Abstract
G4C2 repeat expansions within the C9ORF72 gene are the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). These bidirectionally transcribed expansions lead to (1) the accumulation of sense G4C2 and antisense G2C4 repeat-containing RNA, (2) the production of proteins of repeating dipeptides through unconventional translation of these transcripts, and (3) decreased C9ORF72 mRNA and protein expression. Consequently, there is ample opportunity for the C9ORF72 mutation to give rise to a spectrum of clinical manifestations, ranging from muscle weakness and atrophy to changes in behavior and cognition. It is thus somewhat surprising that investigations of these three seemingly disparate events often converge on similar putative pathological mechanisms. This review aims to summarize the findings and questions emerging from the field’s quest to decipher how C9ORF72 repeat expansions cause the devastating diseases collectively referred to as “c9ALS/FTD.”
Amyotrophic lateral sclerosis (ALS), the most common form of motor neuron disease (MND), is characterized by the progressive degeneration of upper and lower motor neurons, leading to muscle weakness, atrophy, and spasticity. In addition, ALS involves several nonmotor systems and subcortical structures (Lowe 1994). In fact, cognitive and behavioral impairments reminiscent of frontotemporal dementia (FTD) are present in up to 50% of ALS patients (Lomen-Hoerth et al. 2003). FTD, second only to Alzheimer’s disease as a cause of dementia in patients under 65 (Neary et al. 1998), encompasses a group of disorders neuropathologically characterized by degeneration of the frontal and temporal lobes. Most commonly, patients present with behavioral variant FTD, which is marked by changes in personality and behavior. Other syndromes falling under the FTD umbrella are categorized by changes in language function. The heterogeneous nature of FTD is further underscored by the fact that a proportion of patients with FTD develop ALS (Snowden et al. 2002). A better understanding for the basis of the clinical overlap between ALS and FTD came about in 2006 with the discovery that TDP-43, an RNA/DNA-binding protein, forms inclusions in the central nervous system (CNS) of the majority of ALS cases and the most common pathological subtype of FTD, frontotemporal lobar degeneration with TDP-43-positive inclusions (FTLD-TDP) (Arai et al. 2006; Neumann et al. 2006). Relatively soon thereafter, another major discovery connecting ALS and FTD was made: G4C2 hexanucleotide repeat expansions in chromosome 9 open reading frame 72 (C9ORF72) were identified as the most common known genetic cause of ALS and FTD (DeJesus-Hernandez et al. 2011; Renton et al. 2011). C9ORF72 expansions account for ∼10%–50% of familial ALS, 5%–7% of sporadic ALS, 12%–25% of familial FTD, and 6%–7% of sporadic FTD (Majounie et al. 2012; Cruts et al. 2013; van der Zee et al. 2013), and are also found in related clinical phenotypes, including Alzheimer’s disease and parkinsonism (Cooper-Knock et al. 2014a; also see Ghasemi and Brown 2017 and Gijselinck et al. 2017).
Clinical heterogeneity is associated with the C9ORF72 expansion, with the majority of affected patients displaying symptoms of either ALS or FTD or mixed features of both (referred to as FTD-MND). There is also great variability among patients with regard to age of disease onset and duration (Boeve et al. 2012; Byrne et al. 2012; Chio et al. 2012; Cooper-Knock et al. 2012; Hsiung et al. 2012; Mahoney et al. 2012; Simon-Sanchez et al. 2012; Snowden et al. 2012). The molecular basis for this clinical variation, seen both among and within families, is not yet known. Upon first consideration, expansion length would seem a likely cause. Although the lower limit for pathological G4C2 expansions is not yet definitely established, unaffected individuals typically have two to 30 repeats, whereas tens to even thousands of repeats can be present in patients with ALS, FTD, and FTD-MND (DeJesus-Hernandez et al. 2011; Renton et al. 2011; Beck et al. 2013; van Blitterswijk et al. 2013; Dols-Icardo et al. 2014). Repeat length not only varies among family members, including monozygotic twins, but also differs among tissues of the same individual owing to somatic instability (Beck et al. 2013; Buchman et al. 2013; van Blitterswijk et al. 2013; Dols-Icardo et al. 2014; Waite et al. 2014; Fratta et al. 2015; Gijselinck et al. 2015; Nordin et al. 2015). At present, the influence of repeat length on clinical phenotype has not been resolved (van Blitterswijk et al. 2013; Benussi et al. 2014; Dols-Icardo et al. 2014; Cooper-Knock et al. 2015a; Nordin et al. 2015). Some studies found that repeat size had no effect on clinical presentation (van Blitterswijk et al. 2013; Gijselinck et al. 2015; Nordin et al. 2015), whereas others report that patients with ALS harbor a higher number of repeats than FTD patients (Dols-Icardo et al. 2014; Suh et al. 2015). The reported effects of repeat size on age at disease onset, age at death, and disease duration have also been discordant (Beck et al. 2013; van Blitterswijk et al. 2013; Benussi et al. 2014; Dols-Icardo et al. 2014; Gijselinck et al. 2015; Nordin et al. 2015; Suh et al. 2015), perhaps because of the different methodologies used to estimate repeat length, as well as differences in the tissues analyzed and the age of patients at their collection, factors that may influence repeat size.
Neuropathologically, all ALS, FTD, and FTD-MND cases caused by the C9ORF72 repeat expansion, collectively referred to as c9ALS/FTD, have TDP-43 pathology consisting of the abnormal accumulation of neuronal and oligodendroglial TDP-43 inclusions in the frontal and temporal cortex, hippocampus, and pyramidal motor system (Mackenzie et al. 2014). The underlying cause of TDP-43 pathology in C9ORF72 repeat expansion carriers, and in sporadic forms of ALS and FTD for that matter, nevertheless remains unknown. In addition to TDP-43 pathology, a characteristic finding of c9ALS/FTD is the presence of TDP-43-negative, p62-positive neuronal inclusions in the cerebellum, hippocampus, and neocortex regions (Pikkarainen et al. 2010; Al-Sarraj et al. 2011). These inclusions, also immunoreactive for ubiquitin and select ubiquitin-binding proteins, most notably ubiquilin-2 (Brettschneider et al. 2012), contain dipeptide repeat (DPR) proteins synthesized from sense and antisense expanded repeat-containing transcripts (r(G4C2)exp and r(G2C4)exp) through repeat associated non-ATG (RAN) translation. r(G4C2)exp and r(G2C4)exp also form nuclear RNA foci throughout the CNS, another consistent feature of c9ALS/FTD. Both RAN translation and foci formation are thought to contribute to disease pathogenesis. Furthermore, because repeat expansions cause a decrease in C9ORF72 mRNA and protein expression, a loss of C9ORF72 function may too be involved. Each of these events and their possible contribution to disease are discussed below, as are potential biomarkers and therapeutic strategies for these devastating diseases.
C9ORF72 LOSS OF FUNCTION
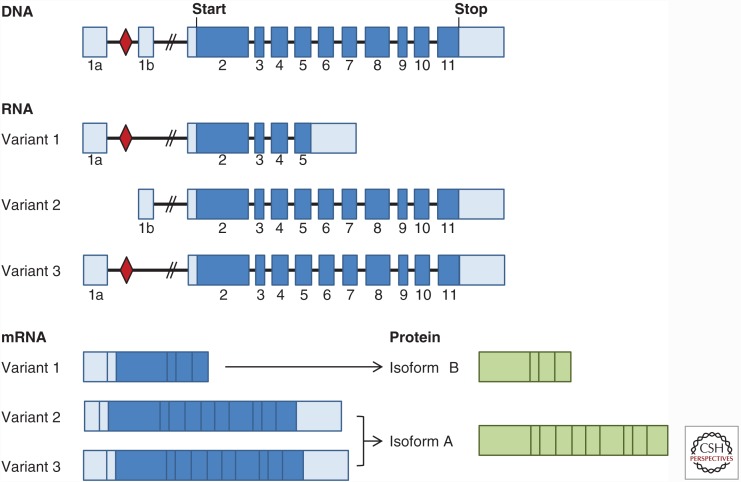
There are two C9ORF72 protein isoforms—a short isoform of 222 amino acids (isoform B) and a long isoform of 481 amino acids (isoform A)—derived from three alternatively spliced C9ORF72 transcripts. The alternative splicing of C9ORF72 pre-mRNA transcripts occurs within two noncoding first exons (exons 1a and 1b) upstream of the translation start site (Fig. 1). The G4C2 repeat is located between exons 1a and 1b, and on either side of the repeat are two putative CpG islands (stretches of DNA with a high frequency of CG sequence that regulate transcription at their associated promoters). If exon 1a is used, the repeat is located in intron 1 and is transcribed (variant 1, NM_145005.6; variant 3, NM_001256054.2); if exon 1b is used, the repeat is located in the promoter region and is not transcribed (variant 2, NM_018325.4) (Fig. 1). Depending on the location of the G4C2 repeat (within the promoter or intron 1), it is conceivable that an expansion of the repeat would influence transcription of C9ORF72 or the processing of its pre-mRNA. Indeed, decreased transcript levels for all C9ORF72 mRNA variants have been observed in C9ORF72 patient CNS tissues, lymphoblastoid cell lines, and some lines of neurons differentiated from patient-derived induced pluripotent stem cells (iPSCs) (DeJesus-Hernandez et al. 2011; Gijselinck et al. 2012; Belzil et al. 2013; Ciura et al. 2013; Donnelly et al. 2013; Fratta et al. 2013; Mori et al. 2013c; Xi et al. 2013; Waite et al. 2014; van Blitterswijk et al. 2015). In the largest case series of C9ORF72 expansion carriers analyzed to date, a significant decrease in variants 1 and 2, but not variant 3, was observed in the frontal cortex and cerebellum of expansion carriers compared with patients without the expansion or control subjects (van Blitterswijk et al. 2015). Notably, an association between variant 1 and survival after disease onset was found, suggesting that higher C9ORF72 levels may have beneficial effects. This study additionally reported that levels of transcripts containing intron 1 (and thus the repeat) were elevated in expansion carriers (van Blitterswijk et al. 2015), in agreement with previous reports (Donnelly et al. 2013; Mori et al. 2013c; Sareen et al. 2013; Zu et al. 2013; Haeusler et al. 2014; Liu et al. 2014).
Figure 1.
The C9ORF72 gene encodes two C9ORF72 protein isoforms derived from three alternatively spliced C9ORF72 transcripts. The G4C2 repeat (red diamond) is located between two noncoding exons of the C9ORF72 gene (exon 1a and exon 1b, light blue). Thus, the repeat is located in the promoter region of transcript variant 2 (NM_018325.4) or in the first intron of variant 1 (NM_145005.6) and variant 3 (NM_001256054.2). Variant 1 is predicted to result in a short C9ORF72 protein of 222 amino acids (exons 2–5, isoform B), whereas variants 2 and 3 encode a long C9ORF72 protein of 481 amino acids (exons 2–11, isoform A).
The reduced expression of C9ORF72 mRNA in c9ALS/FTD patients may be caused by several factors, including the structure of G4C2 repeat DNA and RNA. G4C2 DNA forms G-quadruplex structures (stacks of planar tetramers consisting of four guanines connected by Hoogsteen hydrogen bonds), whereas G4C2 repeat RNA forms G-quadruplex structures and hairpins that can bind to repeat DNA to form RNA–DNA hybrids (Fratta et al. 2012; Reddy et al. 2013; Haeusler et al. 2014; Su et al. 2014). Using in vitro transcription assays of G4C2 repeats of varying lengths (three to 70 repeats), Haeusler et al. observed that the generation of these distinct polymorphic DNA and RNA structures causes, in a repeat length-dependent manner, the abortive transcription of the repeat. It is thus noteworthy that, in the frontal cortex of C9ORF72 expansion carriers, there is an increase in transcripts containing sequences upstream of the intronic repeat but not sequences downstream from the repeat, suggesting that some repeat-containing transcripts are indeed truncated in c9ALS/FTD brain tissues (van Blitterswijk et al. 2015).
Epigenetic processes have also been associated with decreased C9ORF72 expression (Xi et al. 2013, 2014, 2015a,b; Belzil et al. 2013, 2014; Liu et al. 2014; Russ et al. 2014; Gijselinck et al. 2015; He et al. 2015; McMillan et al. 2015), in line with studies showing that expanded repeats in microsatellite expansion disorders dysregulate mRNA expression through changes in DNA and histone methylation (Sutcliffe et al. 1992; Greene et al. 2007; Al-Mahdawi et al. 2008; Todd et al. 2010). Trimethylated histones (H3K9me3, H3K27me3, H3K79me3, and H4K20me3) are known to repress gene expression (Barski et al. 2007). Chromatin immunoprecipitation studies revealed that trimethylated histones bound strongly to expanded G4C2 repeats but not to nonpathogenic repeats in the frontal cortex and cerebellum, and this binding is associated with decreased C9ORF72 mRNA levels in c9ALS/FTD patients (Belzil et al. 2013). A causal effect between histone trimethylation and decreased C9ORF72 expression was supported by the findings that treatment of c9ALS/FTD-derived fibroblasts with a DNA and histone-demethylating agent not only decreased trimethylated histone binding to C9ORF72 but also increased C9ORF72 mRNA expression (Belzil et al. 2013). In addition to histone methylation, repeat and CpG island hypermethylation may decrease C9ORF72 mRNA expression in c9ALS/FTD. Methylation of the repeat was found to occur in virtually all individuals with alleles of more than 90 repeats and associated with reduced C9ORF72 expression (Xi et al. 2015a,b). The CpG island 5′ of the repeat was also hypermethylated in an expansion-specific manner, albeit only in a proportion of carriers (Xi et al. 2013, 2014; Belzil et al. 2014; Liu et al. 2014; Russ et al. 2014). Nonetheless, Liu et al. (2014) show that, for the subset of cases for which this CpG island was hypermethylated, transcriptional silencing of C9ORF72 was observed. Somewhat counterintuitively, rather than being detrimental, epigenetic silencing of the mutant C9ORF72 allele may be protective, as it was shown to associate with decreased accumulation of intronic repeat-containing RNA, RNA foci formation, and RAN translation. To test this hypothesis further, the relationship between C9ORF72 promoter hypermethylation and clinical features was investigated (Russ et al. 2014). C9ORF72 hypermethylation did not significantly differ between ALS and FTD cases, nor did it predict age of onset. Nonetheless, in c9FTD cases, C9ORF72 hypermethylation in both the cerebellum and peripheral blood associated with later age at death, and C9ORF72 hypermethylation in the blood also associated with longer disease duration. Furthermore, neuroimaging and neuropathological studies showed that C9ORF72 promoter hypermethylation measured in the blood associated with less atrophy and neuronal loss in several brain regions, and with slower rates of verbal recall decline on neuropsychological testing (McMillan et al. 2015). Taken together, these results suggest that epigenetic silencing of C9ORF72 may represent a protective response by mitigating toxic events caused by the accumulation of repeat-containing RNA, but also may promote a putatively detrimental loss of C9ORF72 function. To establish a better understanding of the latter, it is imperative to thoroughly evaluate C9ORF72 protein expression in c9ALS/FTD, as well as determine the functions of C9ORF72 and how well loss of these functions is tolerated.
With the first in vivo model of C9ORF72 loss of function, Ciura et al. (2013) showed that downregulation of the C9ORF72 ortholog, zC9orf72, in zebra fish led to altered morphology of motor neuron axons and locomotor deficits, and that these features were reversed upon expression of human C9ORF72. Similarly, deletion of the C9orf72 ortholog, alfa-1, in Caenorhabditis elegans caused the degeneration of GABAergic motor neurons, age-dependent motility defects leading to paralysis, and sensitivity to environmental stress (Therrien et al. 2013). In mice, C9orf72 is enriched in neurons known to degenerate in ALS and FTD, perhaps shedding mechanistic light on the selective vulnerability of these neuronal populations (Suzuki et al. 2013). However, knockdown of C9ORF72 was well tolerated in primary cortical and motor neurons of rats (Wen et al. 2014), mice (Lagier-Tourenne et al. 2013; Koppers et al. 2015), and neurons differentiated from c9ALS/FTD iPSC (iPSNs) (Donnelly et al. 2013; Sareen et al. 2013). It nonetheless remains unknown how sustained loss of C9ORF72 in the context of the aging brain would influence neuronal function and survival. Studies on C9ORF72 function are adding much-needed perspective on this question.
Until recently, there has been little consensus on whether C9ORF72 protein expression is appreciably decreased in c9ALS/FTD (DeJesus-Hernandez et al. 2011; Renton et al. 2011; Cooper-Knock et al. 2012; Gijselinck et al. 2012; Hsiung et al. 2012; Simon-Sanchez et al. 2012; Snowden et al. 2012; Stewart et al. 2012). With the generation of new antibodies, C9ORF72 depletion in specific neuroanatomical regions has been demonstrated. Though studies on larger case series are needed, data indicate that the long C9ORF72 isoform is downregulated in the frontal cortex and temporal cortex of c9ALS/FTD cases, but not in the motor cortex or cerebellum (Waite et al. 2014; Xiao et al. 2015). Intriguingly, the short C9ORF72 isoform is concomitantly upregulated in the temporal and frontal cortices (Xiao et al. 2015). Using isoform-specific antibodies, the Robertson group also discovered that the biochemical profile and intracellular distribution of long and short C9ORF72 isoforms differ from one another in human brain tissues. This suggests that the functions of each isoform are distinct (Xiao et al. 2015), and similar findings were reported for C9orf72 protein isoforms in mice (Atkinson et al. 2015). In ALS cases with or without C9ORF72 expansions, and in healthy controls, the long C9ORF72 isoform is diffusely distributed in the cytoplasm of neurons and also labels large speckles in the cytoplasm and dendrites of cerebellar Purkinje cells. In healthy controls, the short isoform localizes to the nuclear membrane where it interacts with importin-B1 and Ran-GTPase. These are components of the nuclear pore complex, a channel used for the active transport of large proteins between the cytoplasm and the nucleus. Compared with control cases, the nuclear membrane localization of the short C9ORF72 isoform, as well as of importin-B1 and Ran-GTPase, is diminished in ALS cases with or without C9ORF72 expansions. Importantly, this associates with the mislocalization of TDP-43 from the nucleus to the cytoplasm and TDP-43 pathology. Consequently, C9ORF72 may play a role in nucleocytoplasmic transport, and defects in this pathway may account, at least in part, for nuclear TDP-43 depletion and TDP-43 pathology in ALS.
In addition to a potential role in nucleocytoplasmic shuttling, homology searches suggest that C9ORF72 is a member of the DENN (differentially expressed in normal and neoplasia)-like superfamily (Zhang et al. 2012; Levine et al. 2013). DENN domain proteins are highly conserved Rab-GEFs, GDP/GTP exchange factors that activate Rab-GTPases, the master regulators for intracellular membrane trafficking (Hutagalung and Novick 2011). Protein trafficking through the endosomal system is required for sorting and degrading proteins through autophagy or the ubiquitin–proteasome system (UPS) (Korolchuk et al. 2010). In neuronal-like cell lines, primary neurons, and human spinal cord sections, C9ORF72 colocalizes and/or interacts with Rab-GTPases implicated in endosomal transport and autophagy (Farg et al. 2014). When C9ORF72 was knocked down, endocytosis was impaired and autophagosome formation dysregulated. Studies also suggest that C9ORF72 interacts with ubiquilin-2, a member of the ubiquilin family that regulates the degradation of ubiquitinated proteins, and with heterogeneous nuclear ribonucleoproteins (hnRNP) A1 and hnRNPA2/B1 (Farg et al. 2014). hnRNPs, which include TDP-43, are RNA-binding proteins involved in many aspects of mRNA metabolism. Although hnRNP A1 and hnRNP A2/B1 reside predominantly in the nucleus, exposure to stress can trigger their recruitment to cytoplasmic stress granules, which assemble to facilitate cell survival by triaging RNAs not required for coping with the stress (Guil et al. 2006).
Additional studies are needed to fully decipher the potential role of C9ORF72 in nucleocytoplasmic transport, stress granule dynamics, and autophagy. However, with regard to the latter, it is conceivable that perturbations in C9ORF72 function contribute to the increased sensitivity of c9ALS/FTD iPSNs to chloroquine and 3-MA, two inhibitors of autophagy (Almeida et al. 2013). This increased sensitivity suggests that proper autophagic processing was compromised, which is further supported by the finding that levels of p62, a known substrate of the autophagy pathway, were significantly higher in iPSNs from C9ORF72 repeat expansion carriers compared with iPSNs from noncarriers (Almeida et al. 2013). Nevertheless, it is not yet known whether loss of C9ORF72 function and/or the presence of DPR proteins, which have also been linked to defects in protein degradation (May et al. 2014; Zhang et al. 2014; Yamakawa et al. 2015), cause the enhanced vulnerability of c9ALS/FTD iPSNs to autophagy inhibitors.
TOXICITY MEDIATED THROUGH (G4C2)exp OR (G2C4)exp TRANSCRIPTS
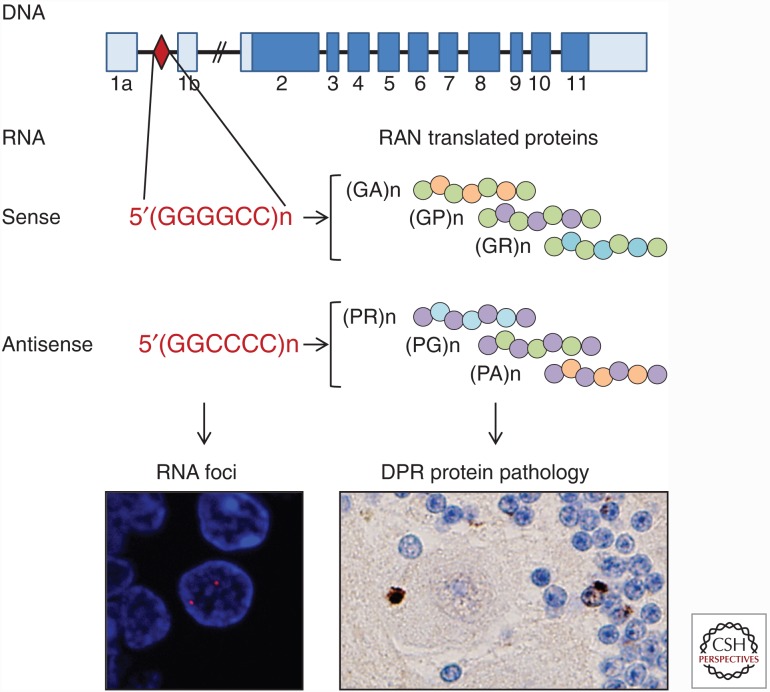
Repeat-containing RNA transcribed from C9ORF72 expansions, which leads to RNA foci formation and the production of DPR proteins through RAN translation, has been heavily implicated in c9ALS/FTD pathogenesis (Fig. 2). Exogenous expression of expanded sense G4C2 or antisense G2C4 repeats alone (i.e., not in the context of the full C9ORF72 gene) leads to the development of various aberrant phenotypes and toxicity in cultured cell models (Lee et al. 2013; Xu et al. 2013; Zu et al. 2013; Wen et al. 2014; Rossi et al. 2015), Drosophila (Xu et al. 2013; Mizielinska et al. 2014; Freibaum et al. 2015; Tran et al. 2015; Zhang et al. 2015), zebra fish embryos (Lee et al. 2013), and mice (Hukema et al. 2014; Chew et al. 2015). Indeed, using somatic brain transgenesis to induce (G4C2)66 expression throughout the murine CNS, the Petrucelli group reported the first mouse model to mimic neuropathological and behavioral phenotypes of c9ALS/FTD (Chew et al. 2015). The brains of (G4C2)66 mice exhibited nuclear RNA foci, inclusions of DPR proteins, and neurodegeneration. Furthermore, these mice developed TDP-43 pathology, as evidenced by inclusions of phosphorylated endogenous TDP-43 in the hippocampus and cortex. The latter is especially remarkable given that most rodent TDP-43 transgenic models fail to show appreciable TDP-43 pathology (Gendron and Petrucelli 2011). These findings also place TDP-43 abnormalities downstream from the accumulation of r(G4C2)exp, although whether RNA foci per se are involved is not yet known. In the c9ALS/FTD frontal cortex, hippocampus, and cerebellum, the presence of sense or antisense foci does not predict TDP-43 pathology (Mizielinska et al. 2013; Cooper-Knock et al. 2014b), but antisense foci have been shown to associate with nuclear loss of TDP-43 in c9ALS motor neurons (Cooper-Knock et al. 2015b). Yet another possibility is that TDP-43 pathology develops in (G4C2)66 mice as a consequence of DPR protein expression and subsequent downstream events. Because the expression of expanded repeats in (G4C2)66 mice and the above-mentioned models give rise to the accumulation of repeat-containing RNA, RNA foci, and RAN-translated DPR proteins, it is difficult to definitively ascribe the emergence of c9ALS/FTD-associated phenotypes to a given event. In the sections below, the neurotoxic potential mediated specifically by RNA toxicity or DPR proteins is further discussed.
Figure 2.
The repeat expansion in C9ORF72 is bidirectionally transcribed and can lead to foci formation and RAN translation. Sense and antisense RNA transcribed from the C9ORF72 repeat expansion accumulate as foci in the nucleus of cells and also serve as templates for RAN translation, thereby resulting in the production of six dipeptide repeat proteins that aggregate into neuronal inclusions.
RNA TOXICITY
RNA-mediated toxicity, first described for myotonic dystrophy type 1, is a putative pathomechanism involved in various microsatellite expansion disorders (Belzil et al. 2012; Gendron et al. 2014). It is believed to arise from the abnormal interaction between essential RNA-binding proteins and repeat expansion RNA transcripts, and the sequestration of these proteins into nuclear RNA foci. In c9ALS/FTD, r(G4C2)exp and r(G2C4)exp foci are detected throughout the CNS, being present mostly in neurons but also in astrocytes, oligodendrocytes, and microglia (DeJesus-Hernandez et al. 2011; Donnelly et al. 2013; Gendron et al. 2013; Lagier-Tourenne et al. 2013; Lee et al. 2013; Mizielinska et al. 2013; Zu et al. 2013; Cooper-Knock et al. 2014b, 2015b). Foci are also present in c9ALS/FTD lymphoblastoid cell lines (Lagier-Tourenne et al. 2013; Cooper-Knock et al. 2014b), peripheral blood leukocytes (Zu et al. 2013), human neurons differentiated directly from fibroblasts (iNeurons) or from iPSC (Almeida et al. 2013; Donnelly et al. 2013; Sareen et al. 2013; Su et al. 2014), and fibroblasts (Almeida et al. 2013; Donnelly et al. 2013; Lagier-Tourenne et al. 2013; Cooper-Knock et al. 2014b). In fact, foci are detected in fibroblasts of both symptomatic and asymptomatic carriers of the C9ORF72 expansion (Lagier-Tourenne et al. 2013; Cooper-Knock et al. 2014b).
Though inconsistencies exist among studies, the percentage of cells bearing sense or antisense foci appears to vary among neuroanatomical regions and could thus potentially influence disease phenotype. Mizielinska and colleagues report that the highest burden of foci was consistently found in the frontal cortex, the region that suffers the greatest neuronal loss in FTLD. However, a separate study observed a higher frequency of foci in the cerebellum of c9ALS/FTD cases compared with the frontal cortex and temporal lobes (Lee et al. 2013). That foci burden inversely correlated with age of onset in a cohort of eight c9FTD cases is of interest, yet needs validation in a larger series of cases (Mizielinska et al. 2013). Also intriguing, but in need of confirmation in additional cases, was the observation that, in three c9ALS patients, RNA foci were present in a higher proportion of motor neurons of the ventral horn compared with cerebellar granule cells; conversely, in a patient who first presented with FTD, more foci were detected in the cerebellum than in the ventral horn (Cooper-Knock et al. 2014b).
Heterogeneity in clinical phenotype among C9ORF72 repeat expansion carriers might be influenced not only by regional differences in foci frequency but also by the differential binding and/or sequestration of select RNA-binding proteins in various neuroanatomical regions. Indeed, irrespective of whether the binding of proteins to r(G4C2)exp or r(G2C4)exp causes their sequestration into foci, a potentially reversible event (Cooper-Knock et al. 2014b), these abnormal interactions are likely to adversely influence their function. It is thus not surprising that the transcriptome in fibroblasts (Donnelly et al. 2013; Lagier-Tourenne et al. 2013), iPSNs (Donnelly et al. 2013; Sareen et al. 2013), laser-captured motor neurons (Cooper-Knock et al. 2015a), the motor cortex (Donnelly et al. 2013), the frontal cortex (Prudencio et al. 2015), and especially the cerebellum (Prudencio et al. 2015) of C9ORF72 repeat expansion carriers differs from that in respective models/tissues from control subjects, ostensibly as a consequence of aberrant binding of repeat-containing transcripts to proteins that regulate gene expression and splicing. In line with this notion, prediction studies for regulators of alternative splicing events in c9ALS brain tissues identified hnRNP H (Prudencio et al. 2015), a protein binder of r(G4C2) previously implicated in r(G4C2)exp toxicity (Lee et al. 2013; Cooper-Knock et al. 2014b). Additional members of the hnRNP family have been identified in screens to determine protein binders of r(G4C2) and r(G2C4), as have many other proteins. These include hnRNP A3, hnRNP A1, serine–arginine-rich splicing factor 1 (SF2), serine–arginine-rich splicing factor 2 (SC35), Pur α, Aly/REF export factor (ALYREF), eIF2α, eIF2β, nucleolin, ADARB2, and RanGAP1 (Almeida et al. 2013; Donnelly et al. 2013; Mori et al. 2013b; Reddy et al. 2013; Sareen et al. 2013; Xu et al. 2013; Cooper-Knock et al. 2014b, 2015b; Haeusler et al. 2014; Rossi et al. 2015). The interactions between G4C2-repeat RNA and some of these proteins are discussed in more detail below.
The binding of nucleolin, a principal component of the nucleolus, to r(G4C2) when in a G-quadruplex structure (Haeusler et al. 2014) implicates nucleolar stress as a potential pathomechanism of c9ALS/FTD. In lymphoblastoid cell lines, fibroblasts, and iPSNs from c9ALS patients, the nucleolus appeared more fractured and the nucleolin more dispersed throughout the nucleus compared with cells from ALS controls. In the c9ALS/FTD motor cortex, nucleolin frequently colocalized with r(G4C2)exp foci, and there was evidence of impaired rRNA processing. In accordance with chronic nucleolar stress, an increase in processing bodies (P bodies), which are composed of ribonucleoprotein complexes involved in the degradation of untranslated mRNA, were significantly increased in c9ALS iPSNs (Haeusler et al. 2014).
The binding of r(G4C2) to mRNA export adapters, such as ALYREF, may provide a means by which r(G4C2)exp makes its way from the nucleus to the cytoplasm (Cooper-Knock et al. 2014b). It has also been speculated that the binding of (G4C2)exp pre-mRNA to hnRNP A3, which is also involved in mRNA export (Ma et al. 2002), would result in the export of r(G4C2)exp to the cytoplasm where it could be RAN translated (Mori et al. 2013b). Consistent with this, there is a significant reduction of intranuclear hnRNP A3 staining in the hippocampus of c9ALS/FTD patients along with the formation of cytoplasmic and intranuclear hnRNP A3 inclusions (Mori et al. 2013c). Pur α, shown to target specific mRNA molecules to sites of dendritic translation (Johnson et al. 2006), may also influence the localization of r(G4C2)exp or vice versa. For instance, in cultured cell models, (G4C2)31 markedly affected the distribution of Pur α and its binding partner, fragile X mental retardation proteins, leading to their accumulation in stress granules (Rossi et al. 2015). It is also notable that Pur α formed inclusions in the eyes of (G4C2)30-expressing flies, and that eye degeneration was suppressed upon Pur α overexpression. These data suggest that binding of Pur α by r(G4C2)exp causes a toxic loss of Pur α function, a theory that is strengthened by the fact that depletion of Pur α was sufficient to reduce N2A cell viability, which was also rescued by Pur α overexpression (Xu et al. 2013).
In a screen for r(G4C2) binders, the Rothstein and Sattler groups identified 19 proteins and initially focused their attention on ADARB2, a member of the ADAR family of proteins involved in RNA editing. ADARB2 abnormally accumulated in the nucleus and colocalized with r(G4C2)exp foci in iPSNs and the motor cortex of c9ALS patients (Donnelly et al. 2013). The resulting loss of ADARB2 function is believed to have contributed to the heightened vulnerability of c9ALS iPSNs to glutamate as knockdown of ADARB2 in control iPSNs enhanced their susceptibility to glutamate toxicity to levels comparable to those observed in c9ALS iPSNs (Donnelly et al. 2013). Furthermore, knockdown of ADARB2 decreased the number of c9ALS iPSNs with RNA foci by half, suggesting that ADARB2 is involved in the formation or maintenance of sense RNA foci, and that the interaction between ADARB2 and (G4C2)exp RNA plays a role in RNA-mediated toxicity. To determine which of the remaining 19 protein binders of r(G4C2) modify r(G4C2)-mediated neurodegeneration, genetic screens were performed in Drosophila that express 30 repeats and develop RNA foci but show no detectable signs of RAN translation (Zhang et al. 2015). This led to the identification of RanGAP as a potent suppressor of G4C2 repeat toxicity in fly eyes and motor neurons. RanGAP, the fly ortholog of human RanGAP1, is a key regulator of nucleocytoplasmic transport. Confirming that G4C2 repeat toxicity in Drosophila involves disrupted nucleocytoplasmic transport, a decrease in nuclear import was observed in (G4C2)30-expressing flies, and enhancing nuclear import or suppressing nuclear export of proteins suppressed (G4C2)30-mediated neurodegeneration. Importantly, RanGAP1 and components of the nuclear pore complex were mislocalized in c9ALS iPSNs and motor cortex, and c9ALS iPSNs showed signs of impaired nuclear import and disrupted TDP-43 localization. These defects, as well as foci formation, were rescued in c9ALS iPSNs by ASOs targeting G4C2 repeat RNA, suggesting that sense r(G4C2)exp causes nucleocytoplasmic transport deficits. It would thus appear that aberrations in the short C9ORF72 protein isoform and G4C2 RNA-mediated toxicity converge on a similar mechanism of toxicity. Remarkably, this same pathway has been implicated in poly(PR) and poly(GR)-mediated toxicity, as discussed below.
RAN TRANSLATION
DPR Protein Pathology: A Neuropathological Hallmark of c9ALS/FTD
RAN translation is a noncanonical form of translation discovered by the Ranum group while investigating the microsatellite expansion disorders myotonic dystrophy type 1 and spinocerebellar ataxia type 8 (Zu et al. 2011). RAN translation can occur across an entire expanded repeat without frameshifting or prematurely stopping, despite the absence of an ATG start codon. In addition, because RAN translation can initiate in multiple frames, various products can be synthesized from a given transcript. In c9ALS/FTD, both sense r(G4C2)exp and antisense r(G2C4)exp are RAN translated, leading to the synthesis of poly(GA), poly(GP), and poly(GR) proteins from r(G4C2)exp, and poly(PR), poly(PG), and poly(PA) proteins from r(G2C4)exp (Fig. 2) (Ash et al. 2013; Gendron et al. 2013; Mann et al. 2013; Mori et al. 2013a,c; Zu et al. 2013). Immunohistochemical analyses using multiple antibodies against the various DPR proteins have demonstrated highly specific staining of cytoplasmic inclusions, nuclear inclusions, or dystrophic neurites in c9ALS/FTD patients (Ash et al. 2013; Gendron et al. 2013; Mackenzie et al. 2013, 2015; Mann et al. 2013; Mori et al. 2013a,c; Zu et al. 2013; Zhang et al. 2014; Schipper et al. 2015; Schludi et al. 2015). These inclusions, which are present in neurons throughout the CNS (Ash et al. 2013; Mackenzie et al. 2013) and in ependymal cells of the spinal cord central canal and lateral ventricles (Schludi et al. 2015), can contain multiple DPR proteins (Mori et al. 2013a), are positive for p62 (Mann et al. 2013; Mori et al. 2013a), and, at least in the case of poly(GA) inclusions, contain filaments (Zhang et al. 2014). The majority of DPR protein inclusions are devoid of TDP-43, yet inclusions positive for both poly(GA) and TDP-43 have been observed on rare occasions (Mackenzie et al. 2013; Mori et al. 2013c). In such instances, poly(GA) formed the core of the inclusion and was surrounded by TDP-43, suggesting that DPR protein aggregation precedes TDP-43 pathology (Mackenzie et al. 2013; Mori et al. 2013c).
Poly(GA), poly(GP), and poly(GR) inclusions are numerous in neocortical regions, the hippocampus, and the cerebellum but are infrequently observed in the lower motor neurons of the spinal cord (Ash et al. 2013; Mackenzie et al. 2013, 2015; Mann et al. 2013; Mori et al. 2013a,c; Gomez-Deza et al. 2015; Schludi et al. 2015). Compared with these DPR proteins, inclusions of poly(PR) and poly(PA), which are uniquely RAN translated from antisense repeats, are far less abundant (Gendron et al. 2013; Mann et al. 2013; Mori et al. 2013a; Mackenzie et al. 2015; Schludi et al. 2015), perhaps because RAN translation is less efficient from antisense r(G2C4)exp or because r(G2C4)exp levels in the cytoplasm are relatively low. Alternatively, poly(PR) and/or poly(PA) proteins may be inherently more toxic than the other DPR proteins, or their biochemical properties (e.g., half-life and solubility) may not be conducive to their aggregation. With regard to the latter, a larger proportion of total poly(GP) is soluble in c9ALS/FTD cerebellar homogenates, whereas a larger proportion of total poly(GA) is insoluble (Gendron et al. 2015), consistent with poly(GA) inclusions being more frequent than those composed of poly(GP) (Mackenzie et al. 2015). Investigating these possibilities will shed light on the paucity of poly(PR) and poly(PA) inclusions in c9ALS/FTD, whether soluble DPR proteins may be toxic entities, and will also offer mechanistic insight on the factors that govern RAN translation in c9ALS/FTD. Studies to date show that cerebellar poly(GP) and poly(GA) expression levels associate with C9ORF72 transcript variant 3, the pre-mRNA of which contains the expanded repeat that serves as a template for RAN translation, but do not associate with repeat length (Gendron et al. 2015).
DPR Protein Pathology and Clinical/Neuropathological Correlations
The low abundance of DPR protein inclusions in the spinal cord, which shows marked motor neuron loss in c9ALS, renders the contribution of RAN translation to disease pathogenesis unclear (Ash et al. 2013; Gendron et al. 2013; Zu et al. 2013; Gomez-Deza et al. 2015; Schludi et al. 2015). In addition, reports on clinical and neuropathological correlations with DPR protein pathology have been conflicting, perhaps because different methods to estimate DPR protein pathology were used (e.g., semiquantitative versus quantitative measures), different neuroanatomical regions and DPR proteins were evaluated, and the staging of cases also differed (i.e., clinically versus neuropathologically) (Mackenzie et al. 2013, 2015; Mann et al. 2013; Davidson et al. 2014; Schludi et al. 2015). Although Mackenzie and colleagues found TDP-43 pathology to closely parallel neurodegeneration, they detected only moderate associations between the amount of poly(GA) dystrophic neurites and degeneration in the frontal cortex, and between total poly(GA) pathology and disease onset (Mackenzie et al. 2013, 2015). Moreover, they observed no other relationship between DPR protein pathology and clinical phenotype or neurodegeneration in their cohort of 35 cases representing the clinical spectrum associated with the C9ORF72 mutation (Mackenzie et al. 2013, 2015). Likewise, the Mann group found no significant differences in the pattern or frequency of poly(GA) inclusions among FTD, ALS, and FTD-MND cases caused by C9ORF72 repeat expansions, nor did they observe differences in the extent of TDP-43 pathology between FTD patients bearing an expansion and those who did not (Davidson et al. 2014). Conversely, the Edbauer group discovered that, in a cohort of 14 C9ORF72 expansion carriers, poly(PR) aggregates, despite being present in very low frequency, were more common in the hippocampus of FTLD cases compared with MND cases, and that poly(GA) inclusions in cerebellar granule cells were significantly more abundant in FTLD cases compared with MND or FTLD-MND cases (Schludi et al. 2015). Finally, taking a departure from traditional immunohistochemical approaches and instead using immunoassays to quantitatively measure poly(GP) and poly(GA) levels in various brain regions from 55 C9ORF72 mutation carriers, the Petrucelli group observed that poly(GP) levels in the cerebellum were significantly lower in patients with ALS compared with patients with FTLD or FTLD-MND. Furthermore, although cerebellar poly(GP) did not associate with age of disease onset or survival after onset in this large cohort, it did associate with cognitive impairment in 15 c9ALS patients for whom neuropsychological data were available (Gendron et al. 2015). Taken together, the latter two studies implicate cerebellar abnormalities as a contributor to the neuropathological and clinical heterogeneity associated with C9ORF72 repeat expansions. Given that DPR protein expression is highest in the cerebellum (Gendron et al. 2015), that robust transcriptome changes occur in the cerebellum of c9ALS patients (Prudencio et al. 2015), that cerebellar atrophy is reported in C9ORF72 expansion carriers (Mahoney et al. 2012; Sha et al. 2012; Whitwell et al. 2012), and that the growing body of neuroimaging and clinical evidence supports cerebellar involvement in cognitive and affective regulation (Koziol et al. 2014), more thorough investigations on the role of the cerebellum in c9ALS/FTD pathogenesis are warranted. In addition, the findings above collectively suggest that DPR protein pathology/expression may influence clinical features of C9ORF72 repeat expansion despite a lack of association with TDP-43 pathology or neurodegeneration. Along this line of thought, the discovery of a c9FTD kindred with early intellectual disability and extensive poly(GA) inclusions but little, if any, TDP-43 pathology could indicate that DPR proteins are harmful or, at the very least, that clinical symptoms can manifest in C9ORF72 expansion carriers prior to TDP-43 pathology (Proudfoot et al. 2014). This is further supported by three C9ORF72 mutation carriers who developed fairly rapid cognitive decline but died prematurely because of unrelated illness; these patients had abundant poly(GA) pathology but only sparse TDP-43 pathology (Baborie et al. 2015). Despite the marked poly(GA) pathology in these patients, it must nonetheless be kept in mind that their clinical features could also have been caused by C9ORF72 loss of function and/or RNA toxicity. The same is true for findings emerging from the above-described correlation studies between DPR protein pathology and clinical phenotypes. Nevertheless, findings from several groups that specifically examined DPR proteins in a variety of models provide compelling evidence that certain DPR proteins are toxic (Zu et al. 2013; Kwon et al. 2014; May et al. 2014; Mizielinska et al. 2014; Wen et al. 2014; Zhang et al. 2014; Freibaum et al. 2015; Jovicic et al. 2015; Tao et al. 2015; Tran et al. 2015; Yamakawa et al. 2015; Yang et al. 2015).
In Vitro and In Vivo Models of DPR Protein Toxicity
To distinguish between repeat RNA and DPR protein toxicity, the Isaacs group used an elegant strategy to generate either “pure repeats,” which could form foci and be RAN translated, or “RNA-only” repeats, which formed foci but were not RAN translated because of stop codons interrupting the repeats. Expression of the pure repeats in the Drosophila eye caused eye degeneration not observed when RNA-only repeats were expressed. Similarly, the expression of pure repeats, but not RNA-only repeats, in adult neurons caused early lethality of flies (Mizielinska et al. 2014). These in vivo experiments demonstrated that the C9ORF72 repeat expansion likely causes toxicity through the production of DPR proteins in this model. The Gao group reached a similar conclusion using their Drosophila model, which expressed a C9ORF72 minigene that contained 160 G4C2 repeats flanked by human intronic and exonic sequences. Although abundant nuclear RNA foci formed in the neurons and glia of these flies, DPR protein levels were extremely low and virtually no toxicity ensued (Tran et al. 2015). By comparing poly(GP) protein levels between their (G4C2)160 flies and the Isaacs fly model that expressed 36 highly toxic G4C2 repeats in the context of a poly(A)+ mRNA, the Gao group found poly(GP) levels to be ∼100-fold higher in (G4C2)36 flies than in (G4C2)160 flies, presumably because the poly(A) tail allowed (G4C2)36 to be efficiently exported to the cytoplasm for RAN translation. Furthermore, the Gao group observed that the modest toxicity that emerged in (G4C2)160 flies when subjected to a higher temperature correlated with an increase in DPR protein production but not in RNA foci (Tran et al. 2015). These data suggest that the marked and mild toxicity observed in flies expressing (G4C2)36 and (G4C2)160, respectively, was caused by DPR proteins. In fact, the Gao group postulated that nuclear foci are neutral intermediates or possibly even protective, as they prevent the export of r(G4C2)exp to the cytoplasm and subsequent RAN translation.
To determine which DPR proteins were toxic in Drosophila, Mizielinska et al. took advantage of “protein-only” constructs that use alternative codons to those found within the G4C2 repeat to express poly(GR), poly(PR), poly(GA), or poly(PA). When these DPR proteins were expressed in the eye, only the arginine-containing DPR proteins were toxic. When expressed in adult neurons, (GR)100 and (PR)100 caused a substantial decrease in survival, (GA)100 caused a late-onset reduction in survival, and (PA)100 had no effect (Mizielinska et al. 2014). Several other groups have shown poly(PR) and/or poly(GR) to be toxic in Drosophila (Wen et al. 2014; Freibaum et al. 2015; Yang et al. 2015). For instance, the Trotti group demonstrated that (PR)50, but not (GA)50 or (PA)50, caused severe neurodegeneration in the fly eye and early lethality when expressed in motor neurons (Wen et al. 2014). This group also showed that exogenous poly(PR) proteins dramatically decreased primary cortical neuron survival, with the risk of death being considerably increased if nuclear aggregates were present. Interestingly, the Trotti group found poly(PR) aggregates to be very stable and to remain present even after the neurons harboring them had died (Wen et al. 2014). Together, these findings suggest that poly(GR) and poly(PR) contribute to neurodegeneration; because both of these DPR proteins are arginine-rich, they may even share pathological mechanisms. Multiple studies provide evidence that one such mechanism may involve impaired RNA biogenesis and nucleolar stress (Kwon et al. 2014; Wen et al. 2014; Jovicic et al. 2015; Tao et al. 2015; Yang et al. 2015), and others suggest these arginine-rich proteins impair the dynamics of RNA granules (i.e., stress granules and P bodies) (Wen et al. 2014; Tao et al. 2015; Yamakawa et al. 2015). Another likely mechanism of disease has emerged through genetic modifier screens using yeast and Drosophila models—impaired nucleocytoplasmic transport (Freibaum et al. 2015; Jovicic et al. 2015). Using codon-optimized constructs to express a given DPR protein without the use of G4C2 repeat sequences, the Gitler group found that arginine-rich DPR proteins, especially poly(PR), were toxic in yeast. Subsequent unbiased screens to identify genetic modifiers that suppressed or enhanced poly(PR) toxicity uncovered genes involved in nucleocytoplasmic transport; these included members of the karyopherin family of nuclear import proteins, components of the nuclear pore complex, and regulators of the RAN-GTPase cycle, which generates the energy needed to power nuclear import. Notably, upregulating karyopherin expression rescued poly(PR) toxicity (Jovicic et al. 2015). A genetic screen to find modifiers of (G4C2)58 toxicity in Drosophila similarly led to the identification of genes encoding proteins with nucleocytoplasmic transport functions, including several components of the nuclear pore complex and the machinery that coordinates the import of nuclear proteins and the export of nuclear RNA (Freibaum et al. 2015). With regard to the latter, cells of (G4C2)58-expressing Drosophila and c9FTD iPSNs showed evidence of nuclear RNA retention, consistent with defects in RNA export. Overall, there is a compelling body of work supporting the toxicity of arginine-rich DPR proteins.
Based on data from multiple models investigating poly(GA) (in the absence of the confounding contribution of RNA toxicity or the expression of other DPR proteins), a case in favor of poly(GA) toxicity is also mounting (May et al. 2014; Mizielinska et al. 2014; Zhang et al. 2014; Jovicic et al. 2015; Yamakawa et al. 2015). When expressed in cultured cell lines, rodent primary neurons, and P10 mice, poly(GA) formed soluble and insoluble high-molecular-weight species, as well as inclusions positive for p62 and ubiquitin, the majority of which were cytoplasmic, although nuclear inclusions were also observed (May et al. 2014; Zhang et al. 2014; Schludi et al. 2015; Yamakawa et al. 2015). Zhang et al. found that expression of GFP-(GA)50 induced caspase 3 activation and increased extracellular lactate dehydrogenase (LDH) levels in HEK293T cells. Although May et al. failed to see similar poly(GA)-induced toxicity in their HEK293FT cultures, both groups found that poly(GA) proteins were neurotoxic to primary rodent neuronal cultures. As in cultured cells, poly(GA) proteins formed p62- and ubiquitin-positive inclusions in neuronal cultures, which was associated with the activation of caspase 3, DNA fragmentation, impaired neurite outgrowth or branching, and enhanced LDH release (May et al. 2014; Zhang et al. 2014). Poly(GA) expression was also associated with increased p62 expression (May et al. 2014) and the accumulation of ubiquitinated proteins (Zhang et al. 2014), which suggests impaired activity of the UPS. Consistent with this finding, Zhang et al. found that proteasome activity decreased in neurons expressing GFP-(GA)50, which, in turn, led to activation of endoplasmic reticulum (ER) stress, the inhibition of which provided rescue from poly(GA)-induced neurotoxicity. Importantly, mRNA levels of ER stress markers ATF4 and CHOP were significantly increased in the frontal cortex of c9ALS cases compared with sporadic ALS cases (Zhang et al. 2014), and c9ALS iPSNs exhibited an increased vulnerability to tunicamycin, an ER stress inducer (Haeusler et al. 2014), offering compelling evidence that UPS dysfunction and ER stress may be pathogenic mechanisms in c9ALS/FTD.
Offering additional support that the UPS is influenced by poly(GA) proteins, Yamakawa et al. reported that poly(GA), as well as poly(GP) and poly(GR), caused UPS dysfunction in cultured cells and also rendered cells more vulnerable to UPS inhibition. Furthermore, through an unbiased approach to identify poly(GA)-interacting proteins, May et al. found that proteasomal subunits and other ubiquitin-related proteins (e.g., ubiquilin 1 and 2) were enriched in the poly(GA) interactome. Another interacting partner of interest is Unc119, a protein required for the maintenance of the nervous system architecture in C. elegans that, when mutated, causes movement, sensory, and behavioral abnormalities in this model (Maduro et al. 2000; Knobel et al. 2001). When coexpressed with poly(GA) in HEK293 cells or primary neurons, Unc119 colocalized with poly(GA) inclusions and became insoluble, in contrast to its normal diffuse cytoplasmic localization. As the sequestration of Unc119 could cause its loss of function and contribute to neurotoxicity, this potential pathomechanism was examined. Knocking down Unc119 in primary neurons led to dendritic withering, akin to that observed upon poly(GA) expression, and neuronal death. Furthermore, Unc119 overexpression was able to partially reduce toxicity associated with poly(GA) expression, again suggesting that Unc119 loss of function contributes to poly(GA)-induced neurotoxicity. It is thus of interest that inclusions of Unc119 were observed in tandem with a loss of cytosolic Unc119 staining in the brain of C9ORF72 expansion carriers. Overall, multiple lines of evidence suggest that poly(GA) proteins, the most abundantly expressed DPR protein in c9FTD/ALS, are detrimental.
c9ALS/FTD THERAPEUTICS AND BIOMARKERS
The substantial advances made toward elucidating how C9ORF72 repeat expansions contribute to c9ALS/FTD provide insight into potential therapeutics and diagnostics. Based on the assumption that r(G4C2)exp and r(G2C4)exp are at the crux of c9ALS/FTD pathogenesis, as they are required for foci formation and RAN translation, strategies that target these transcripts and result in their neutralization or degradation could effectively block early culprits of disease. It is therefore promising that several groups have identified C9ORF72-targeting antisense oligonucleotides (ASOs) that mitigate r(G4C2)exp foci formation, gene expression alterations, enhanced susceptibility to excitotoxicity, and/or nucleocytoplasmic transport deficits in c9ALS/FTD fibroblasts, iPSN, and/or (G4C2)30-expressing Drosophila (Donnelly et al. 2013; Lagier-Tourenne et al. 2013; Sareen et al. 2013; Zhang et al. 2015). Boding well for the potential use of ASOs to treat c9ALS/FTD, intrathecal administration of ASOs against SOD1 was well tolerated and showed no significant adverse events in a phase 1 study, and ASOs are moving toward a phase 3 trial for spinal muscular atrophy (Miller et al. 2013; Reddy and Miller 2015; Wirth et al. 2015). In addition to ASOs, small molecules offer an attractive approach for targeting r(G4C2)exp and r(G2C4)exp given pharmacological advantages, such as their small molecular weight, which is important for blood–brain barrier permeability. Indeed, the Disney and Petrucelli groups recently identified small molecules that bind the hairpin structure of r(G4C2)exp in vitro and in cells, and that significantly decrease RAN translation and foci formation in cultured cells overexpressing r(G4C2)66 and iNeurons derived from C9ORF72 repeat expansion carriers (Su et al. 2014). On an encouraging note, data from the Petrucelli (G4C2)66 mouse model imply that therapeutics targeting G4C2 repeat RNA to mitigate foci formation and RAN translation should also attenuate TDP-43-mediated toxicity (Chew et al. 2015).
Should findings in yeast and Drosophila that implicate defective nucleocytoplasmic transport as a disease mechanism bear out in mammalian models and patient-derived cells/tissues, this pathway would offer additional prospective therapeutic targets for c9ALS/FTD. It is thus noteworthy that TMPyP4, a porphyrin compound that destabilizes RNA G-quadruplex structures and that blocks in vitro interactions between r(G4C2)8 and its protein-binding partners (Zamiri et al. 2014), was found to similarly reduce the affinity of RanGAP1 for r(G4C2)10 (Zhang et al. 2015). Moreover, TMPyP4, as well as a small-molecule inhibitor of nuclear export, rescued nuclear import defects and eye neurodegeneration in (G4C2)30-expressing Drosophila (Zhang et al. 2015).
For all c9ALS/FTD therapeutic strategies being investigated, there are considerations to keep in mind. Among them, their effect on C9ORF72 mRNA expression should be carefully monitored. Although the exact contribution of C9ORF72 loss of function must still be deciphered, causing a further decrease in C9ORF72 expression by a purportedly protective intervention could have adverse effects, especially in light of the finding that higher levels of C9ORF72 variant 1 are associated with prolonged survival after disease onset (van Blitterswijk et al. 2015). Therapeutics that increase C9ORF72 expression may reverse harmful effects that could occur as a consequence of C9ORF72 loss of function, although it would be important that they not do so at the expense of increasing r(G4C2)exp and r(G2C4)exp. Because epigenetic processes mediate the decrease in C9ORF72 mRNA expression in c9ALS/FTD, they may offer a unique opportunity for therapeutic development; however, caution is needed given findings suggesting that transcriptional silencing of C9ORF72 through CpG island hypermethylation is protective by virtue of decreasing RNA foci formation and RAN translation (Belzil et al. 2013, 2014; Xi et al. 2013, 2014, 2015a,b; Liu et al. 2014; Russ et al. 2014; Gijselinck et al. 2015; He et al. 2015; McMillan et al. 2015).
To increase the probability of success in developing efficient therapeutic interventions, biomarkers and pharmacodynamic measures of disease activity and drug action must be identified. These biomarkers could encompass epigenetic, RNA, or protein signatures unique to C9ORF72 repeat expansion carriers that can be easily detected (e.g., in cerebrospinal fluid [CSF], blood, skin, and other biofluids or peripheral tissues). One such potential biomarker recently came to light with the discovery that poly(GP) DPR proteins are detectable in the CSF of c9ALS patients but not sporadic ALS patients (Su et al. 2014). This discovery could not only facilitate the identification of C9ORF72 repeat expansion carriers in the course of diagnostic workups, but may also pave the way in determining whether changes in DPR protein levels in CSF correlate with clinical phenotype or disease severity and progression. Importantly, CSF DPR proteins, especially poly(GP) proteins that are synthesized from both sense and antisense transcripts, may prove useful as a pharmacodynamic biomarker to assess the efficacy of r(G4C2)exp- or r(G2C4)exp-targeting therapies. The existence of DPR proteins in CSF also raises the possibility of their propagation from neuron to neuron, a phenomenon that is becoming more widely recognized in neurodegenerative diseases. Whether DPR proteins are released from cells by regulated processes or as a result of degenerating neurons, and whether pathology may be propagated in this fashion are questions that are worthy of investigation.
CONCLUSION
Since the relatively recent discovery of C9ORF72 repeat expansions, an impressive and fruitful effort has been put toward investigating the mechanisms of disease. The normal functions of C9ORF72 are being pieced together, the interacting partners of r(G4C2)exp and r(G2C4)exp identified, the toxic potential of DPR proteins evaluated, and many new models developed. In addition to these studies, therapeutic approaches are being explored and biomarkers sought. Nevertheless, the exact pathomechanisms of C9ORF72 expansions remain unknown, and much work lies ahead. Indeed, it is critical to confirm findings from experimental models in large series of patient-derived cells and tissues. It is nonetheless interesting to note that certain abnormalities, such as nucleolar stress, RNA dysregulation, nucleocytoplasmic transport deficits, and impaired protein degradation, have been uncovered by studies investigating RNA and DPR protein toxicity, as well as C9ORF72 loss of function. Also presently obscure, despite recent clues, is how the same repeat expansion, albeit with varying repeat lengths, causes a wide spectrum of clinical phenotypes. Overall, much about c9ALS/FTD remains to be untangled, but should the current pace of research on these devastating diseases continue, our understanding of c9ALS/FTD is expected to grow swiftly.
ACKNOWLEDGMENTS
This work is supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke [R21NS089979 (T.F.G.), R21NS084528 (L.P.), R01NS088689 (L.P.), R01NS063964 (L.P.), R01NS077402 (L.P.), P01NS084974 (L.P.)], Department of Defense [ALSRP AL130125 (L.P.)], Mayo Clinic Foundation (L.P.), ALS Association (T.F.G.), Target ALS (L.P.), and the Robert Packard Center for ALS Research at Johns Hopkins (L.P.).
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Diseases available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D, Pook M. 2008. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet 17: 735–746. [DOI] [PubMed] [Google Scholar]
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, et al. 2013. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol 126: 385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sarraj S, King A, Troakes C, Smith B, Maekawa S, Bodi I, Rogelj B, Al-Chalabi A, Hortobagyi T, Shaw CE. 2011. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol 122: 691–702. [DOI] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, et al. 2006. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 351: 602–611. [DOI] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW III, Rademakers R, et al. 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RA, Fernandez-Martos CM, Atkin JD, Vickers JC, King AE. 2015. C9ORF72 expression and cellular localization over mouse development. Acta Neuropathol Commun 3: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baborie A, Griffiths TD, Jaros E, Perry R, McKeith IG, Burn DJ, Masuda-Suzukake M, Hasegawa M, Rollinson S, Pickering-Brown S, et al. 2015. Accumulation of dipeptide repeat proteins predates that of TDP-43 in frontotemporal lobar degeneration associated with hexanucleotide repeat expansions in C9ORF72 gene. Neuropathol Appl Neurobiol 41: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, Campbell T, Uphill J, Borg A, Fratta P, et al. 2013. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am J Hum Genet 92: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Gendron TF, Petrucelli L. 2012. RNA-mediated toxicity in neurodegenerative disease. Mol Cell Neurosci: S1044–S7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, et al. 2013. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol 126: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Bauer PO, Gendron TF, Murray ME, Dickson D, Petrucelli L. 2014. Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res 1584: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benussi L, Rossi G, Glionna M, Tonoli E, Piccoli E, Fostinelli S, Paterlini A, Flocco R, Albani D, Pantieri R, et al. 2014. C9ORF72 hexanucleotide repeat number in frontotemporal lobar degeneration: A genotype-phenotype correlation study. J Alzheimers Dis 38: 799–808. [DOI] [PubMed] [Google Scholar]
- Boeve BF, Boylan KB, Graff-Radford NR, Dejesus-Hernandez M, Knopman DS, Pedraza O, Vemuri P, Jones D, Lowe V, Murray ME, et al. 2012. Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain 135: 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettschneider J, Van Deerlin VM, Robinson JL, Kwong L, Lee EB, Ali YO, Safren N, Monteiro MJ, Toledo JB, Elman L, et al. 2012. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol 123: 825–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman VL, Cooper-Knock J, Connor-Robson N, Higginbottom A, Kirby J, Razinskaya OD, Ninkina N, Shaw PJ. 2013. Simultaneous and independent detection of C9ORF72 alleles with low and high number of GGGGCC repeats using an optimised protocol of Southern blot hybridisation. Mol Neurodegener 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, Heverin M, Jordan N, Kenna K, Lynch C, et al. 2012. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: A population-based cohort study. Lancet Neurol 11: 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, et al. 2015. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348: 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Borghero G, Restagno G, Mora G, Drepper C, Traynor BJ, Sendtner M, Brunetti M, Ossola I, Calvo A, et al. 2012. Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain 135: 784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, Kabashi E. 2013. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann Neurol 74: 180–187. [DOI] [PubMed] [Google Scholar]
- Cooper-Knock J, Hewitt C, Highley JR, Brockington A, Milano A, Man S, Martindale J, Hartley J, Walsh T, Gelsthorpe C, et al. 2012. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain 135: 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Shaw PJ, Kirby J. 2014a. The widening spectrum of C9ORF72-related disease; genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol 127: 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J, et al. 2014b. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 137: 2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Bury JJ, Heath PR, Wyles M, Higginbottom A, Gelsthorpe C, Highley JR, Hautbergue G, Rattray M, Kirby J, et al. 2015a. C9ORF72 GGGGCC expanded repeats produce splicing dysregulation which correlates with disease severity in amyotrophic lateral sclerosis. PLoS ONE 10: e0127376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Higginbottom A, Stopford MJ, Highley JR, Ince PG, Wharton SB, Pickering-Brown S, Kirby J, Hautbergue GM, Shaw PJ. 2015b. Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol 130: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, Van Langenhove T, van der Zee J, Van Broeckhoven C. 2013. Current insights into the C9orf72 repeat expansion diseases of the FTLD/ALS spectrum. Trends Neurosci 36: 450–459. [DOI] [PubMed] [Google Scholar]
- Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, Smith B, Al-Saraj S, Shaw C, Rollinson S, et al. 2014. Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dols-Icardo O, Garcia-Redondo A, Rojas-Garcia R, Sanchez-Valle R, Noguera A, Gomez-Tortosa E, Pastor P, Hernandez I, Esteban-Perez J, Suarez-Calvet M, et al. 2014. Characterization of the repeat expansion size in C9orf72 in amyotrophic lateral sclerosis and frontotemporal dementia. Hum Mol Genet 23: 749–754. [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. 2013. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80: 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, et al. 2014. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 23: 3579–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. 2012. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep 2: 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Poulter M, Lashley T, Rohrer JD, Polke JM, Beck J, Ryan N, Hensman D, Mizielinska S, Waite AJ, et al. 2013. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol 126: 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Polke JM, Newcombe J, Mizielinska S, Lashley T, Poulter M, Beck J, Preza E, Devoy A, Sidle K, et al. 2015. Screening a UK amyotrophic lateral sclerosis cohort provides evidence of multiple origins of the C9orf72 expansion. Neurobiol Aging 36: 546 e541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. 2015. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525: 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Petrucelli L. 2011. Rodent models of TDP-43 proteinopathy: Investigating the mechanisms of TDP-43-mediated neurodegeneration. J Mol Neurosci 45: 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. 2013. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126: 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. 2014. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol 127: 359–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, van Blitterswijk M, Bieniek KF, Daughrity LM, Jiang J, Rush BK, Pedraza O, Lucas JA, Murray ME, Desaro P, et al. 2015. Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol 130: 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ghasemi M, Brown RH Jr. 2017. Genetics of amyotrophic lateral sclerosis. Cold Spring Harb Perspect Med 10.1101/cshperspect.a024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, et al. 2012. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: A gene identification study. Lancet Neurol 11: 54–65. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Engelborghs S, De Bleecker J, Ivanoiu A, Deryck O, Edbauer D, Zhang M, et al. 2015. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol Psychiatry 21: 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Gijselinck I, Cruts M, Van Broeckhoven C. 2017. The genetics of C9orf72 expansions. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Deza J, Lee YB, Troakes C, Nolan M, Al-Sarraj S, Gallo JM, Shaw CE. 2015. Dipeptide repeat protein inclusions are rare in the spinal cord and almost absent from motor neurons in C9ORF72 mutant amyotrophic lateral sclerosis and are unlikely to cause their degeneration. Acta Neuropathol Commun 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E, Mahishi L, Entezam A, Kumari D, Usdin K. 2007. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res 35: 3383–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Long JC, Caceres JF. 2006. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol 26: 5744–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. 2014. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Tang L, Benyamin B, Shah S, Hemani G, Liu R, Ye S, Liu X, Ma Y, Zhang H, et al. 2015. C9orf72 hexanucleotide repeat expansions in Chinese sporadic amyotrophic lateral sclerosis. Neurobiol Aging 36: 2660 e2661–2668. [DOI] [PubMed] [Google Scholar]
- Hsiung GY, Dejesus-Hernandez M, Feldman HH, Sengdy P, Bouchard-Kerr P, Dwosh E, Butler R, Leung B, Fok A, Rutherford NJ, et al. 2012. Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p. Brain 135: 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukema RK, Riemslagh FW, Melhem S, van der Linde HC, Severijnen L, Edbauer D, Maas A, Charlet-Berguerand N, Willemsen R, van Swieten JC. 2014. A new inducible transgenic mouse model for C9orf72-associated GGGGCC repeat expansion supports a gain-of-function mechanism in C9orf72 associated ALS and FTD. Acta Neuropathol Commun 2: 166. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hutagalung AH, Novick PJ. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91: 119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, Khalili K, Winckler B, Gordon J. 2006. Role of Pur α in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res 83: 929–943. [DOI] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G, et al. 2015. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18: 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobel KM, Davis WS, Jorgensen EM, Bastiani MJ. 2001. UNC-119 suppresses axon branching in C. elegans. Development 128: 4079–4092. [DOI] [PubMed] [Google Scholar]
- Koppers M, Blokhuis AM, Westeneng HJ, Terpstra ML, Zundel CA, Vieira de Sa R, Schellevis RD, Waite AJ, Blake DJ, Veldink JH, et al. 2015. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann Neurol 78: 426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC. 2010. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 584: 1393–1398. [DOI] [PubMed] [Google Scholar]
- Koziol LF, Budding D, Andreasen N, D’Arrigo S, Bulgheroni S, Imamizu H, Ito M, Manto M, Marvel C, Parker K, et al. 2014. Consensus paper: The cerebellum’s role in movement and cognition. Cerebellum 13: 151–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. 2014. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345: 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, et al. 2013. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci 110: E4530–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. 2013. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5: 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. 2013. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29: 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, Irwin DJ, Van Deerlin VM, Lee EB. 2014. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol 128: 525–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomen-Hoerth C, Murphy J, Langmore S, Kramer JH, Olney RK, Miller B. 2003. Are amyotrophic lateral sclerosis patients cognitively normal? Neurology 60: 1094–1097. [DOI] [PubMed] [Google Scholar]
- Lowe J. 1994. New pathological findings in amyotrophic lateral sclerosis. J Neurol Sci 124: 38–51. [DOI] [PubMed] [Google Scholar]
- Ma AS, Moran-Jones K, Shan J, Munro TP, Snee MJ, Hoek KS, Smith R. 2002. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J Biol Chem 277: 18010–18020. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D, et al. 2013. Dipeptide repeat protein pathology in C9ORF72 mutation cases: Clinico-pathological correlations. Acta Neuropathol 126: 859–879. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Frick P, Neumann M. 2014. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta Neuropathol 127: 347–357. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D, et al. 2015. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol 130: 845–861. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Gordon M, Jacobs R, Pilgrim DB. 2000. The UNC-119 family of neural proteins is functionally conserved between humans, Drosophila and C. elegans. J Neurogenet 13: 191–212. [DOI] [PubMed] [Google Scholar]
- Mahoney CJ, Beck J, Rohrer JD, Lashley T, Mok K, Shakespeare T, Yeatman T, Warrington EK, Schott JM, Fox NC, et al. 2012. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: Clinical, neuroanatomical and neuropathological features. Brain 135: 736–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, et al. 2012. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol 11: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M, et al. 2013. Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 1: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf-Strathmann J, et al. 2014. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol 128: 485–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan CT, Russ J, Wood EM, Irwin DJ, Grossman M, McCluskey L, Elman L, Van Deerlin V, Lee EB. 2015. C9orf72 promoter hypermethylation is neuroprotective: Neuroimaging and neuropathologic evidence. Neurology 84: 1622–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, Andres PL, Mahoney K, Allred P, Alexander K, et al. 2013. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol 12: 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, Isaacs AM. 2013. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol 126: 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. 2014. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345: 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, et al. 2013a. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol 126: 881–893. [DOI] [PubMed] [Google Scholar]
- Mori K, Lammich S, Mackenzie IR, Forne I, Zilow S, Kretzschmar H, Edbauer D, Janssens J, Kleinberger G, Cruts M, et al. 2013b. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol 125: 413–423. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. 2013c. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339: 1335–1338. [DOI] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, et al. 1998. Frontotemporal lobar degeneration: A consensus on clinical diagnostic criteria. Neurology 51: 1546–1554. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133. [DOI] [PubMed] [Google Scholar]
- Nordin A, Akimoto C, Wuolikainen A, Alstermark H, Jonsson P, Birve A, Marklund SL, Graffmo KS, Forsberg K, Brannstrom T, et al. 2015. Extensive size variability of the GGGGCC expansion in C9orf72 in both neuronal and non-neuronal tissues in 18 patients with ALS or FTD. Hum Mol Genet 24: 3133–3142. [DOI] [PubMed] [Google Scholar]
- Pikkarainen M, Hartikainen P, Alafuzoff I. 2010. Ubiquitinated p62-positive, TDP-43-negative inclusions in cerebellum in frontotemporal lobar degeneration with TAR DNA binding protein 43. Neuropathology 30: 197–199. [DOI] [PubMed] [Google Scholar]
- Proudfoot M, Gutowski NJ, Edbauer D, Hilton DA, Stephens M, Rankin J, Mackenzie IR. 2014. Early dipeptide repeat pathology in a frontotemporal dementia kindred with C9ORF72 mutation and intellectual disability. Acta Neuropathol 127: 451–458. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Belzil V, Batra R, Ross CA, Gendron TF, Pregent L, Murray ME, Overstreet K, Piazza-Johnston A, Desaro P, et al. 2015. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18: 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy LV, Miller TM. 2015. RNA-targeted therapeutics for ALS. Neurotherapeutics 12: 424–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB Jr, Pearson CE. 2013. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem 288: 9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S, Serrano A, Gerbino V, Giorgi A, Di Francesco L, Nencini M, Bozzo F, Schinina ME, Bagni C, Cestra G, et al. 2015. Nuclear accumulation of mRNAs underlies G4C2-repeat-induced translational repression in a cellular model of C9orf72 ALS. J Cell Sci 128: 1787–1799. [DOI] [PubMed] [Google Scholar]
- Russ J, Liu EY, Wu K, Neal D, Suh E, Irwin DJ, McMillan CT, Harms MB, Cairns NJ, Wood EM, et al. 2014. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol 129: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. 2013. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 5: 208ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper LJ, Raaphorst J, Aronica E, Baas F, de Haan R, de Visser M, Troost D. 2015. Prevalence of brain and spinal cord inclusions, including dipeptide repeat proteins, in patients with the C9ORF72 hexanucleotide repeat expansion: A systematic neuropathological review. Neuropathol Appl Neurobiol 42: 547–560. [DOI] [PubMed] [Google Scholar]
- Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D. 2015. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol 130: 537–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha SJ, Takada LT, Rankin KP, Yokoyama JS, Rutherford NJ, Fong JC, Khan B, Karydas A, Baker MC, DeJesus-Hernandez M, et al. 2012. Frontotemporal dementia due to C9ORF72 mutations: Clinical and imaging features. Neurology 79: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Dopper EG, Cohn-Hokke PE, Hukema RK, Nicolaou N, Seelaar H, de Graaf JR, de Koning I, van Schoor NM, Deeg DJ, et al. 2012. The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain 135: 723–735. [DOI] [PubMed] [Google Scholar]
- Snowden JS, Neary D, Mann DM. 2002. Frontotemporal dementia. Br J Psychiatry 180: 140–143. [DOI] [PubMed] [Google Scholar]
- Snowden JS, Rollinson S, Thompson JC, Harris JM, Stopford CL, Richardson AM, Jones M, Gerhard A, Davidson YS, Robinson A, et al. 2012. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain 135: 693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart H, Rutherford NJ, Briemberg H, Krieger C, Cashman N, Fabros M, Baker M, Fok A, Dejesus-Hernandez M, Eisen A, et al. 2012. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathol 123: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. 2014. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 83: 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh E, Lee EB, Neal D, Wood EM, Toledo JB, Rennert L, Irwin DJ, McMillan CT, Krock B, Elman LB, et al. 2015. Semi-automated quantification of C9orf72 expansion size reveals inverse correlation between hexanucleotide repeat number and disease duration in frontotemporal degeneration. Acta Neuropathol 130: 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. 1992. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet 1: 397–400. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Maroof AM, Merkle FT, Koszka K, Intoh A, Armstrong I, Moccia R, Davis-Dusenbery BN, Eggan K. 2013. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci 16: 1725–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Wang H, Xia Q, Li K, Jiang X, Xu G, Wang G, Ying Z. 2015. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet 24: 2426–2441. [DOI] [PubMed] [Google Scholar]
- Therrien M, Rouleau GA, Dion PA, Parker JA. 2013. Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS ONE 8: e83450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, Pandey UB, Di Prospero NA, Min KT, Taylor JP, Paulson HL. 2010. Histone deacetylases suppress CGG repeat-induced neurodegeneration via transcriptional silencing in models of fragile X tremor ataxia syndrome. PLoS Genet 6: e1001240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z, et al. 2015. Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a Drosophila model of C9ORF72 FTD/ALS. Neuron 87: 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Dejesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO, et al. 2013. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): A cross-sectional cohort study. Lancet Neurol 12: 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Gendron TF, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Daughrity LM, Murray ME, Heckman MG, Jiang J, et al. 2015. Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathol 130: 863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Engelborghs S, Philtjens S, Vandenbulcke M, Sleegers K, Sieben A, et al. 2013. A pan-European study of the C9orf72 repeat associated with FTLD: Geographic prevalence, genomic instability, and intermediate repeats. Hum Mutat 34: 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ. 2014. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging 35: 1779.e5–1779.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. 2014. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Weigand SD, Boeve BF, Senjem ML, Gunter JL, Dejesus-Hernandez M, Rutherford NJ, Baker M, Knopman DS, Wszolek ZK, et al. 2012. Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain 135: 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B, Barkats M, Martinat C, Sendtner M, Gillingwater TH. 2015. Moving towards treatments for spinal muscular atrophy: Hopes and limits. Expert Opin Emerg Drugs 20: 353–356. [DOI] [PubMed] [Google Scholar]
- Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, Zheng Y, Ghani M, Dib S, Keith J, et al. 2013. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet 92: 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Rainero I, Rubino E, Pinessi L, Bruni AC, Maletta RG, Nacmias B, Sorbi S, Galimberti D, Surace EI, et al. 2014. Hypermethylation of the CpG-island near the C9orf72 G4C2-repeat expansion in FTLD patients. Hum Mol Genet 23: 5630–5637. [DOI] [PubMed] [Google Scholar]
- Xi Z, van Blitterswijk M, Zhang M, McGoldrick P, McLean JR, Yunusova Y, Knock E, Moreno D, Sato C, McKeever PM, et al. 2015a. Jump from pre-mutation to pathologic expansion in C9orf72. Am J Hum Genet 96: 962–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Zhang M, Bruni AC, Maletta RG, Colao R, Fratta P, Polke JM, Sweeney MG, Mudanohwo E, Nacmias B, et al. 2015b. The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol 129: 715–727. [DOI] [PubMed] [Google Scholar]
- Xiao S, MacNair L, McGoldrick P, McKeever PM, McLean JR, Zhang M, Keith J, Zinman L, Rogaeva E, Robertson J. 2015. Isoform-specific antibodies reveal distinct subcellular localizations of C9orf72 in amyotrophic lateral sclerosis. Ann Neurol 78: 568–583. [DOI] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, et al. 2013. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci 110: 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa M, Ito D, Honda T, Kubo K, Noda M, Nakajima K, Suzuki N. 2015. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet 24: 1630–1645. [DOI] [PubMed] [Google Scholar]
- Yang D, Abdallah A, Li Z, Lu Y, Almeida S, Gao FB. 2015. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol 130: 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamiri B, Reddy K, Macgregor RB Jr, Pearson CE. 2014. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J Biol Chem 289: 4653–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, He F, Aravind L. 2012. Discovery of novel DENN proteins: Implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet 3: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, et al. 2014. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol 128: 505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. 2015. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. 2011. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci 108: 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. 2013. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci 110: E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]