Abstract
Ischemic heart disease (IHD) is a leading cause of morbidity and mortality in developed countries. Current pharmacological and interventional therapies provide significant improvement in the life quality of patient; however, they are mostly symptom-oriented and not curative. A high disease and economic burden of IHD requires the search for new therapeutic strategies to significantly improve patients’ prognosis and quality of life. One of the main challenges during IHD is the massive loss of cardiomyocytes that possess minimal regenerative capacity. Recent understanding of the pathophysiological mechanisms underlying IHD, as well as new therapeutic approaches provide new hope for patients suffering from IHD. Synthetic modified mRNA (modRNA) is a new gene delivery vector that is increasingly used in in vivo applications. modRNA is a relatively stable, non-immunogenic, highly-expressed molecule that has been shown to mediate high and transient expression of proteins in different type of cells and tissues including cardiomyocytes. modRNA properties, together with its expression kinetics in the heart make it an attractive option for the treatment of IHD, especially after myocardial infarction. In this review we discuss the role of gene therapy in cardiac regeneration as an approach to treat IHD; traditional and innovative gene delivery methods; and focus specifically on modRNA structure, mode of delivery, and its use for the induction of endogenous regenerative capacity, mainly in the context of IHD.
INTRODUCTION
Ischemic heart disease (IHD) is a leading cause of morbidity and mortality in developed countries. Acute myocardial infarction (MI) affects more than one million Americans yearly, and IHD is accountable for approximately 90% of the deaths due to cardiac causes.1 Despite advances in coronary interventional procedures, IHD remains the main cause of disability and health care expenditure around the world.1
IHD includes several related syndromes caused by acute and chronic myocardial ischemia. After acute myocardial ischemia, a series of adverse processes takes place that includes the formation of scar tissue in the infarcted zone, and an increase in pressure and volume load that leads to a remodeling process of the ventricles in proportion to infarct size. In most patients with acute coronary syndrome, early reperfusion improves myocardial salvage and significantly reduces mortality. Revascularization strategies include fibrinolytic therapy, catheter-based reperfusion and coronary artery bypass surgery (CABG). However, despite the introduction of advanced drug-eluting stents and minimally invasive CABG, the failure rate as a result or coronary restenosis and graft occlusion remains relatively high. Furthermore, 30–35% of patients cannot undergo full revascularization due to suboptimal anatomy or for genetic and molecular reasons, and eventually develop chronic IHD with end-stage heart failure.2 Since IHD includes many pathophysiological mechanisms, it requires a combined treatment approach. Pharmacological management recommended by American Heart Association has an important impact on the mid and long-term survival post-MI. Current medical treatments include the use of (1) angiotensin-converting-enzyme inhibitors which exert a positive effect on cardiac function by reducing blood preload and afterload; (2) beta-blockers that trigger cellular changes by multiple mechanisms and prevent the loss of cardiac cells; and (3) angiotensin II receptor blockers which inhibit stimulation of the renin-angiotensin-aldosterone system. However, a study that investigated the improvement in survival rate when a full complex of medical treatments was used in cases of ischemic heart failure, has found that 5-year survival did not exceed 40–50%.3
Additional treatment modality is heart transplantation. The number of heart transplantations world-wide is relatively low (~5000 heart transplants yearly), especially when compared to the demand. The main reasons for low number of transplantations are the limited numbers of donors and many serious complications associated with immunosuppressive therapy. Therefore, new therapeutic approaches to treat acute transmural MI, chronic IHD post-MI conditions, and ischemic heart failure are needed with the hope of significantly improving patients’ quality of life and survival.
ROLE OF GENE THERAPY IN CARDIAC REGENERATION AND IHD
Advances in the understanding of the molecular basis of heart dysfunction during IHD and specifically during and after MI, have created an opportunity for the development of gene-based therapies. Gene therapy goals in treating of IHD are: (1) activation of adult cardiomyocytes proliferation, (2) attenuating the innate and adaptive immune response, (3) induction of angiogenesis (4) preventing cardiac cells apoptosis and necrosis (see Figure 1).4 Currently there are several studies that utilize gene therapy strategy to investigate and treat IHD: (1) enhancing cardiac muscle contractility by over expression of adenylyl cyclase type 6, SERCA2a, SUMO1, I1C, S100A1 or R1R2; (2) enhancing angiogenesis by over expression of VEGF-A; (3) reducing cell death by overexpression of haem oxygenase-1 and VEGF-B, and (4) recruiting stem cells to the injury site by overexpression of SDF-1.5
FIGURE 1.
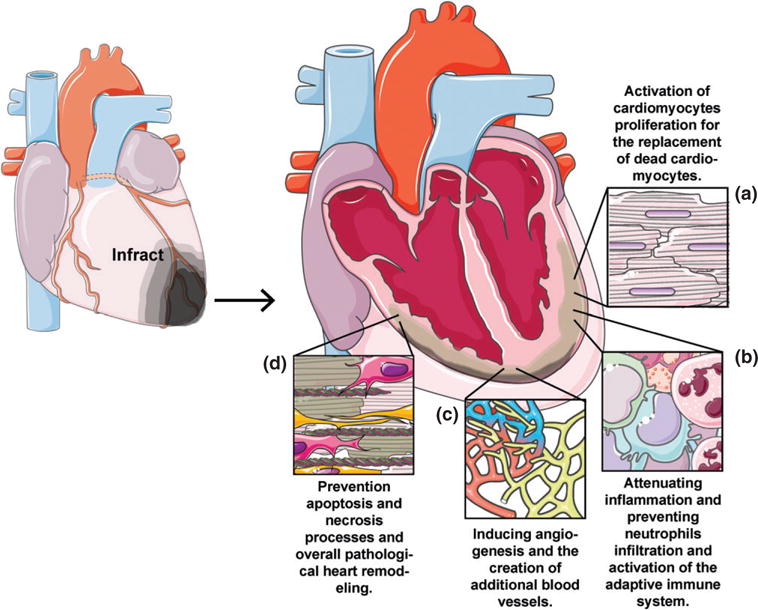
Ideal treatment for ischemic heart disease. Gene therapy goals in treating of IHD are in prevention of pathological heart remodeling by: (a) activation of adult cardiomyocytes proliferation (b) attenuating the innate and adaptive immune response (c) induction of angiogenesis (d) preventing cardiac cells apoptosis and necrosis.
The notion that structural recovery of ischemia-related damaged heart can be achieved through cell regeneration has attracted attention of scientists in recent years. During embryonic period, cardiac growth is carried out only through hyperplasia, but after birth, cardiac cells lose their capacity to proliferate, and growth occurs by hypertrophy. Currently, it is generally accepted that cardiomyocytes have an ability to minimally renew throughout human life with a rate of approximately 1% per year.6 Thus, in recent years, the concept that the heart is unable to regenerate has changed. Moreover, it has been demonstrated that myocytes are able to reenter the cell cycle and undergo nuclear mitotic division early after infarction.7,8 Those studies have shown that the number of dividing myocytes was larger especially in the infarct and border zone than in the remote myocardium.9 However, the presence of cell division in the remote myocardium suggests continuous cells processes in the healthy parts of the heart as well.
MECHANISMS OF HEART REGENERATION
Endogenous regenerative capacity of the adult heart is currently not being the goal of existing available treatments for IHD. One of the proposed mechanisms that could enhance cell regeneration in the infarcted myocardium is myocyte proliferation.10,11 It was shown that surgical resection of the ventricular apex in 1-day-old mice stimulates a regenerative response to restore the damaged heart. This regeneration was characterized by cardiomyocyte proliferation with minimal hypertrophy or fibrosis. However, the mouse heart loses this potential within the first week of postnatal life.12 Cell transplantation offered a new promise for regeneration of the damaged myocardium. Supplying new cardiomyocytes differentiated from multipotent progenitor cells, pluripotent stem cells, embryonic stem cells, or induced pluripotent stem cells is a promising approach.13–17 Importantly, the engrafted cells must be compatible with living myocytes, not only in anatomic structure and maturation state, but also, they should be able to contract via excitation-contraction coupling and to convert an electrical stimulus into a mechanical response. It was previously demonstrated that stem cells can differentiate into cardiomyocytes and partially restore their functional capacity,18 supporting the concept that different stem or progenitor cells could be used for heart regeneration. However, for clinical cell transplantation, many basic issues should be solved such as engraftment, survival, cell homing, differentiation and the role of paracrine factors release.
Recent advances in cell reprogramming technologies offer new approaches to generate cardiomyocytes. It has been shown that direct cell reprogramming into myocytes can be achieved by forced expression of the transcription factors Gata4, Mef2c, and Tbx5.19,20 The majority of the adult heart cell types are cardiac fibroblasts which account for up to two-thirds of cells. They provide a mechanical support for cardiomyocytes and coordinate excitation-contraction coupling.21 After MI, cardiac fibroblasts proliferate, mobilize and mature to create scar tissue that replaces the dead cardiomyocytes. A critical step in this process is the phenotype conversion of cardiac fibroblasts to myofibroblasts. The large population of cardiac fibroblasts is a potential source of regenerative cardiomyocytes. Reprogramming of cardiac fibroblasts to cardiomyocytes is currently being actively researched.22,23 However, the efficiency of this process in vivo remains low in means of absolute number of converted cardiomyocyte-like cells, their contraction, and their electrical properties.20 In order to reactivate the proliferative potential of cardiomyocytes or to reprogram fibroblasts into cardiomyocytes, an efficient strategy for exogenous gene delivery into the myocardium is required.
GENE DELIVERY METHODS
There are several ways to introduce exogenous proteins to the cells of a living organism. The most straightforward way is to directly inject the desired peptide or protein to the body. This approach maintains high level of control over protein dosage, with immediate effects, and with no need for cellular translation. However, the half-life of many proteins in the physiological environment is very short and may require repetitive administration in order to maintain the desired active or therapeutic concentration.
The use of nucleic acids (DNA or mRNA) for gene delivery is an effective approach to overcome protein stability challenges. The two main strategies to introduce DNA to cells are viral vectors or plasmids.24 The most common viral vector is adenovirus that can infect a wide range of cells types. Adenovirus mediates transient expression with high efficiency; however it is highly immunogenic.25,26 Additional commonly-used viral vector is a lentivirus that efficiently infects a broad range of dividing and non-dividing cells. This vector is less immunogenic than adenovirus, but unlike adenovirus, it mediates stable integration of the exogenous gene into the genome of the host cell, enabling stable expression of the gene. This property is beneficial in certain circumstances but also holds the risk for malignant transformation, depending on the location and nature of the integration site.27 Currently, the most promising viral vector is associated adeno virus (AAV). This relatively small virus has low immunogenicity and the ability to mediate a long-term expression of genes with minimal frequency of integration into the host genome.28 AAV mediates high expression levels of protein starting a few days after infection. Protein levels elevate and reach a plateau after a few weeks (depending on the AAV serotype), and last for at least 11 months.29 Importantly, 20–60% of the world population has neutralizing antibodies against different serotype of AAV, making it a less attractive therapeutic agent for more than a quarter of patients.30,31
Non-viral vectors include plasmid DNA and mRNA. Plasmid DNA is minimally immunogenic, but suffers from poor transduction capacity and thus, mediates low levels of gene expression.32 Additionally, even though naked DNA is missing the ability to integrate into the host genome as lentiviral vectors, there is still a small risk of exogenous DNA integration and malignant transformation due to endogenous homologues recombination mechanism of the host cells.33 (For a summary of the current gene delivery methods see Table 1.)
TABLE 1.
Vectors for Gene Therapy
| Vector | Expression Levels | Expression Kinetics | Risk of Integration in Host DNA and Malignant Transformation | Immunogenicity | Advantages and Drawbacks |
|---|---|---|---|---|---|
| Protein | High, depending on the given dosage | High level from the time of administration with very fast decline | No | No | Protein level can be tightly controlled Protein is not stable and has very short half life |
| Adenovirus | High | Reach a pick level within hours and decline for more than 14 days | Low | High | Mediates high expression level shortly after infection Highly immunogenic |
| Lentivirus | High | Reach a pick level few days after infection. Then decline until reaching stable and permanent levels | High | Moderate | Mediates high and permanent expression Integrates into the host genome |
| AAV | High | Expression starts a few days after infection and elevates until reaching a plateau after a few weeks | Low | Low | Mediates high expression level few days after infection for long period of time (≥11 months) Over 60% of the world population has neutralizing antibodies against AAV |
| Plasmid DNA | Moderate | Reach to a pick level after 48 h and start to decline after 5 days | Low | Very low | Not immunogenic Mediates relatively low transfection and expression |
| mRNA | Low | Reach a pick level within 18 h and then gradually decline in the next few days | No | High | Highly immunogenic and unstable, prone for RNase activity |
| modRNA | High | Reach pick levels within 18 h and then gradually decline in the next few days, depending on the amount of cellular proliferation | No | Very low | Mediates high and transient expression within hours after transfection Very low immunogenicity Stable and not prone for RNase activity Transient expression makes it unsuitable for the treatment of genetic and chronic conditions that require stable expression of deficient or mutated genes |
MODIFIED mRNA
mRNA was first suggested to serve as a vector for gene therapy more than 20 years ago. Wolff et al. have demonstrated for the first time, the use of mRNA as a vector for gene expression in mammalian cells in vivo. In this pivotal work, the authors generated synthetic mRNAs encoding the genes chloramphenicol acetyltransferase and luciferase. The mRNA was injected into mouse skeletal muscle and resulted in high expression levels of both proteins 18 h post injection, that gradually decreased for the next 42 h.34 In 1992, synthetic mRNA was used to treat Brattleboro rats suffering from diabetes insipidus. Those rats have a mutation that prevents the translation of the hormones arginine vasopressin (AVP) in the hypothalamus. After injection of a synthetic AVP mRNA into the lateral hypothalamus, the authors reported a temporary reversal of diabetes insipidus symptoms in the treated animals.35 Since then synthetic mRNA has been widely used for vaccination to treat of infectious diseases and cancer.36–43 However, the development of synthetic mRNA for gene replacement therapy was delayed due to two critical limitations. First, mRNA is not stable in physiological conditions and is rapidly degraded by cellular and extracellular Ribonucleases (RNase). Different RNases are abundant in mammals’ plasma and cells, and degrade mRNA as a host defense mechanism against pathogens,44 as well as part of a quality control step for endogenous mRNA.45 As a consequence, the half-life of exogenous mRNA in the body is very short and the amount of proteins produced using exogenous mRNA template is limited. Second, mRNA is highly immunogenic and can trigger the innate immune response by activating Toll-like receptors (TLRs) RIG-1 and MDA-5 in the transfected cells. Exogenous RNA is recognized by TLR3, TLR7 and TLR8 in the endosome. The activation of these receptors results in the induction of inflammation and inhibition of protein translation.46–49 In non-immune cells, the exogenous mRNA is recognized by RIG-1 and MDA5 and induces inhibition of protein translation, mRNA degradation and inflammation.50–53 Each receptor recognizes different features of the exogenous mRNA, providing the organism with reliable protection against a wide range of viruses. TLR3 recognizes double-strand RNA,49 TLR7 and TLR8 recognize single strand RNA,47 and TLR7 also recognizes poly(U).46 RIG-1 and MDA-5 are activated by short and long double-strand RNA respectively.50,51
About a decade ago, Katalin Karik’o and Drew Weissman et al. demonstrated that in vitro synthesis of mRNA with naturally occurring chemically modified nucleotides, produces a more stable, minimally immunogenic mRNA that mediates rapid and strong expression of genes.54–56 Based on the findings that chemical modification of DNA can attenuate immune response to exogenous DNA,57 and the fact that there are at least 100 known naturally occurring modification of ribonucleotides,58 they have used exogenous synthetic mRNA with different modified nucleotides to study immune recognition and response by cells54 and organisms.55 They have found that incorporation of modified nucleotides, including 5-methylcytidine (m5C), N6-methyladenosine (m6A), 5-methyluridine (m5U), 2-thiouridine (s2U) and pseudouridine (Ψ) to synthetic mRNA, dramatically reduces the recognition of RNA by TLR3, 7 and 8, and results in the inhibition of innate immune response of dendritic cells.54 Following experiments have shown that Ψ modification yields the highest translational capacity and biological stability in vivo compared to unmodified mRNA or m5C, m6A, m5U, or s2U modified mRNAs (mod-RNAs).55 The enhancement in translation is attributed, at least to some extent, to reduced activation of RNA-dependent protein kinase (PKR),56 and high resistance to RNaseL.59 The combination of more than one nucleotide modification may have a variable effect on the translation efficiency, depending on the type of the transfected cell.60
Translation efficiency of synthetic mRNA can be further enhanced by using stable cap analog, optimization of the 5ʹUTR and 3ʹUTR vector sequences, as well as by optimization of codon usage61–64 (see Figure 2).
FIGURE 2.
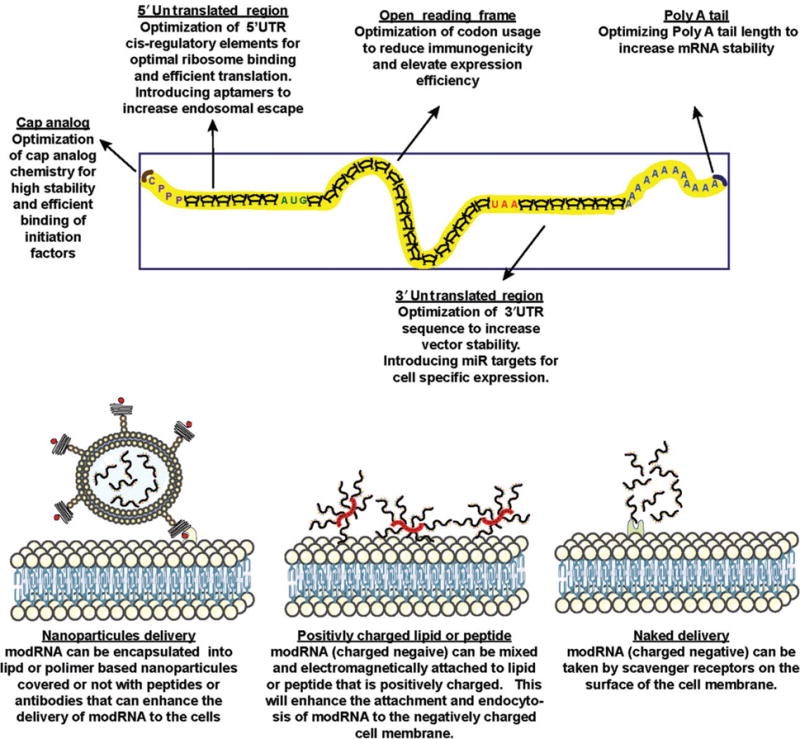
Possible modification of mRNA structure and delivery vehicle for optimal gene expression in the heart. Stability and expression level of proteins can be optimize by modifying the mRNA composition and structure. The modRNA can be delved naked or after complexing it with biomaterials to achieve optimal stability and penetration to the cardiac cells.
CURRENT USES OF modRNA
Currently, modRNA is widely used in generating induced pluripotent stem cells by over expression of dedifferentiation factors in somatic cells,65–67 and as a vaccination for infectious diseases40–43 and cancer36–39 (by direct injection of mRNA encoding specific epitopes to induce immune response against the pathogen or malignant cells or by injection of transfected dendritic cells). modRNA is also used for enhanced engraftment and direct differentiation of ex vivo treated cells prior to transplantation.68
A few studies have demonstrated the potential of modRNA for protein replacement. In 2011, Korman et al. used erythropoietin modRNA to increase hematocrit levels in mice. They also showed the therapeutic efficiency of surfactant protein B modRNA in a mouse model of lethal congenital lung disease.69 In 2013, Zangi et al. showed for the first time that mod-RNA with cap analog and optimized 5ʹ and 3ʹ UTRs can induce expression of genes in heart cells, including cardiomyocytes.70 Additional information about the study can be found in the next sections.
Following that pivotal work, the use of mRNA-based gene replacement was demonstrated in other diseases using different genes: (1) Bcl-2 mod-RNA was used to reduce hepatocytes apoptosis in a mouse model of fulminant hepatitis;71 (2) Intrathecal injection of mRNA, with optimized codon usage, encoding frataxin was used to treat Friedreich’s ataxia in mice;72 (3) The use of modRNA encoding TLR1 TLR2 and TLR6 has been shown to improve lung function as well as reduced airway inflammation in a mouse model of asthma;73 (4) ModRNA encoding the transcription factor RUNEX1 was used to treat osteoarthritis in mice;74 (5) IGF-1 modRNA was shown to reduce apoptosis in mice model of MI;75 (6) In 2015, Thess et al. demonstrated efficient delivery and physiological effects of modRNA encoding for erythropoietin in large animals including pigs and non-human primates.61
ROUTES OF modRNA DELIVERY TO THE HEART
Transduction of foreign RNA into cardiac cells has a great potential value for basic science and for clinical applications. However, the success of gene therapy depends largely on the use of a reliable and efficient delivery methods.
An effective cardiac gene therapy delivery platform must have the following attributes: (a) ability to transfer the gene to different areas of the myocardium including infarct, border and remote zones; (b) transduce sufficient numbers of myocytes of the left and right ventricles, and (c) establish a predictable relationship between the amount of cardiac genome-copy number and cardiac function. Additionally, there is a need to determine whether regional gene expression is sufficient to treat underlying cardiac disease or whether a global distribution is required.76 Cardiac gene delivery techniques consider, among others, variations of heart perfusion during gene transfer, site and method of gene administration, and interventional approach like surgical, catheter-based77 (see Figure 3). The different approaches have been tested in many completed and ongoing clinical trials in cardiology and cardiac surgery. Results from those trials indicate that all three routes of gene delivery are safe, feasible and potentially efficacious. However, so far, a comparative clinical evaluation has not been conducted. Nevertheless, it assumed that the best method is to combine minimization of technique-associated morbidity and extend vector residence time in coronary circulation.76
FIGURE 3.
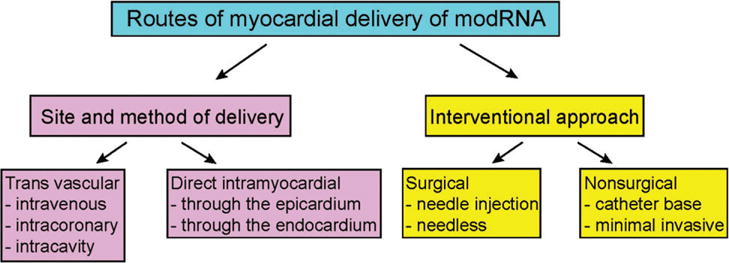
Routes of modRNA delivery to the myocardium. A current diagram of existing cardiac gene delivery techniques considers site and method of administration and, interventional approach. Methods for direct gene delivery include injection from epicardial or endocardial layers into myocardium. A global transduction of the myocardium can only be achieved with intravascular transfer, which includes simple intravenous injection, intracavitary administration and intracoronary arteries route of infusion.
Globally, it is possible to divide all transfer routes to direct myocardial or transvascular. Each strategy has its own barriers that must be overcome for successful delivery. The key transfer steps in transvascular delivery are migration through the blood vessel compartment, transit via the endothelial barrier, extracellular matrix (ECM) that is composed of extracellular fluid and protein structures, navigation in the interstitial environment, and, uptake through the cell membrane. During these steps, RNAses are major impediments to mRNA delivery.78 During direct myocardial delivery, mRNA is exposed to various critical extracellular and intracellular components. For both delivery systems mRNA must navigate through the ECM in order to reach cardiomyocytes plasma membrane. This process is regulated by tissue collagen and hyaluronic acid.79 After passing through the ECM, mRNA must be taken up by cells via receptors or nonspecific binding mechanisms. Nucleic acids typically enter cells through endocytosis, phagocytosis, scavenger receptors or micropinocytosis. The cellular endosome is one of the most difficult biological obstacles to overcome. The endosome contains a number of TLRs and enzymes that recognize foreign internalized molecules or ligands and send them to degradation, or for recycling back to the plasma membrane. Another obstacle of transduction is the ability of the nucleic acids to properly bind to the ribosome and be translated to functional protein after their release from the endosome. Additionally, the ischemic conditions present after MI dramatically reduce the translation of mRNA. One of the important advantages of direct myocardial approach over transvascular approach is the minimization of undesirable systemic effects of introducing foreign genes to the body.
To improve the stability of mRNA in physiological conditions, cell penetration, and endosomal escape, mRNA can be complexed with biomaterials such as protamine, lipid nanoparticles or polymeric nanoparticles80 (see Figure 2). Each class of biomaterials has advantages and limitations. Protamin is a cationic protein that can complex with negatively-charged nucleic acids and increase the transfection ability of cells. However this complex may increase the immune response to the transfected cells.81 Lipid nanoparticles are very efficient and widely used for in vitro transfection, however they have been found to be toxic for cells, which makes them problematic as a vehicle for in vivo transfection.82 Polymeric nanoparticles are relatively new and promising biomaterials as vehicles for mRNA delivery, and can mediate an efficient transfection with low cytotoxicity.83 More research and optimization of vehicles is needed to fully exploit the potential of modRNA as a vector for gene therapy.
modRNA USE FOR THE INDUCTION OF ENDOGENOUS REGENERATIVE CAPACITY
As mentioned above, MI occurs as a result of ischemia that leads to the death of a significant portion of cardiomyocytes. The limited capacity of the heart to regenerate reduces the heart function and leads to high morbidity and mortality.
The properties of modRNA as a delivery method in terms of protein expression efficiency and kinetics make it an attractive option for enhancing the endogenous regenerative capacity (see Figure 4). After intramyocardial injection of luciferase modRNA into the left ventricle, high luciferase activity can be detected within 3 h post injection. The activity levels reach to a pick 18 h after injection and decline gradually for 6 days. This time frame is compatible with the timing of therapeutic opportunity during and after MI. The timeline of MI development includes the induction of cell death less than an hour after occlusion, secretion of pro-inflammatory chemokines 4 h after occlusion, followed by the development of inflammation. Two days post MI, a rapid proliferation of fibroblasts and angiogenesis can be observed, followed by vascular maturation and scar formation 2–3 weeks from MI.84–86 Owing to its expression kinetics, modRNA serves as an optimal vector to quickly and efficiently express gene or genes combinations to minimize heart injury and/or induce regeneration. For example, anti-apoptotic and antioxidant proteins, pro-survival genes, inflammation modulators, angiogenic factors and genes that may induce cardiomyocyte proliferation, or reprograming factors to convert fibroblasts into cardiomyocytes.
FIGURE 4.
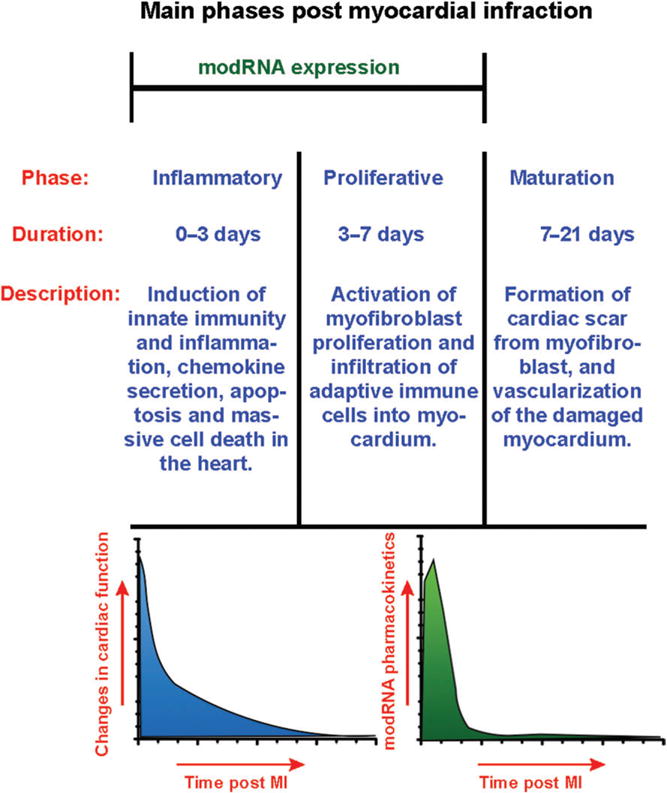
Main phases of Post-MI myocardial remodeling in mouse. modRNA expression occurs during inflammatory and proliferative phases, when most of the changes in cardiac function take place.
As discussed previously, while overexpression of the proper exogenous genes in the heart can reduce injury and induce regeneration, prolonged expression of those genes might have undesirable long-term effects such as malignant transformation by inhibiting apoptosis and suppressing the immune system, combined with uncontrolled proliferation.
Pioneering work in the field has demonstrated that direct injection of VEGF-A modRNA to the mice’ infarcted heart mediates ‘pulse-like’ expression of VEGF-A that resulted in reduced infarct size, enhanced myocardial perfusion and improved survival. In part, this effect was due to a previously unknown effect of VEGF-A on epicardial progenitors. VEGF-A modRNA amplified these progenitors, mobilized their migration into the myocardium and redirected their differentiation toward cardiovascular lineages.70
Huang et al. injected IGF-1 to the infarcted heart of mice and showed a reduction of apoptosis in the infarct border zone. The reduction of apoptosis was attributed to an increase in the level of Akt and Erk phosphorylation as well as expression of miR-1 and miR-133.75 However, our own unpublished data suggest that using IGF-1 modRNA induces adipogenic differentiation of epicardial progenitor cells, and the formation of epicardial adipose tissue in mouse heart after MI (Zangi et al.; under revision). Epidemiological studies have demonstrated that epicardial adipose tissue is closely related to coronary artery disease. Moreover, infiltration of the myocardium by adipocytes is also a hallmark of arrhythmogenic cardiomyopathy. To overcome this issue, we designed a dominant negative IGF1R modRNA, that was applied to the heart after MI, and was able to block epicardial fat formation. This shows that modified mRNA can be used not only to activate signaling pathways but also to block signaling pathways, with physiological impact.
The above-described studies demonstrate the complexity of using paracrine factors for the treatment of MI, and highlight the need for careful interpretation of the results. Remaining challenges are to find the ideal delivery route (intramyocardial or transvascular), or other types of delivery route), and to increase clinical applicability by reducing the cost of modRNA production and handling, or alternatively, the amount of modRNA needed for each in vivo delivery. Additionally, it is not yet clear if a short, transient expression of a gene is enough to induce substantial cardiac regenerative changes. Improved modRNA with longer, but still relatively short expression period of 2–3 weeks, or alternatively, the ability to perform repeated modRNA transfections/injection to maintain effective protein levels for a longer, but still controlled period, may be needed in order to achieve a long-lasting therapeutic effect.
As research in the modRNA field rapidly accumulates, technical aspects to this promising technology will improve, enabling the in-depth, and clinically-applicable investigation of several mod-RNAs candidates for the treatment of IHD, as well as other morbidities.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al. Heart disease and stroke statistics—2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox K, Garcia MA, Ardissino D, Buszman P, Camici PG, Crea F, Daly C, De Backer G, Hjemdahl P, Lopez-Sendon J, et al. Guidelines on the management of stable angina pectoris: executive summary: the task force on the management of stable angina pectoris of the European Society of Cardiology. Eur Heart J. 2006;27:1341–1381. doi: 10.1093/eurheartj/ehl001. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg RJ, Ciampa J, Lessard D, Meyer TE, Spencer FA. Long-term survival after heart failure: a contemporary population-based perspective. Arch Intern Med. 2007;167:490–496. doi: 10.1001/archinte.167.5.490. [DOI] [PubMed] [Google Scholar]
- 4.Lavu M, Gundewar S, Lefer DJ. Gene therapy for ischemic heart disease. J Mol Cell Cardiol. 2011;50:742–750. doi: 10.1016/j.yjmcc.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hulot JS, Ishikawa K, Hajjar RJ. Gene therapy for the treatment of heart failure: promise postponed. Eur Heart J. 2016;37:1651–1658. doi: 10.1093/eurheartj/ehw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anversa P, Leri A, Kajstura J. Cardiac regeneration. J Am Coll Cardiol. 2006;47:1769–1776. doi: 10.1016/j.jacc.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by preexisting cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senyo SE, Lee RT, Kuhn B. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. 2014;13:532–541. doi: 10.1016/j.scr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urbanek K, Torella D, Sheikh F, De Angelis A, Nurzynska D, Silvestri F, Beltrami CA, Bussani R, Beltrami AP, Quaini F, et al. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc Natl Acad Sci USA. 2005;102:8692–8697. doi: 10.1073/pnas.0500169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, Nadal-Ginard B, Silvestri F, Leri A, Beltrami CA, et al. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 12.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz MG, Fargnoli AS, Kendle AP, Hajjar RJ, Bridges CR. The role of microRNAs in cardiac development and regenerative capacity. Am J Physiol Heart Circ Physiol. 2016;310:H528–H541. doi: 10.1152/ajpheart.00181.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei H, Tan G, Manasi QS, Kong G, Yong P, Koh C, Ooi TH, Lim SY, Wong P, et al. One-step derivation of cardiomyocytes and mesenchymal stem cells from human pluripotent stem cells. Stem Cell Res. 2012;9:87–100. doi: 10.1016/j.scr.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, Cashman J, Mercola M. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL, Robbins RC. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature. 2004;428:668–673. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- 19.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. in vivo repro-gramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spach MS. Mounting evidence that fibrosis generates a major mechanism for atrial fibrillation. Circ Res. 2007;101:743–745. doi: 10.1161/CIRCRESAHA.107.163956. [DOI] [PubMed] [Google Scholar]
- 22.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibro-blasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 23.Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53:323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Hadas Y, Etlin A, Falk H, Avraham O, Kobiler O, Panet A, Lev-Tov A, Klar A. A ’tool box’ for deciphering neuronal circuits in the developing chick spinal cord. Nucleic Acids Res. 2014;42:e148. doi: 10.1093/nar/gku750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatsis N, Ertl HC. Adenoviruses as vaccine vectors. Mol Ther. 2004;10:616–629. doi: 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnell MA, Zhang Y, Tazelaar J, Gao GP, Yu QC, Qian R, Chen SJ, Varnavski AN, LeClair C, Raper SE, et al. Activation of innate immunity in nonhuman primates following intraportal administration of adenoviral vectors. Mol Ther. 2001;3:708–722. doi: 10.1006/mthe.2001.0330. [DOI] [PubMed] [Google Scholar]
- 27.Persons DA. Lentiviral vector gene therapy: effective and safe? Mol Ther. 2010;18:861–862. doi: 10.1038/mt.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz BR, Chamberlain JS. Recombinant adeno-associated virus transduction and integration. Mol Ther. 2008;16:1189–1199. doi: 10.1038/mt.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 30.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 31.Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawabata K, Takakura Y, Hashida M. The fate of plasmid DNA after intravenous injection in mice: involvement of scavenger receptors in its hepatic uptake. Pharm Res. 1995;12:825–830. doi: 10.1023/a:1016248701505. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Troilo PJ, Wang X, Griffiths TG, Pacchione SJ, Barnum AB, Harper LB, Pauley CJ, Niu Z, Denisova L, et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11:711–721. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- 34.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 35.Jirikowski GF, Sanna PP, Maciejewski-Lenoir D, Bloom FE. Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 36.Rittig SM, Haentschel M, Weimer KJ, Heine A, Muller MR, Brugger W, Horger MS, Maksimovic O, Stenzl A, Hoerr I, et al. Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol Ther. 2011;19:990–999. doi: 10.1038/mt.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weide B, Pascolo S, Scheel B, Derhovanessian E, Pflugfelder A, Eigentler TK, Pawelec G, Hoerr I, Rammensee HG, Garbe C. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 38.Su Z, Dannull J, Yang BK, Dahm P, Coleman D, Yancey D, Sichi S, Niedzwiecki D, Boczkowski D, Gilboa E, et al. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J Immunol. 2005;174:3798–3807. doi: 10.4049/jimmunol.174.6.3798. [DOI] [PubMed] [Google Scholar]
- 39.Heiser A, Coleman D, Dannull J, Yancey D, Maurice MA, Lallas CD, Dahm P, Niedzwiecki D, Gilboa E, Vieweg J. Autologous dendritic cells transfected with prostate-specific antigen RNA stimulate CTL responses against metastatic prostate tumors. J Clin Invest. 2002;109:409–417. doi: 10.1172/JCI14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleeton MN, Chen M, Berglund P, Rhodes G, Parker SE, Murphy M, Atkins GJ, Liljestrom P. Self-replicative RNA vaccines elicit protection against influenza A virus, respiratory syncytial virus, and a tick-borne encephalitis virus. J Infect Dis. 2001;183:1395–1398. doi: 10.1086/319857. [DOI] [PubMed] [Google Scholar]
- 41.Van Gulck E, Vlieghe E, Vekemans M, Van Tendeloo VF, Van De Velde A, Smits E, Anguille S, Cools N, Goossens H, Mertens L, et al. mRNA-based dendritic cell vaccination induces potent antiviral T-cell responses in HIV-1-infected patients. AIDS. 2012;26:F1–F12. doi: 10.1097/QAD.0b013e32834f33e8. [DOI] [PubMed] [Google Scholar]
- 42.Hekele A, Bertholet S, Archer J, Gibson DG, Palladino G, Brito LA, Otten GR, Brazzoli M, Buccato S, Bonci A, et al. Rapidly produced SAM((R)) vaccine against H7N9 influenza is immunogenic in mice. Emerg Microbes Infect. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allard SD, De Keersmaecker B, de Goede AL, Verschuren EJ, Koetsveld J, Reedijk ML, Wylock C, De Bel AV, Vandeloo J, Pistoor F, et al. A phase I/IIa immunotherapy trial of HIV-1-infected patients with Tat, Rev and Nef expressing dendritic cells followed by treatment interruption. Clin Immunol. 2012;142:252–268. doi: 10.1016/j.clim.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Dyer KD, Rosenberg HF. The RNase a superfamily: generation of diversity and innate host defense. Mol Divers. 2006;10:585–597. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 45.Rigby RE, Rehwinkel J. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 2015;36:179–188. doi: 10.1016/j.it.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y, Reis e Sousa C. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur J Immunol. 2006;36:3256–3267. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 47.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 48.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 49.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 50.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5ʹ tri-phosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 53.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 54.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 55.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Kariko K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 58.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2ʹ-5ʹ-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchida S, Kataoka K, Itaka K. Screening of mRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics. 2015;7:137–151. doi: 10.3390/pharmaceutics7030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thess A, Grund S, Mui BL, Hope MJ, Baumhof P, Fotin-Mleczek M, Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jemielity J, Fowler T, Zuberek J, Stepinski J, Lewdorowicz M, Niedzwiecka A, Stolarski R, Darzynkiewicz E, Rhoads RE. Novel “anti-reverse” cap analogs with superior translational properties. RNA. 2003;9:1108–1122. doi: 10.1261/rna.5430403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaw G, Kamen R. A conserved AU sequence from the 3ʹ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 64.Kariko K, Kuo A, Barnathan E. Overexpression of urokinase receptor in mammalian cells following administration of the in vitro transcribed encoding mRNA. Gene Ther. 1999;6:1092–1100. doi: 10.1038/sj.gt.3300930. [DOI] [PubMed] [Google Scholar]
- 65.Mandal PK, Rossi DJ. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat Pro-toc. 2013;8:568–582. doi: 10.1038/nprot.2013.019. [DOI] [PubMed] [Google Scholar]
- 66.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luni C, Giulitti S, Serena E, Ferrari L, Zambon A, Gagliano O, Giobbe GG, Michielin F, Knobel S, Bosio A, et al. High-efficiency cellular reprogramming with microfluidics. Nat Methods. 2016;13:446–452. doi: 10.1038/nmeth.3832. [DOI] [PubMed] [Google Scholar]
- 68.Lui KO, Zangi L, Silva EA, Bu L, Sahara M, Li RA, Mooney DJ, Chien KR. Driving vascular endothelial cell fate of human multipotent Isl1+ heart progenitors with VEGF modified mRNA. Cell Res. 2013;23:1172–1186. doi: 10.1038/cr.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kormann MS, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, Huppmann M, Mays LE, Illenyi M, Schams A, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 70.Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Spater D, Xu H, Tabebordbar M, Gorbatov R, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsui A, Uchida S, Ishii T, Itaka K, Kataoka K. Messenger RNA-based therapeutics for the treatment of apoptosis-associated diseases. Sci Rep. 2015;5:15810. doi: 10.1038/srep15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nabhan JF, Wood KM, Rao VP, Morin J, Bhamidipaty S, LaBranche TP, Gooch RL, Bozal F, Bulawa CE, Guild BC. Intrathecal delivery of frataxin mRNA encapsulated in lipid nanoparticles to dorsal root ganglia as a potential therapeutic for Friedreich’s ataxia. Sci Rep. 2016;6:20019. doi: 10.1038/srep20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeyer F, Mothes B, Will C, Carevic M, Rottenberger J, Nurnberg B, Hartl D, Handgretinger R, Beer-Hammer S, Kormann MS. mRNA-mediated gene supplementation of toll-like receptors as treatment strategy for asthma in vivo. PLoS One. 2016;11:e0154001. doi: 10.1371/journal.pone.0154001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aini H, Itaka K, Fujisawa A, Uchida H, Uchida S, Fukushima S, Kataoka K, Saito T, Chung UI, Ohba S. Messenger RNA delivery of a cartilage-anabolic transcription factor as a disease-modifying strategy for osteoarthritis treatment. Sci Rep. 2016;6:18743. doi: 10.1038/srep18743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang CL, Leblond AL, Turner EC, Kumar AH, Martin K, Whelan D, O’Sullivan DM, Caplice NM. Synthetic chemically modified mrna-based delivery of cytoprotective factor promotes early cardiomyocyte survival post-acute myocardial infarction. Mol Pharm. 2015;12:991–996. doi: 10.1021/mp5006239. [DOI] [PubMed] [Google Scholar]
- 76.Katz MG, Swain JD, White JD, Low D, Stedman H, Bridges CR. Cardiac gene therapy: optimization of gene delivery techniques in vivo. Hum Gene Ther. 2010;21:371–380. doi: 10.1089/hum.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katz MG, Fargnoli AS, Williams RD, Bridges CR. The road ahead: working towards effective clinical translation of myocardial gene therapies. Ther Deliv. 2014;5:39–51. doi: 10.4155/tde.13.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase. J Nat Struct Mol Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- 79.Escoffre JM, Teissie J, Rols MP. Gene transfer: how can the biological barriers be overcome? J Membr Biol. 2010;236:61–74. doi: 10.1007/s00232-010-9275-0. [DOI] [PubMed] [Google Scholar]
- 80.Islam MA, Reesor EK, Xu Y, Zope HR, Zetter BR, Shi J. Biomaterials for mRNA delivery. Biomater Sci. 2015;3:1519–1533. doi: 10.1039/c5bm00198f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skold AE, van Beek JJ, Sittig SP, Bakdash G, Tel J, Schreibelt G, de Vries IJ. Protamine-stabilized RNA as an ex vivo stimulant of primary human dendritic cell subsets. Cancer Immunol Immunother. 2015;64:1461–1473. doi: 10.1007/s00262-015-1746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31:6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 83.Li L, Wei Y, Gong C. Polymeric Nanocarriers for Non-Viral Gene Delivery. J Biomed Nanotechnol. 2015;11:739–770. doi: 10.1166/jbn.2015.2069. [DOI] [PubMed] [Google Scholar]
- 84.Turillazzi E, Di Paolo M, Neri M, Riezzo I, Fineschi V. A theoretical timeline for myocardial infarction: immunohistochemical evaluation and western blot quantification for Interleukin-15 and monocyte chemotactic protein-1 as very early markers. J Transl Med. 2014;12:188. doi: 10.1186/1479-5876-12-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frangogiannis NG. Regulation of the inflammatory response in cardiac repair. Circ Res. 2012;110:159–173. doi: 10.1161/CIRCRESAHA.111.243162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liehn EA, Postea O, Curaj A, Marx N. Repair after myocardial infarction, between fantasy and reality: the role of chemokines. J Am Coll Cardiol. 2011;58:2357–2362. doi: 10.1016/j.jacc.2011.08.034. [DOI] [PubMed] [Google Scholar]


