Abstract
Animal models are an essential feature of the vaccine design toolkit. Although animal models have been invaluable in delineating the mechanisms of immune function, their precision in predicting how well specific vaccines work in humans is often suboptimal. There are, of course, many obvious species differences that may limit animal models from predicting all details of how a vaccine works in humans. However, careful consideration of which animal models may have limitations should also allow more accurate interpretations of animal model data and more accurate predictions of what is to be expected in clinical trials. In this article, we examine some of the considerations that might be relevant to cross-species extrapolation of vaccine-related immune responses for the prediction of how vaccines will perform in humans.
Great Debates
What are the most interesting topics likely to come up over dinner or drinks with your colleagues? Or, more importantly, what are the topics that don't come up because they are a little too controversial? In Immune Memory and Vaccines: Great Debates, Editors Rafi Ahmed and Shane Crotty have put together a collection of articles on such questions, written by thought leaders in these fields, with the freedom to talk about the issues as they see fit. This short, innovative format aims to bring a fresh perspective by encouraging authors to be opinionated, focus on what is most interesting and current, and avoid restating introductory material covered in many other reviews.
The Editors posed 13 interesting questions critical for our understanding of vaccines and immune memory to a broad group of experts in the field. In each case, several different perspectives are provided. Note that while each author knew that there were additional scientists addressing the same question, they did not know who these authors were, which ensured the independence of the opinions and perspectives expressed in each article. Our hope is that readers enjoy these articles and that they trigger many more conversations on these important topics.
The ability to induce protection against pathogens is the hallmark of vaccinology. There are a variety of ways to define whether a vaccine “works,” but these definitions may vary in specifics. For example, one may measure immunogenicity, reduction of pathogen replication on experimental exposure, or reduced acquisition of infection, all of which may depend on a different immune mechanism. In humans, vaccine efficacy (VE) is the gold standard for testing how well a candidate vaccine “works.” VE is defined as the proportional reduction in a specified outcome in a vaccinated group compared with an unvaccinated group (Guinovart and Alonso 2007). VE studies in humans are costly, complex, prone to geographic or demographic biases, require large numbers of participants, and may fail for unexpected reasons (Beran et al. 2009). Moreover, it is unethical to challenge volunteers with certain pathogens such as HIV, so vaccine studies usually must follow large cohorts of subjects for years to ask whether a particular vaccine candidate has protective efficacy. In contrast, animal models permit faster evaluation of promising vaccine candidates at lower risk to humans and lower cost, while at the same time permitting mechanistic study of the immune response. Ideally, an animal model should be permissive to infection by a target pathogen, should mimic pathophysiology of human disease, should have reagents sufficiently available to permit mechanistic dissection of the immune response, should be easy to maintain, and should sustain immune responses that closely resemble human immune responses. Animal models that fulfill these criteria have been the holy grail of vaccine research for decades.
Overall, studies in animal models have had mixed success in predicting outcomes in human studies. A systematic review of six interventions found only partial concordance between results in animal models and humans (Perel et al. 2007). A stark example is therapeutic antibodies against CD28 that proved highly toxic in humans. Monkeys treated with anti-CD28 monoclonal antibodies did not show the dramatic pathogenic responses observed in human volunteers, despite high similarity for CD28 in monkeys and humans (Attarwala 2010). In other settings, vaccines such as early-generation DNA vaccines or other viral vector-based approaches yielded promising results in animal models only to induce much weaker responses in humans (Bainbridge et al. 2015). In other cases, however, results of testing in animal models foreshadowed similar results in human trials. For example, preclinical studies of adenoviral vector-based vaccines showed tepid results in nonhuman primate (NHP) models of simian immunodeficiency virus (SIV) infection (Shiver et al. 2002; Casimiro et al. 2005) and, indeed, subsequently failed to protect humans against HIV in the Step study (Buchbinder et al. 2008). Similarly, the MVA85A candidate vaccine against tuberculosis (TB) showed relatively modest effects in animal models (McShane and Williams 2014) and similarly failed to elicit strong protective efficacy for TB infection in humans compared with placebo (Tameris et al. 2013; Ndiaye et al. 2015). Thus, although in some cases modest responses in preclinical animal studies were associated with poor human vaccination, strategies to improve the predictive accuracy of animal models are needed to permit an understanding of the mechanisms involved in protection and extrapolation to outcomes in humans.
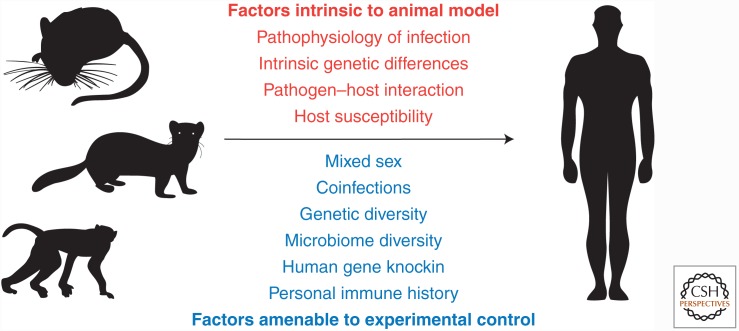
There are a number of aspects about how animal models relate to human disease that should be considered (Fig. 1). Discrepancies between animal models and humans can be fundamentally classified as nonmodifiable or modifiable. Examples of nonmodifiable discrepancies between animal models and humans include genetic differences, lack of natural infection of the animal model by a pathogen of interest, pathophysiologic differences in response to the pathogen (e.g., human CD8 T cells express the cytotoxic molecule granulysin, whereas mouse CD8 T cells do not), and preconditioning. Modifiable discrepancies include controlled genetic and microbiome variability, better simulation of human immune responses in animal models, knowledge of infection history and concurrent infections, and choice of alternative end points in animal studies. Here, we discuss how considerations of these differences between animal models and humans may be able to improve prediction of VE in humans.
Figure 1.
Discrepancies between animal models and humans can be classified as intrinsic to the animal model or amenable to experimental control. Factors that are intrinsic to the animal model are typically nonmodifiable, whereas factors amenable to control may have significant use in helping to make results from animal models better predictors of results in subsequent human studies.
DISCREPANCIES INTRINSIC TO THE ANIMAL MODEL
Lack of “Natural” Infection in the Animal Model
Many animal models do not represent the natural host for the pathogen of interest. For example, HIV does not naturally replicate in small animal models, whereas NHPs can sustain HIV replication but do not have the same immunopathology as occurs in human infection. In most cases, SIV or SIV/HIV hybrid viruses are used in NHPs. Alternatively, an animal model may be able to sustain infection but only if the pathogen is delivered in a specific way. For example, several million colony-forming units of Bordetella pertussis are instilled into the lungs or inoculated into nares of mice to establish infection (Melvin et al. 2014). In contrast, human infection is often initiated by small amounts of bacteria in the upper respiratory tract or nasopharynx. A candidate vaccine that performs well in mice could conceivably lead to robust intrapulmonary immunity but fail to establish the upper airway protection that would be critical for protection in humans. A similar scenario may exist for respiratory viral infections (see below). Establishment of infection in an animal model may allow testing of some concepts of protection but may not be sufficient for studying all aspects of the immune response. Study of pathogens that do not naturally infect the animal model used may yield interesting and important data on the immune response, but if these mechanisms are distinct from those that occur in the natural host, then the predictive capacity may be low. Models can, of course, be adapted. For example, study of polio in mice required development of a transgenic model engineered with the human poliovirus receptor, although even then the transgenic mouse was susceptible only to certain viral strains (Ren et al. 1990). Nevertheless, the greater the knowledge of the pathogenesis of infection in humans, the more likely that animal models can be developed that will lead to accurate predictions of protection and VE.
Pathophysiology of the Infection May Be Discrepant
Differences in the pathophysiology of an infection can underlie discrepancies between immune responses in animal model and humans and thus affect protective immunity. Influenza is an informative example. Mice are not susceptible to human influenza strains (Ilyushina et al. 2010) and most work in this species uses mouse-adapted influenza viruses. Moreover, influenza infection in humans is typically associated with fever, minimal hypoxia, and sometimes secondary bacterial infections (Bouvier and Lowen 2010). In contrast, influenza infection in mice is not associated with fever but rather hypothermia, induces interstitial pneumonia and diffuse alveolar damage leading to hypoxia (Fukushi et al. 2011), and is not associated with secondary bacterial infections. In humans, influenza virus infection tends to initially occur predominantly in the upper respiratory tract and later enters the lower respiratory tract (Bouvier and Lowen 2010). This ability to descend into the lower respiratory tract is associated with features of the viral hemagglutinin (HA) that allow switching to the use of different sialic acid (SA) linkages in different parts of the respiratory tract. In mice, these SA linkages are distinct from humans, and most (although not all) experimental infections in mice are initiated in both the upper and lower respiratory tract with mouse-adapted influenza virus strains. Ferrets, on the other hand, are capable of direct infection by primary human strains of influenza and develop upper respiratory tract disease. However, some strains, such as H5N1, lead to prominent central nervous system infection in ferrets, which is uncommon in humans (Plourde et al. 2012). Even when a natural mouse pathogen exists for a particular microbe that infects humans, key features of disease may differ. For example, both human and mouse noroviruses (NVs) exist that are species specific. However, although human NVs cause acute gastroenteritis and diarrhea, mouse NV infection in immunocompetent mice is largely asymptomatic (Karst et al. 2014). There is little doubt that these models have usage in understanding principles of antipathogen immunity. However, direct extrapolation of data from studies of vaccines based on animal models whose pathophysiology of infection differ greatly from humans could lead to suboptimal predictions of VE in human trials unless underlying pathophysiological differences are carefully considered.
FACTORS AMENABLE TO MODIFICATION
Despite the discrepancies described above, vaccine candidate studies in animal models can yield highly useful information, particularly if some key variables are amenable to modification. Modifiable features may include choosing the “right” animal model, emphasizing development of biomarkers and correlates of protection, increasing the similarity between the animal model and humans, and adding “controlled” variability, including previous immunological conditioning of the host (Fig. 1). These modifications may improve the accuracy of predictions for human study outcomes based on animal models and could even shed light on the generalizability of certain human study results.
Choosing the Model
First, attention needs to be given to the choice of pathogen and the question(s) being asked. Some scenarios will be best served by use of a pathogen naturally found in the animal model. For example, it may be highly relevant to study murine cytomegalovirus (CMV) in a mouse for questions of long-term impact of this pathogen on immune responses because natural host–pathogen interactions may be of key importance. Vaccine studies in which reduction of pathogen transmissibility is an important element need to be conducted in animal models in which transmissibility occurs and shows similar features to what occurs in humans. For these reasons, studies of influenza vaccine strategies in ferrets and, more recently, guinea pigs (Lowen et al. 2009; Thangavel and Bouvier 2014) have yielded valuable, highly relevant information that predicts outcomes in human (Bouvier and Lowen 2010; Thangavel and Bouvier 2014).
Improve the Similarity between Animal Model and Human
There are several ways in which the immune response in animal models could better mimic what occurs in humans. Attempts to more closely simulate the human immune system by incorporating human elements into the animal model may yield more human-relevant data, as has been shown for humanized mice (Garcia 2016). Introduction of human human leukocyte antigen (HLA) alleles can aid in tracking responses to the same T-cell epitopes recognized in humans (Ovsyannikova et al. 2014; Prentice et al. 2015) and, of course, other human immune genes can be “knocked in” to mice to study specific human immune molecules or pathways. Use of large animal models can also aid in testing vaccine candidates. NHPs, for example, are naturally susceptible to many human pathogens and have informed our understanding of vaccine candidates against pertussis, dengue, and HIV (Rivera-Hernandez et al. 2014), although feasibility unfortunately limits use of large animal models to late-stage vaccine candidates. Nonetheless, strategies to introduce human variables may increase relevance of the results to later human studies.
Preconditioning and Coinfections Affect Immune Response
The degree to which animal models mimic human inflammatory responses has been controversial (Seok et al. 2013; Takao and Miyakawa 2015). Although there are clear genetic differences between mouse and human immunology (e.g., natural killer [NK] cell killer inhibitory receptor [KIR] versus Ly49 receptors), at least some discrepant results could be the result of infection or immunological history. To deduce mechanistic insight into the immune response to a vaccine, a reductionist approach is often used to limit the variables that could affect the result. Experiments involving animal models are initiated with specific pathogen-free animals so that the immune response can be attributed only to the vaccine administered. However, the pattern, order, and timing of infection differs for each person. Better prediction of vaccine outcomes in humans based on studies in animal models requires better understanding of the immunological imprint of resolved and concurrent infections.
Recent work in mice clearly shows the importance of immunological history on subsequent immune responses, and so-called “dirty mice” that have likely experienced more human-like previous (and possibly concurrent) infections have a more human-like immune profile (Beura et al. 2016). Moreover, mice preconditioned with helminth and viral infections before vaccination with yellow fever vaccine had lower levels of vaccine-induced immunoglobulin G (IgG) than age-matched specific pathogen-free control mice (Reese et al. 2016). Repetitive immune responses to the same or similar pathogens over time can also alter immune responses. Dengue hemorrhagic fever (DHF) is observed in humans in the setting of sequential heterotypic dengue infection. Thus, vaccine candidates should aim to induce concurrent immunity against all four circulating viral strains, but incomplete strain coverage could impact perceived VE on challenge with a poorly matched viral strain resulting in DHF. Together, these concepts underscore the importance of the individual immunological history in the later immune response.
Concurrent chronic infections can also alter immune responses in ways that are highly relevant to human studies (Stelekati and Wherry 2012). However, few vaccination studies in animal models mimic the potential real-world impact of coinfections. Chronic helminth infections, prevalent in hundreds of millions of people worldwide, altered the host response to HIV, TB, and malaria (Salgame et al. 2013), and administration of antihelminthics leading to resolution of chronic infection restores some immune responses (Elias et al. 2001). Chronic herpesvirus infections in mice limited susceptibility to Yersinia pestis and Listeria monocytogenes (Barton et al. 2007), indicating a beneficial effect on limiting pathogen replication. This same effect, mediated by low-level interferon γ (IFN-γ), may limit the “take” of some live attenuated vaccines that need to replicate to induce immunity. Despite virological suppression of HIV by antiretroviral therapy, rates of seroprotective neutralizing antibody titers following influenza vaccine in HIV-infected adults are lower compared with historical healthy controls (Tebas et al. 2010). At least one mechanistic explanation may involve reduced generation of long-term protective immunity caused by skewing of immune memory by chronic inflammation (Stelekati and Wherry 2012; Stelekati et al. 2014). Studies of vaccine candidates in animal models may be aided by incorporating the possibility and impact of chronic concurrent infections on the desired immune response to help yield generalizable predictions for human studies.
Controlled Variability
Attention to genetics, microbiome, sex, and immunological history all have the potential to improve our ability to extrapolate data from animal models to humans. Increased genetic diversity, typically introduced by use of outbred animals, could substantially reduce discrepancies of animal model studies with later human results in some settings. For example, use of mice from the Collaborative Cross panel yielded Ebola infection results that better mimicked outcomes of human infection than when inbred mice were used (Rasmussen et al. 2014). Next, the impact of the microbiome is beginning to be appreciated as an important factor in understanding vaccine responses (Valdez et al. 2014). Animals obtained from different commercial vendors can have meaningful differences in microbiota that alter immune responses (Villarino et al. 2016). Certainly, sex has a far-reaching but poorly understood impact on immune responses (Klein and Flanagan 2016), and new National Institutes of Health (NIH) guidelines reinforce the importance of obtaining data from both male and female mice. Finally, as described above, individual immunological history should be considered when comparing results in animal models to humans. Addition of controlled variability may be able to better replicate at least some of the inherent diversity in humans and yield more accurate expectations of vaccine outcomes in humans.
Emphasize Development of Biomarkers and Correlates of Protection
Development of suitable biomarkers and robust correlates of protection can greatly enhance the translatability of findings from animal models to patients (Plotkin 2010). Understanding of the critical aspects of the immune response to a given pathogen and distinguishing correlates versus mechanisms of protective immunity is essential to development of optimal animal models. Correlates may or may not be causally linked to mechanisms of protection and incomplete information about these features may limit the ability to extrapolate across species. For example, nearly all licensed vaccines are assessed for immunogenicity based on induction of antibodies. However, immunogenicity and protective immunity can often reflect unrelated, or at least partially nonoverlapping, responses. Some biomarkers, such as vaccine-induced antibodies against rotavirus, are primarily informative for the specific model animal (Desselberger and Huppertz 2011), whereas other biomarkers can reflect general immunologic activity, such as active germinal centers (Havenar-Daughton et al. 2016). Even animal models that do not fully recapitulate human illness can be used to derive immunologic or other signatures of response and could yield useful markers of protection. Appropriate biomarkers can provide insight into the underlying mechanism of protection, and use of surrogate end points may permit more rapid vaccine development. Identification of robust correlates of protection may lead to more accurate predictions for VE in human use.
CONCLUSION
Ultimately, animal models offer a tantalizing opportunity to test and understand mechanisms of vaccine-mediated protection against pathogens. Although true VE in humans may be difficult to accurately estimate from animal models because of the variety of differences described earlier, careful study of animal models can yield insights into the likelihood of success of a vaccine candidate. Animal models may be most useful in stratifying vaccine candidates to facilitate more careful evaluation in an efficient manner for later studies in humans.
ACKNOWLEDGMENTS
R.S.H. is supported by the National Institutes of Health (NIH) Grants AI114852 and AG047773. Work in the Wherry Laboratory is supported by the Penn Center for AIDS Research (P30 AI045008), the NIH Grants AI112521, AI11 7950, AI2010085, as well as the U.S. Broad Agency Announcements Grant HHSN2722011 00018C. E.J.W. is also supported by the Parker Institute for Cancer Immunotherapy.
Footnotes
Editors: Shane Crotty and Rafi Ahmed
Additional Perspectives on Immune Memory and Vaccines: Great Debates available at www.cshperspectives.org
REFERENCES
- Attarwala H. 2010. TGN1412: From discovery to disaster. J Young Pharm 2: 332–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge JWB, Mehat MS, Sundaram V, Robbie SJ, Barker SE, Ripamonti C, Georgiadis A, Mowat FM, Beattie SG, Gardner PJ, et al. 2015. Long-term effect of gene therapy on Leber’s congenital amaurosis. N Engl J Med 372: 1887–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton ES, White DW, Cathelyn JS, Brett-McClellan K, Engle M, Diamond MS, Miller VL, Virgin HW. 2007. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447: 326–329. [DOI] [PubMed] [Google Scholar]
- Beran J, Wertzova V, Honegr K, Kaliskova E, Havlickova M, Havlik J, Jirincova H, Van Belle P, Jain V, Innis B, et al. 2009. Challenge of conducting a placebo-controlled randomized efficacy study for influenza vaccine in a season with low attack rate and a mismatched vaccine B strain: A concrete example. BMC Infect Dis 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, et al. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532: 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM, Lowen AC. 2010. Animal models for influenza virus pathogenesis and transmission. Viruses 2: 1530–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, et al. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372: 1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro DR, Wang F, Schleif WA, Liang X, Zhang ZQ, Tobery TW, Davies ME, McDermott AB, O’Connor DH, Fridman A, et al. 2005. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with DNA and recombinant adenoviral vaccine vectors expressing Gag. J Virol 79: 15547–15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U, Huppertz HI. 2011. Immune responses to rotavirus infection and vaccination and associated correlates of protection. J Infect Dis 203: 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias D, Wolday D, Akuffo H, Petros B, Bronner U, Britton S. 2001. Effect of deworming on human T cell responses to mycobacterial antigens in helminth-exposed individuals before and after bacille Calmette-Guérin (BCG) vaccination. Clin Exp Immunol 123: 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi M, Ito T, Oka T, Kitazawa T, Miyoshi-Akiyama T, Kirikae T, Yamashita M, Kudo K. 2011. Serial histopathological examination of the lungs of mice infected with influenza A virus PR8 strain. PLoS ONE 6: e21207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JV. 2016. Humanized mice for HIV and AIDS research. Curr Opin Virol 19: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinovart C, Alonso PL. 2007. Methods for determining vaccine efficacy and effectiveness and the main barriers to developing a fully deployable malaria vaccine. Am J Trop Med Hyg 77: 276–281. [PubMed] [Google Scholar]
- Havenar-Daughton C, Lindqvist M, Heit A, Wu JE, Reiss SM, Kendric K, Bélanger S, Kasturi SP, Landais E, Akondy RS, et al. 2016. CXCL13 is a plasma biomarker of germinal center activity. Proc Natl Acad Sci 113: 2702–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyushina NA, Khalenkov AM, Seiler JP, Forrest HL, Bovin NV, Marjuki H, Barman S, Webster RG, Webby RJ. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J Virol 84: 8607–8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. 2014. Advances in norovirus biology. Cell Host Microbe 15: 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Flanagan KL. 2016. Sex differences in immune responses. Nat Rev Immunol 16: 626–638. [DOI] [PubMed] [Google Scholar]
- Lowen AC, Steel J, Mubareka S, Carnero E, García-Sastre A, Palese P. 2009. Blocking interhost transmission of influenza virus by vaccination in the guinea pig model. J Virol 83: 2803–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane H, Williams A. 2014. A review of preclinical animal models utilised for TB vaccine evaluation in the context of recent human efficacy data. Tuberculosis (Edinb) 94: 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin JA, Scheller EV, Miller JF, Cotter PA. 2014. Bordetella pertussis pathogenesis: Current and future challenges. Nat Rev Microbiol 12: 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye BP, Thienemann F, Ota M, Landry BS, Camara M, Dièye S, Dieye TN, Esmail H, Goliath R, Huygen K, et al. 2015. Safety, immunogenicity, and efficacy of the candidate tuberculosis vaccine MVA85A in healthy adults infected with HIV-1: A randomised, placebo-controlled, phase 2 trial. Lancet Respir Med 3: 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Pankratz VS, Larrabee BR, Jacobson RM, Poland GA. 2014. HLA genotypes and rubella vaccine immune response: Additional evidence. Vaccine 32: 4206–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perel P, Roberts I, Sena E, Wheble P, Briscoe C, Sandercock P, Macleod M, Mignini LE, Jayaram P, Khan KS. 2007. Comparison of treatment effects between animal experiments and clinical trials: Systematic review. BMJ 334: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA. 2010. Correlates of protection induced by vaccination. Clin Vaccine Immunol 17: 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plourde JR, Pyles JA, Layton RC, Vaughan SE, Tipper JL, Harrod KS. 2012. Neurovirulence of H5N1 infection in ferrets is mediated by multifocal replication in distinct permissive neuronal cell regions. PLoS ONE 7: e46605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice HA, Tomaras GD, Geraghty DE, Apps R, Fong Y, Ehrenberg PK, Rolland M, Kijak GH, Krebs SJ, Nelson W, et al. 2015. HLA class II genes modulate vaccine-induced antibody responses to affect HIV-1 acquisition. Sci Transl Med 7: 296ra112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AL, Okumura A, Ferris MT, Green R, Feldmann F, Kelly SM, Scott DP, Safronetz D, Haddock E, LaCasse R, et al. 2014. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 346: 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beura LK, Bürger MC, Pulendran B, Sekaly RP, Jameson SC, Masopust D, et al. 2016. Sequential infection with common pathogens promotes human-like immune gene expression and altered vaccine response. Cell Host Microbe 19: 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren RB, Costantini F, Gorgacz EJ, Lee JJ, Racaniello VR. 1990. Transgenic mice expressing a human poliovirus receptor: A new model for poliomyelitis. Cell 63: 353–362. [DOI] [PubMed] [Google Scholar]
- Rivera-Hernandez T, Carnathan DG, Moyle PM, Toth I, West NP, Young PR, Silvestri G, Walker MJ. 2014. The contribution of non-human primate models to the development of human vaccines. Discov Med 18: 313–322. [PMC free article] [PubMed] [Google Scholar]
- Salgame P, Yap GS, Gause WC. 2013. Effect of helminth-induced immunity on infections with microbial pathogens. Nat Immunol 14: 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. 2013. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci 110: 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiver JW, Fu TM, Chen L, Casimiro DR, Davies ME, Evans RK, Zhang ZQ, Simon AJ, Trigona WL, Dubey SA, et al. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415: 331–335. [DOI] [PubMed] [Google Scholar]
- Stelekati E, Wherry EJ. 2012. Chronic bystander infections and immunity to unrelated antigens. Cell Host Microbe 12: 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelekati E, Shin H, Doering T, Dolfi DV, Ziegler CG, Beiting DP, Dawson L, Liboon J, Wolski D, Ali M, et al. 2014. Bystander chronic infection negatively impacts development of CD8+ T cell memory. Immunity 40: 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao K, Miyakawa T. 2015. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci 112: 1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, Shea JE, McClain JB, Hussey GD, Hanekom WA, et al. 2013. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: A randomised, placebo-controlled phase 2b trial. Lancet 381: 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas P, Frank I, Lewis M, Quinn J, Zifchak L, Thomas A, Kenney T, Kappes R, Wagner W, Maffei K, et al. 2010. Poor immunogenicity of the H1N1 2009 vaccine in well controlled HIV-infected individuals. AIDS 24: 2187–2192. [DOI] [PubMed] [Google Scholar]
- Thangavel RR, Bouvier NM. 2014. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods 410: 60–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez Y, Brown EM, Finlay BB. 2014. Influence of the microbiota on vaccine effectiveness. Trends Immunol 35: 526–537. [DOI] [PubMed] [Google Scholar]
- Villarino NF, LeCleir GR, Denny JE, Dearth SP, Harding CL, Sloan SS, Gribble JL, Campagna SR, Wilhelm SW, Schmidt NW. 2016. Composition of the gut microbiota modulates the severity of malaria. Proc Natl Acad Sci 113: 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]