sulfo-NHS-SS-Biotin (Pierce, 21331) in PBS for 1 h at 4 °C. Biotin was quenched by washing with 50 mm glycine and lysates were collected. To assess surface protein internalization, cells were stimulated for appropriate amounts of time with VEGF following biotin labeling. Residual surface biotin was then removed by incubating in a stripping solution consisting of 45 mm l-glutathione, 75 mm NaCl, 75 mm NaOH, 1 mm EDTA, 1% BSA in PBS, followed by a quick wash with 5 mg/ml of iodoacetamide in PBS, and lysates were collected. To determine VEGFR2 levels, 200 μg of protein was incubated overnight at 4 °C with 50 μl of Immobilized NeutrAvidin protein beads (Pierce, 29200) diluted in lysis buffer. Biotin-bound beads were then washed multiple times with lysis buffer, spun down, resuspended in SDS-sample buffer (Boston BioProducts, BP110R), boiled to denature, and assessed by Western blot.
Assessment by qualitative real-time PCR
RNA was isolated using the RNeasy Kit from Qiagen (catalog number 74104). cDNA was obtained by performing reverse transcription using the iScript cDNA Synthesis Kit (Bio Rad, 170–8890). For real-time PCR, iQ SYBR Green Supermix (Bio Rad, 170–8880) and a CFX96 Real Time System (Bio Rad) were employed. To assess VEGFR2 transcript levels, we used QuantiTect primers for human VEGFR2 (KDR) from Qiagen (QT00069818). GAPDH (5′GAGTCAACGGATTTGGTCGT-3′, 5′-GACAAGCTTCCCGTTCTCAG-3′) was used for normalization.
Assessment of internalized VEGFR2 co-localization
HUVEC were plated on fibronectin-coated 35-mm glass-bottom plates (MatTek, P35-1.5-20C). 24 h after transfection, cells were starved overnight in 0.5% FBS and then placed on ice for 20 min to stop constitutive receptor internalization. Cells were then incubated with an antibody against the extracellular domain of VEGFR2 (R&D, AF357, 1:50) diluted in starvation media for 20 min on ice, and then washed with cold PBS. Cells were stimulated with pre-warmed media containing 50 ng/ml of VEGF-A165 at 37 °C for 5, 15, or 30 min to induce antibody-bound VEGR2 internalization and trafficking. After stimulation, residual antibody on the cell surface was removed by washing cells with cold acidic PBS (pH 2.5) and cells were then processed for immunocytochemistry.
Immunoprecipitation
HUVEC were grown to confluence on 10-cm plates, washed twice with PBS, and cell lysates were collected in protein lysis buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 2 mm MGCl2, 1% Nonidet P-40 containing protease and phosphatase inhibitors. To minimize unspecific binding, 500 μg of protein lysate was first cleared by incubation with 100 μl of washed Protein A/G-agarose (Thermo Fisher, 20423) for 1 h at 4 °C on a rotator, samples were spun down and then the cleared protein lysate was transferred to a new tube. Optimized amounts of antibody were added to the cleared lysates and incubated overnight at 4 °C on a rotator. The next day, antibody-bound protein was isolated by incubation with 100 μl of washed Protein A/G-agarose overnight at 4 °C on a rotator. Samples were then washed multiple times with lysis buffer, spun down, and resuspended in SDS-sample buffer, boiled to denature, and assessed by Western blot.
Structural illumination microscopy
Because we were unable to obtain an effective antibody against RABEP2 for immunocytochemistry, we infected HUVEC with adenovirus expressing V5-tagged RABEP2 to visualize RABEP2 cellular localization. We cloned the wildtype C57Bl/6 mouse RABEP2 transcript into pAd/CMV/V5-DEST vector (Invitrogen, V493-20) using the pENTR Directional TOPO Cloning Kit (Invitrogen, K2400-20). This resulted in expression of a adenoviral vector encoding RABEP2 under control of a cytomegalovirus promoter with a C-terminal V5 tag (pAdRABEP2-V5). To produce RABEP2-V5 expressing adenovirus, we transfected 293A cells with pAdRABEP2-V5 using Lipofectamine 2000 (Invitrogen, 11668) according to the manufacturer's instructions. 7 days after transfections, cells and media were collected and lysed by performing several rounds of freeze-thaws using liquid nitrogen. The collected supernatant was passed through a cell strainer and then used to infect an additional plate of 293A cells. 3 days after infection, cells and media where collected, lysed by freeze-thaw, and filtered to be used for infection of HUVEC. For immunocytochemistry assessment, HUVEC were plated on fibronectin-coated 35-mm glass-bottom plates and infected with 10 μl of media from 293A infected with adenovirus expressing RABEP2-V5. 48 h later, cells were stimulated and processed for immunocytochemistry, following the method described earlier. SIM images were acquired on the OMX version 3 system (Applied Precision) using a U-PLANAPO 603/1.42 PSF, oil immersion objective lens (Olympus) and CoolSNAP HQ2 CCD cameras with a pixel size of 0.080 mm (Photometrics). Samples were illuminated with 488, 561, and 642 nm solid-state lasers (coherent, MPB Communications) and acquired as described before (13). Raw images were processed, reconstructed, and aligned to reveal structures with 100–125 nm resolution using Softworx software (Applied Precision) (44).
Statistics
Statistical analysis was performed using a two-tailed Student's t test. Error bars represent mean ± S.E. Statistical significance determined as p > 0.05, p > 0.01, and p > 0.001, as indicated.
Author contributions
N. K. designed, performed, and interpreted experiments. N. K., F. R., D. T., and M. S. conceptualized experiments. F. R., F. C., and Y. D. performed experiments. N. K. and M. S. wrote the manuscript.
Acknowledgments
We thank Dr. Georgia Zarkada, Dr. Alexandre Dubrac, Dr. Gaël Genet, Dr. Irinna Papengeli, Dr. John Rhodes, Dr. Nicolas Ricard, Dr. Pengchun Yu, and Dr. Brian Coon for scientific and experimental advice. We also thank Dorina Defilippo for administrative assistance, and Rita Webber for assistance with mouse colonies. Last, we thank our colleagues at the Yale Cardiovascular Research Center for their endless scientific advice and support.
This work was supported, in part, by National Institutes of Health Training Grant T32 HL007778-20 (to N. K.) and National Institutes of Health Grants R01 HL053793, HL084619, and PO1 5P01HL107205 (to M. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- VEGF2
- vascular endothelial growth factor receptor-2
- HUVEC
- human umbilicus venous endothelial cell
- NRP1
- Neuropilin-1
- EEA-1
- early endosome antigen 1
- SIM
- structural illumination microscopy
- PTP1b
- phosphotyrosine phosphatase 1b
- ICC
- immunocytochemistry.
References
- 1. Shalaby F., Rossant J., Yamaguchi T. P., Gertsenstein M., Wu X. F., Breitman M. L., and Schuh A. C. (1995) Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66 10.1038/376062a0 [DOI] [PubMed] [Google Scholar]
- 2. Ferrara N., Carver-Moore K., Chen H., Dowd M., Lu L., O'Shea K. S., Powell-Braxton L., Hillan K. J., and Moore M. W. (1996) Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature 380, 439–442 10.1038/380439a0 [DOI] [PubMed] [Google Scholar]
- 3. Carmeliet P., Ferreira V., Breier G., Pollefeyt S., Kieckens L., Gertsenstein M., Fahrig M., Vandenhoeck A., Harpal K., Eberhardt C., Declercq C., Pawling J., Moons L., Collen D., Risau W., and Nagy A. (1996) Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380, 435–439 10.1038/380435a0 [DOI] [PubMed] [Google Scholar]
- 4. Bautch V. L., and Caron K. M. (2015) Blood and lymphatic vessel formation. Cold Spring Harb. Perspect. Biol. 7, a008268 10.1101/cshperspect.a008268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Potente M., Gerhardt H., and Carmeliet P. (2011) Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887 10.1016/j.cell.2011.08.039 [DOI] [PubMed] [Google Scholar]
- 6. Olsson A.-K., Dimberg A., Kreuger J., and Claesson-Welsh L. (2006) VEGF receptor signalling: in control of vascular function. Nat. Rev. Mol. Cell Biol. 7, 359–371 10.1038/nrm1911 [DOI] [PubMed] [Google Scholar]
- 7. Jeltsch M., Leppänen V. M., Saharinen P., and Alitalo K. (2013) Receptor tyrosine kinase-mediated angiogenesis. Cold Spring Harb. Perspect Biol. 5, a009183 10.1101/cshperspect.a009183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerber H. P., McMurtrey A., Kowalski J., Yan M., Keyt B. A., Dixit V., and Ferrara N. (1998) Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3'-kinase/Akt signal transduction pathway: requirement for Flk-1/KDR activation. J. Biol. Chem. 273, 30336–30343 10.1074/jbc.273.46.30336 [DOI] [PubMed] [Google Scholar]
- 9. Bazzoni G., and Dejana E. (2004) Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiol. Rev. 84, 869–901 10.1152/physrev.00035.2003 [DOI] [PubMed] [Google Scholar]
- 10. Wheeler-Jones C., Abu-Ghazaleh R., Cospedal R., Houliston R. A., Martin J., and Zachary I. (1997) Vascular endothelial growth factor stimulates prostacyclin production and activation of cytosolic phospholipase A2 in endothelial cells via p42/p44 mitogen-activated protein kinase. FEBS Lett. 420, 28–32 10.1016/S0014-5793(97)01481-6 [DOI] [PubMed] [Google Scholar]
- 11. Koch S., and Claesson-Welsh L. (2012) Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb. Perspect. Med. 2, a006502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eitenmüller I., Volger O., Kluge A., Troidl K., Barancik M., Cai W.-J., Heil M., Pipp F., Fischer S., Horrevoets A. J. G., Schmitz-Rixen T., and Schaper W. (2006) The range of adaptation by collateral vessels after femoral artery occlusion. Circ. Res. 99, 656–662 10.1161/01.RES.0000242560.77512.dd [DOI] [PubMed] [Google Scholar]
- 13. Lanahan A. A., Hermans K., Claes F., Kerley-Hamilton J. S., Zhuang Z. W., Giordano F. J., Carmeliet P., and Simons M. (2010) VEGF receptor 2 endocytic trafficking regulates arterial morphogenesis. Dev. Cell 18, 713–724 10.1016/j.devcel.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ren B., Deng Y., Mukhopadhyay A., Lanahan A. A., Zhuang Z. W., Moodie K. L., Mulligan-Kehoe M. J., Byzova T. V., Peterson R. T., and Simons M. (2010) ERK1/2-Akt1 crosstalk regulates arteriogenesis in mice and zebrafish. J. Clin. Invest. 120, 1217–1228 10.1172/JCI39837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simons M., Gordon E., and Claesson-Welsh L. (2016) Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 17, 611–625 10.1038/nrm.2016.87 [DOI] [PubMed] [Google Scholar]
- 16. Boucher J. M., Clark R. P., Chong D. C., Citrin K. M., Wylie L. A., and Bautch V. L. (2017) Dynamic alterations in decoy VEGF receptor-1 stability regulate angiogenesis. Nat. Commun. 8, 15699 10.1038/ncomms15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lanahan A., Zhang X., Fantin A., Zhuang Z., Rivera-Molina F., Speichinger K., Prahst C., Zhang J., Wang Y., Davis G., Toomre D., Ruhrberg C., and Simons M. (2013) The neuropilin 1 cytoplasmic domain is required for VEGF-A-dependent arteriogenesis. Dev. Cell 25, 156–168 10.1016/j.devcel.2013.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Lessen M., Nakayama M., Kato K., Kim J. M., Kaibuchi K., and Adams R. H. (2015) Regulation of vascular endothelial growth factor receptor function in angiogenesis by numb and numb-like. Arterioscler. Thromb. Vasc. Biol. 35, 1815–1825 10.1161/ATVBAHA.115.305473 [DOI] [PubMed] [Google Scholar]
- 19. Simons M. (2012) An inside view: VEGF receptor trafficking and signaling. Physiology 27, 213–222 10.1152/physiol.00016.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horowitz A., and Seerapu H. R. (2012) Regulation of VEGF signaling by membrane traffic. Cell. Signal. 24, 1810–1820 10.1016/j.cellsig.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dougher M., and Terman B. I. (1999) Autophosphorylation of KDR in the kinase domain is required for maximal VEGF-stimulated kinase activity and receptor internalization. Oncogene 18, 1619–1627 10.1038/sj.onc.1202478 [DOI] [PubMed] [Google Scholar]
- 22. Lampugnani M. G., Orsenigo F., Gagliani M. C., Tacchetti C., and Dejana E. (2006) Vascular endothelial cadherin controls VEGFR-2 internalization and signaling from intracellular compartments. J. Cell Biol. 174, 593–604 10.1083/jcb.200602080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wandinger-Ness A., and Zerial M. (2014) Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 6, a022616 10.1101/cshperspect.a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jopling H. M., Odell A. F., Hooper N. M., Zachary I. C., Walker J. H., and Ponnambalam S. (2009) Rab GTPase regulation of VEGFR2 trafficking and signaling in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 29, 1119–1124 10.1161/ATVBAHA.109.186239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gampel A., Moss L., Jones M. C., Brunton V., Norman J. C., and Mellor H. (2006) VEGF regulates the mobilization of VEGFR2/KDR from an intracellular endothelial storage compartment. Blood 108, 2624–2631 10.1182/blood-2005-12-007484 [DOI] [PubMed] [Google Scholar]
- 26. Ballmer-Hofer K., Andersson A. E., Ratcliffe L. E., and Berger P. (2011) Neuropilin-1 promotes VEGFR-2 trafficking through Rab11 vesicles thereby specifying signal output. Blood 118, 816–826 10.1182/blood-2011-01-328773 [DOI] [PubMed] [Google Scholar]
- 27. Jopling H. M., Odell A. F., Pellet-Many C., Latham A. M., Frankel P., Sivaprasadarao A., Walker J. H., Zachary I. C., and Ponnambalam S. (2014) Endosome-to-plasma membrane recycling of VEGFR2 receptor tyrosine kinase regulates endothelial function and blood vessel formation. Cells 3, 363–385 10.3390/cells3020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sawamiphak S., Seidel S., Essmann C. L., Wilkinson G. A., Pitulescu M. E., Acker T., and Acker-Palmer A. (2010) Ephrin-B2 regulates VEGFR2 function in developmental and tumour angiogenesis. Nature 465, 487–491 10.1038/nature08995 [DOI] [PubMed] [Google Scholar]
- 29. Moraes F., Paye J., Mac Gabhann F., Zhuang Z. W., Zhang J., Lanahan A. A., and Simons M. (2013) Endothelial cell-dependent regulation of arteriogenesis. Circ. Res. 113, 1076–1086 10.1161/CIRCRESAHA.113.301340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chittenden T. W., Claes F., Lanahan A. A., Autiero M., Palac R. T., Tkachenko E. V., Elfenbein A., Ruiz de Almodovar C., Dedkov E., Tomanek R., Li W., Westmore M., Singh J. P., Horowitz A., Mulligan-Kehoe M. J., Moodie K. L., Zhuang Z. W., Carmeliet P., and Simons M. (2006) Selective regulation of arterial branching morphogenesis by synectin. Dev. Cell 10, 783–795 10.1016/j.devcel.2006.03.012 [DOI] [PubMed] [Google Scholar]
- 31. Gournier H., Stenmark H., Rybin V., Lippé R., and Zerial M. (1998) Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J. 17, 1930–1940 10.1093/emboj/17.7.1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sealock R., Zhang H., Lucitti J. L., Moore S. M., and Faber J. E. (2014) Congenic fine-mapping identifies a major causal locus for variation in the native collateral circulation and ischemic injury in brain and lower extremity. Circ. Res. 114, 660–671 10.1161/CIRCRESAHA.114.302931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lucitti J. L., Sealock R., Buckley B. K., Zhang H., Xiao L., Dudley A. C., and Faber J. E. (2016) Variants of Rab GTPase-effector binding protein-2 cause variation in the collateral circulation and severity of stroke. Stroke 47, 3022–3031 10.1161/STROKEAHA.116.014160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chalothorn D., Clayton J. A., Zhang H., Pomp D., and Faber J. E. (2007) Collateral density, remodeling, and VEGF-A expression differ widely between mouse strains. Physiol. Genomics 30, 179–191 10.1152/physiolgenomics.00047.2007 [DOI] [PubMed] [Google Scholar]
- 35. Pardanaud L., Pibouin-Fragner L., Dubrac A., Mathivet T., English I., Brunet I., Simons M., and Eichmann A. (2016) Sympathetic innervation promotes arterial fate by enhancing endothelial ERK activity. Circ. Res. 119, 607–620 10.1161/CIRCRESAHA.116.308473 [DOI] [PubMed] [Google Scholar]
- 36. Heinolainen K., Karaman S., D'Amico G., Tammela T., Sormunen R., Eklund L., Alitalo K., and Zarkada G. (2017) VEGFR3 modulates vascular permeability by controlling VEGF/VEGFR2 signaling. Circ. Res. 120, 1414–1425 10.1161/CIRCRESAHA.116.310477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Benedito R., Rocha S. F., Woeste M., Zamykal M., Radtke F., Casanovas O., Duarte A., Pytowski B., and Adams R. H. (2012) Notch-dependent VEGFR3 upregulation allows angiogenesis without VEGF-VEGFR2 signalling. Nature 484, 110–114 10.1038/nature10908 [DOI] [PubMed] [Google Scholar]
- 38. Lanahan A. A., Lech D., Dubrac A., Zhang J., Zhuang Z. W., Eichmann A., and Simons M. (2014) PTP1b is a physiologic regulator of vascular endothelial growth factor signaling in endothelial cells. Circulation 130, 902–909 10.1161/CIRCULATIONAHA.114.009683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jopling H. M., Howell G. J., Gamper N., and Ponnambalam S. (2011) The VEGFR2 receptor tyrosine kinase undergoes constitutive endosome-to-plasma membrane recycling. Biochem. Biophys. Res. Commun. 410, 170–176 10.1016/j.bbrc.2011.04.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kofler N. M., and Simons M. (2015) Angiogenesis versus arteriogenesis: neuropilin 1 modulation of VEGF signaling. F1000Prime Rep. 7, 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kofler N. M., Shawber C. J., Kangsamaksin T., Reed H. O., Galatioto J., and Kitajewski J. (2011) Notch signaling in developmental and tumor angiogenesis. Genes Cancer 2, 1106–1116 10.1177/1947601911423030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chung A. S., and Ferrara N. (2011) Developmental and pathological angiogenesis. Annu. Rev. Cell Dev. Biol. 27, 563–584 10.1146/annurev-cellbio-092910-154002 [DOI] [PubMed] [Google Scholar]
- 43. Allport J. R., Lim Y. C., Shipley J. M., Senior R. M., Shapiro S. D., Matsuyoshi N., Vestweber D., and Luscinskas F. W. (2002) Neutrophils from MMP-9- or neutrophil elastase-deficient mice show no defect in transendothelial migration under flow in vitro. J. Leukoc. Biol. 71, 821–828 [PubMed] [Google Scholar]
- 44. Gustafsson M. G., Shao L., Carlton P. M., Wang C. J., Golubovskaya I. N., Cande W. Z., Agard D. A., and Sedat J. W. (2008) Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 94, 4957–4970 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]