SUMMARY
Telomeres define the ends of linear eukaryotic chromosomes and are required for genome maintenance and continued cell proliferation. The extreme ends of telomeres terminate in a single-strand protrusion, termed the G-overhang, which, in vertebrates and fission yeast, is bound by evolutionarily conserved members of the POT1 (protection of telomeres) protein family. Unlike most other model organisms, the flowering plant Arabidopsis thaliana encodes two divergent POT1-like proteins. Here we show that the single-strand telomeric DNA binding activity present in A. thaliana nuclear extracts is not dependent on POT1a or POT1b proteins. Furthermore, in contrast to POT1 proteins from yeast and vertebrates, recombinant POT1a and POT1b proteins from A. thaliana, and from two additional Brassicaceae species, Arabidopsis lyrata and Brassica oleracea (cauliflower), fail to bind single-strand telomeric DNA in vitro under the conditions tested. Finally, although we detected four single-strand telomeric DNA binding activities in nuclear extracts from B. oleracea, partial purification and DNA cross-linking analysis of these complexes identified proteins that are smaller than the predicted sizes of BoPOT1a or BoPOT1b. Taken together, these data suggest that POT1 proteins are not the major single-strand telomeric DNA binding activities in A. thaliana and its close relatives, underscoring the remarkable functional divergence of POT1 proteins from plants and other eukaryotes.
Keywords: telomerase, G-overhang, Protection of telomeres 1 (POT1), telomere, Brassica, Arabidopsis
INTRODUCTION
Telomeres cap the ends of eukaryotic chromosomes, protecting the termini from a DNA damage response that would lead to inappropriate recognition of chromosome ends as double-strand DNA breaks. Telomeres also allow for the complete replication of chromosome ends through the action of telomerase. In eukaryotes, telomeric DNA consists of simple G-rich repeat arrays. Most plant telomeres, for example, are composed of TTTAGGG repeats, which in Arabidopsis range in size from 2 to 5 kb (Richards and Ausubel, 1988; Shakirov and Shippen, 2004). A few plants harbor non-canonical telomere sequences. In several cases, human-type TTAGGG repeats have been found, but in other instances, the telomere sequence is unknown (reviewed by Fajkus et al., 2005). Telomeric DNA is comprised of two distinct regions, a double-stranded region accounting for most of the telomeric tract, and a short G-rich single-strand protrusion, termed the G-overhang. In Arabidopsis, the G-overhang is estimated to be approximately 20–30 nucleotides long (Riha et al., 2000).
Vertebrate telomeres are bound by a six-member protein complex termed shelterin, which is in contact with both the double- and single-strand portions of the telomere (de Lange, 2005). Proteins that bind the double-strand region include Rap1p and Taz1p in budding and fission yeast, respectively, and TRF1 and TRF2 in vertebrates (reviewed by Kanoh and Ishikawa, 2003). These proteins are required for chromosome end protection (van Steensel and de Lange, 1997; van Steensel et al., 1998), telomerase regulation (Marcand et al., 1997) and T-loop formation (Griffith et al., 1999). Sequence homologs of the human TRF proteins have also been identified in plants (Karamysheva et al., 2004; Yang et al., 2004; Hong et al., 2007), and appear to constitute a large gene family in Arabidopsis (Karamysheva et al., 2004); however, their exact contributions to telomere biology are unclear.
In budding yeast, the single-strand G-overhang is bound by a trimeric Replication Protein A (RPA)-like complex termed CST, which consists of Cdc13/Stn1/Ten1 proteins (Gao et al., 2007). TEN1 and STN1 orthologs have recently been reported in Schizosaccharomyces pombe (Martin et al., 2007); however POT1 (protection of telomeres 1) appears to be the major factor associated with single-strand telomeric DNA in both fission yeast and vertebrates. POT1 proteins have been implicated in telomere length control and chromosome end protection, and in mediating the DNA damage response (reviewed by Palm and de Lange, 2008; Xin et al., 2008). Structurally, POT1 is defined by the presence of two N-terminal oligosaccharide/oligonucleotide binding folds (OB folds) (Mitton-Fry et al., 2002). The crystal structures of human and S. pombe POT1 reveal that several conserved residues in the first and second OB folds are in contact with telomeric DNA 3′ ends (Lei et al., 2003, 2004). The C-terminus of mammalian POT1 proteins is required for interaction with TPP1 (Liu et al., 2004), another shelterin component that is needed for chromosome end protection and complete shelterin assembly (reviewed by Smogorzewska and de Lange, 2004).
Members of the POT1 gene family are widely dispersed among both higher and lower eukaryotes (Baumann et al., 2002). However, despite sequence conservation among POT1 proteins, their functions have diverged rapidly. For instance, while most vertebrates encode only a single POT1 protein, mouse encodes two divergent proteins, POT1a and POT1b, which have partially non-overlapping functions at the telomere (Hockemeyer et al., 2006; Wu et al., 2006). Similarly, Caenorhabditis elegans, Euplotes crassus and Tetrahymena thermophila harbor at least two POT1-like proteins with functions and interacting partners that are distinct from those of mammalian POT1 (Wang et al., 1992; Jacob et al., 2007; Raices et al., 2008). Intriguingly, the C. elegans protein CeOB-2 binds single-strand DNA corresponding to the C-rich telomere strand instead of the G-rich strand (Raices et al., 2008).
POT1-like proteins have also been identified in Arabidopsis and other plants (Baumann et al., 2002; Kuchar and Fajkus, 2004; Shakirov et al., 2005; Tani and Murata, 2005; Shakirov et al., 2008). Arabidopsis thaliana harbors two highly divergent POT1 paralogs, AtPOT1a and AtPOT1b, which encode proteins with only 50% amino acid similarity (Shakirov et al., 2005). A third, truncated gene, termed AtPOT1c, has also been found (E. Shakirov, A. Nelson (Texas A&M University) and D. Shippen, unpublished data), but its function is unknown. Plants over-expressing the N-terminus of AtPOT1b suffer extensive erosion of telomeric DNA tracts as well as chromosome fusions (Shakirov et al., 2005), suggesting that AtPOT1b may contribute to chromosome end protection in a manner similar to the S. pombe and vertebrate POT1 proteins. In contrast, a null mutation in AtPOT1a leads to a dramatic decrease in telomerase activity in vivo, and results in progressive telomere shortening with each plant generation (Surovtseva et al., 2007). AtPOT1a physically associates with the telomerase enzyme, but shows no detectable telomeric DNA binding in vitro (Surovtseva et al., 2007), despite harboring two characteristic OB-fold domains with structural similarity to those of mammalian and fission yeast POT1 proteins. These findings raise the question of whether the POT1 protein family in Arabidopsis truly represents the major G-overhang binding activity in plants.
Here we examine single-strand telomeric DNA binding activities in Arabidopsis thaliana and two additional members of the Brassicaceae family, Arabidopsis lyrata and Brassica oleracea (cauliflower). We present evidence that Arabidopsis and Brassica nuclear extracts harbor highly specific single-strand telomeric DNA binding activities. Surprisingly, however, analysis of Arabidopsis mutants lacking AtPOT1a and AtPOT1b proteins indicated no obvious impact on the biochemical properties of the telomeric DNA binding protein. Furthermore, we were unable to detect the binding of recombinant Brassicaceae POT1 proteins to single-strand telomeric DNA in vitro. Finally, analysis of the DNA binding component of the most specific telomeric DNA binding complex in B. oleracea did not uncover proteins corresponding to BoPOT1a or BoPOT1b. We conclude that POT1 proteins are unlikely to be the major telomeric G-strand binding factors in Brassicaceae.
RESULTS
POT1-independent single-strand telomeric DNA binding activity in Arabidopsis
In vertebrates and fission yeast, single-strand telomeric DNA binding is a crucial feature of POT1 protein function. The POT1 interaction with telomeric DNA substrates is robust, with apparent Kd values ranging from 0.46 to 9.5 nM depending on the organism (reviewed by Croy and Wuttke, 2006). Our initial attempts to detect recombinant AtPOT1a binding to telomeric DNA in vitro failed (Surovtseva et al., 2007), and therefore we attempted to determine whether endogenous AtPOT1a or AtPOT1b could form a complex with single-strand telomeric DNA in vivo. For these studies, we used electrophoretic mobility gel-shift assays (EMSAs) with nuclear extracts produced from wild-type, POT1a- and POT1b-deficient plants (Surovtseva et al., 2007; E. Shakirov, A. Nelson, D. Shippen, unpublished data).
First, an EMSA was performed using wild-type Arabidopsis extracts incubated with radiolabeled (TTTAGGG)5. A single major shifted band was observed (Figure 1, lanes 2 and 13). Telomeric DNA binding was specific, as 50-fold excess of a non-telomeric cold competitor failed to compete (Figure 1, lanes 3 and 4), while cold telomeric (TTTAGGG)5 oligonucleotide competed well at a fivefold excess (Figure 1, lanes 5 and 6). Telomeric DNA binding was length-dependent. The efficiency of competition decreased for oligonucleotides containing fewer than five telomere repeats (Figure 1, lanes 7–10). An oligonucleotide with only two telomeric repeats failed to compete when supplied in 50-fold excess over the labeled oligonucleotide (Figure 1, lanes 11 and 12). We conclude that Arabidopsis nuclear extracts contain specific and length-dependent G-strand telomeric DNA binding activity.
Figure 1.

Single-strand telomeric DNA binding activity in Arabidopsis is not dependent on POT1 proteins.
EMSAs were performed with radioactively labeled (TTTAGGG)5 oligonucleotide (lane 1) and wild-type Arabidopsis nuclear extract (lanes 2 and 13). Competition assays were performed with the indicated fold excess of a cold non-telomeric oligonucleotide 5′-GACTGAGCTCGTCGGATCCAATAAAACCTTAATGT-3′ (lanes 3 and 4), as well as excess cold oligonucleotides containing five (lanes 5 and 6), four (lanes 7 and 8), three (lanes 9 and 10) or two (lanes 11 and 12) telomeric repeats. EMSAs were also performed with nuclear extracts from pot1a-1 (lane 14) and pot1b-1 (lane 15) mutants. The single shifted band is indicated by the arrow.
To determine whether POT1a makes a significant contribution to the G-strand telomeric DNA binding activity observed with wild-type Arabidopsis, we performed an EMSA using nuclear extracts prepared from mutant plants bearing a T-DNA insertion in the AtPOT1a gene that abolishes AtPOT1a protein production (Surovtseva et al., 2007). No detectable change in the intensity or migration of the shifted band was observed (Figure 1, lane 14).
Next we determined whether the gel-shift pattern changed when using nuclear extracts prepared from AtPOT1b-deficient plants. Recently, we identified a GeneTrap line in the Cold Spring Harbor collection (http://genetrap.cshl.org/) that has a T-DNA insertion in the second exon of the AtPOT1b gene. RT-PCR results confirmed that the expression of AtPOT1b full-length mRNA is abolished, but pot1b-1 mutants are wild-type in appearance and fully fertile. In contrast to pot1a mutants (Surovtseva et al., 2007), the bulk telomere length in pot1b-1 plants is not perturbed (data not shown). A detailed analysis of the Arabidopsis pot1b mutant will be described elsewhere (E. Shakirov, A. Nelson, D. Shippen, unpublished data). As with pot1a mutants, we observed no change in the intensity or migration of the single-strand telomeric DNA–protein complex in pot1b mutants relative to wild-type (Figure 1, lane 15). These data imply that neither POT1a nor POT1b is a major telomeric G-strand binding factor in Arabidopsis.
Recombinant Arabidopsis POT1a and POT1b do not exhibit single-strand telomeric DNA binding in vitro
The inability to detect telomeric DNA binding for endogenous Arabidopsis POT1a and POT1b prompted us to more rigorously examine the biochemical properties of these proteins in vitro. Despite repeated efforts with different expression regimes in Escherichia coli and insect cells, and multiple attempts to improve protein solubility using various peptide tags and re-solubilization protocols, we were unable to identify conditions that produced soluble protein. Therefore, we turned to an in vitro eukaryotic expression system, rabbit reticulocyte lysate (RRL), for protein production. Soluble AtPOT1a and AtPOT1b were obtained (Figure 2b, lanes 2 and 3) and were used in an EMSA. As a positive control, we expressed the OB-fold domains of mouse POT1a (mPOT1a_N) (Figure 2b, lane 1), which displays robust binding to mammalian telomeric DNA in vitro (Wu et al., 2006). As expected, mPOT1a_N bound an oligonucleotide corresponding to two vertebrate telomere repeats (GGTTAG)2 (Figure 2c, lane 1). Under the same conditions, we could not detect AtPOT1a binding to oligonucleotides containing either two or five copies of the plant telomere repeat TTTAGGG (Figure 2c, lane 2 and Figure S1, lane 2) or to a cocktail of seven individual oligonucleotides representing all possible permutations of the plant telomere repeat (data not shown). The gel-shift profile was indistinguishable from that of the RRL-only negative control (Figure 2c, lane 8 and Figure S1, lane 1). As for AtPOT1a, AtPOT1b also failed to bind single-strand G-rich telomeric DNA oligonucleotides under these conditions (Figure 2c, lane 3 and Figure S1, lane 3).
Figure 2.
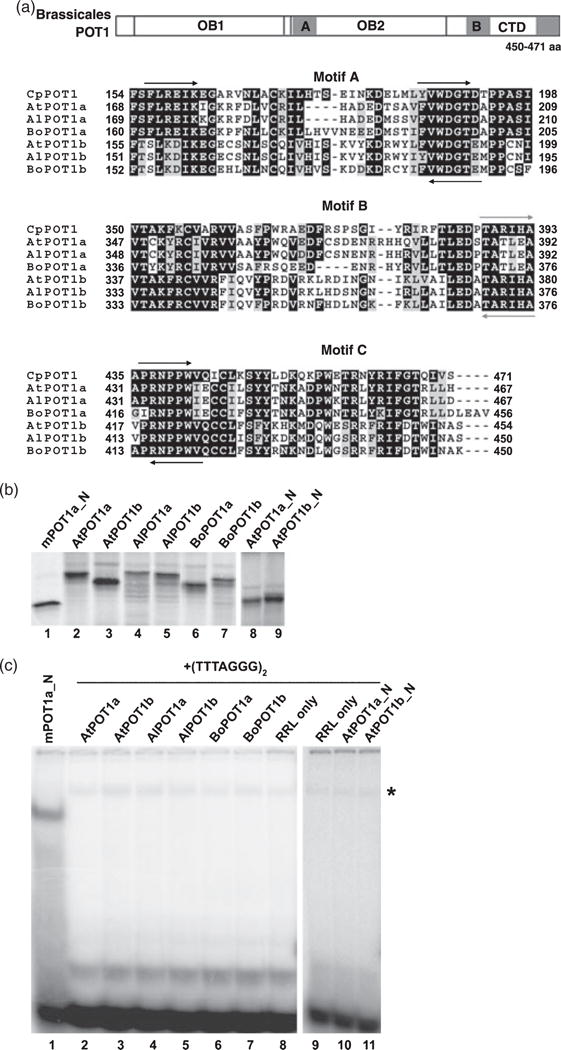
POT1 proteins from Brassicaceae species do not bind telomeric DNA in vitro.
(a) Partial alignment of Brassicaceae POT1a and POT1b proteins with POT1 from Carica papaya. Top panel: generalized diagram of POT1 proteins from Brassicales species. The relative positions of two N-terminal OB folds (OB1 and OB2) and the C-terminal domain (CTD) are shown. Three regions with the highest amino acid conservation (gray squares) were identified in OB2 (motif A) and CTD (motifs B and C). Bottom panels: amino acid alignment of motifs A, B and C in Brassicales POT1 proteins. Arrows indicate the positions of degenerate primers for RT-PCR used for the initial cloning of the partial cDNAs. Black arrows correspond to primers specific for all cDNAs. Gray arrows represent primers specific for the POT1b lineage. Numbers indicate amino acid positions relative to the start codon. The alignment was generated using MEGA 3 software (Kumar et al., 2004) and is visualized in the BOXSHADE format.
(b) SDS–PAGE of in vitro RRL-expressed POT1 proteins assayed in (c). Lane 1, mouse POT1a_N (two N-terminal OB folds); lane 2, full-length Arabidopsis thaliana POT1a; lane 3, full-length Arabidopsis thaliana POT1b; lane 4, full-length Arabidopsis lyrata POT1a; lane 5, full-length Arabidopsis lyrata POT1b; lane 6, full-length Brassica oleracea POT1a; lane 7, full-length Brassica oleracea POT1b; lane 8, At-POT1a_N (two N-terminal OB folds); lane 9, AtPOT1b_N (two N-terminal OB folds).
(c) Electrophoretic mobility shift assay with Brassicaceae POT1 proteins listed in (b) using (TTTAGGG)2 as a probe. Lane 1, mouse POT1a_N and (GGTTTAG)2 oligonucleotide were used as a positive control for the assay. A. thaliana POT1a (lane 2), A. thaliana POT1b (lane 3), A. lyrata POT1a (lane 4), A. lyrata POT1b (lane 5), B. oleracea POT1a (lane 6), B. oleracea POT1b (lane 7), RRL alone (lanes 8 and 9), AtPOT1a_N (lane 10), AtPOT1b_N (lane 11). The asterisk indicates a non-specific band that is also present in negative controls.
Removal of the C-terminus from S. pombe POT1 and mouse POT1a significantly increases their affinity for single-strand telomeric DNA (Baumann and Cech, 2001; Wu et al., 2006). A similar experiment was performed with AtPOT1a and AtPOT1b, but no DNA binding was observed when the C-terminus was deleted from either of the two plant proteins, or when only one of the two OB folds was used for EMSA (Figure 2c, lanes 10, 11, Figure S1, lanes 9, 10, and data not shown). Likewise, co-expression of AtPOT1a and AtPOT1b in the same RRL reaction also failed to generate DNA binding (data not shown).
To test for C-strand telomeric DNA binding by AtPOT1a and AtPOT1b, EMSA was performed using either two or five copies of the C-rich plant telomere repeat. No binding to these oligonucleotides was detected, nor to oligonucleotides corresponding to double-strand plant telomere repeats (data not shown). While it is possible that Arabidopsis POT1 proteins bind telomeric DNA with an affinity below our detection limit, the data suggest that, unlike POT1 proteins from vertebrates and yeast, AtPOT1a and AtPOT1b do not bind single-strand telomeric DNA under standard gel-shift conditions.
Identification and analysis of POT1a and POT1b from Brassicaceae species
We next determined whether the failure to observe single-strand telomeric DNA binding was due to a peculiarity of A. thaliana POT1a or POT1b by examining the DNA binding properties of POT1 proteins from related plants. We cloned POT1a and POT1b cDNAs from two other members of the Brassicaceae family, Arabidopsis lyrata, which shared a last common ancestor with A. thaliana 5.2 million years ago (Koch et al., 2000), and Brassica oleracea (cauliflower), which diverged from the Arabidopsis lineage approximately 20 million years ago (Bailey et al., 2006). Using sequence data for Arabidopsis POT1 genes and the single-copy POT1 gene present in Carica papaya (Shakirov et al., 2008), which belongs to the same Brassicales order and shared a last common ancestor with Brassicaceae approximately 70 million years ago (Wikstrom et al., 2001), we identified three highly conserved motifs in the second OB fold and in the C-terminal domain (CTD) of the Brassicales POT1 proteins, termed A, B and C (Figure 2a). Using these motifs, we designed degenerate primers for RT-PCR experiments and cloned the corresponding Brassicaceae cDNAs. Like A. thaliana, both A. lyrata and B. oleracea harbor POT1a and POT1b homologs, suggesting that the POT1 gene duplication occurred early in evolution of the Brassicaceae lineage. POT1a and POT1b proteins from Brassicaceae species appear to be equally diverged from C. papaya POT1 (53–57% and 57–58% overall amino acid similarity to CpPOT1, respectively), with extensive sequence conservation in the C-terminus and several additional regions throughout the polypeptides (Figure 2a).
Recombinant POT1a and POT1b proteins from A. lyrata and B. oleracea expressed in RRL were soluble (Figure 2b, lanes 4–7) and were analyzed by EMSA. As for A. thaliana, POT1a and POT1b proteins from A. lyrata and B. oleracea did not show detectable telomeric DNA binding in vitro (Figure 2c, lanes 4–7 and Figure S1, lanes 4–7). Furthermore, using a variety of gel-shift conditions and truncated versions of the proteins, we also failed to detect telomeric DNA binding with recombinant proteins encoded by the single-copy POT1 genes in cotton (Gossypium hirsutum), poplar (Populus trichocarpa) and potato (Solanum tuberosum) (data not shown), which diverged from the Arabidopsis lineage 85, 100 and 120 million years ago, respectively (Wikstrom et al., 2001). Thus, the inability of AtPOT1a and AtPOT1b to associate with telomeric DNA under these assay conditions is not a peculiarity of Arabidopsis, but instead appears to be a common feature shared by POT1 proteins from other dicots.
Identification of single-strand telomere binding activities in B. oleracea nuclear extracts
Several candidate single-strand telomeric DNA binding proteins have been identified in Arabidopsis, tobacco and Chlamydomonas through a combination of biochemical purification techniques and MS analysis (Petracek et al., 1994; Hirata et al., 2004; Kwon and Chung, 2004; Yoo et al., 2007). However, none of the factors studied so far correspond to POT1 or any other OB fold-containing proteins. Therefore, we determined whether POT1 could be biochemically purified as the major single-strand telomeric DNA binding activity from a plant that is both closely related to Arabidopsis thaliana and for which a POT1 gene(s) has been cloned. For this test, we chose B. oleracea, as this species is an abundant source of biochemical material. Furthermore, several genome sequencing projects are underway for Brassica species, including B. oleracea (http://www.genomesonline.org), and thus Brassica is poised to become a valuable tool for comparative telomere genomics in Brassicaceae.
EMSAs were performed using G-rich telomeric oligonucleotides and B. oleracea nuclear extracts. Four shifted complexes were found, designated A, B, C and D, from top to bottom (Figure 3, lane 2). An additional signal (asterisk) was also detected, but was less stable and was not observed in some extract preparations. Addition of proteinase K to the binding reaction abolished the formation of all four major complexes, while addition of RNase A had no effect (data not shown), suggesting that the complexes are formed by protein factors. No shifted band was observed with a C-rich telomeric oligonucleotide (C3TA3)5 or with duplex telomere repeats (data not shown). Addition of up to a 100-fold molar excess of cold (T3ACG2)5 oligonucleotide bearing one nucleotide mutation in each telomere repeat did not abolish binding (Figure 3, lanes 11 and 12), indicating that the binding was sequence-specific. This conclusion was supported by competition experiments using human (TTAGGG)5 and ciliate Oxytricha nova (T4G4)5 sequences (Figure 4a). Complexes C and D were abolished when the human telomere repeat competitor was used, while complexes A and B showed only a minor reduction in binding with up to 100-fold excess of (TTAGGG)5 (Figure 4a, lanes 5 and 6). No competition was observed with the ciliate telomeric DNA (Figure 4a, lanes 7 and 8). Thus, the four B. oleracea complexes exhibit distinct DNA binding properties, with the two slower-migrating complexes being the most specific for the plant telomere repeat sequence.
Figure 3.
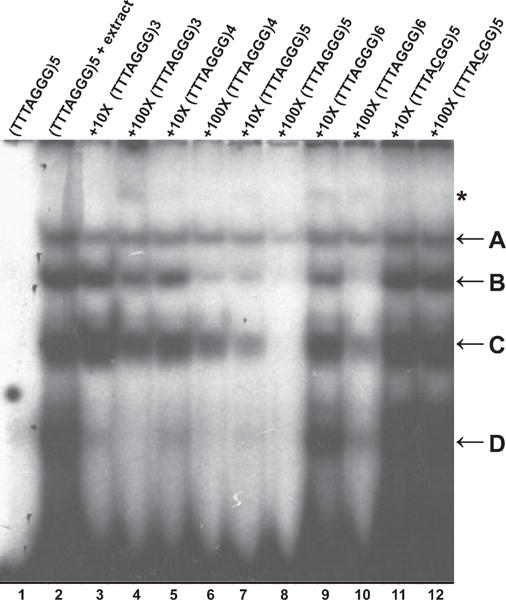
Identification of single-strand telomeric DNA binding complexes in Brassica.
EMSAs were performed using B. oleracea nuclear extracts. Competition was performed with the indicated fold excess of cold oligonucleotides shown above each lane. The four major complexes are designated A–D. The asterisk indicates a minor band of lower mobility. Lane 1, (TTTAGGG)5 oligonucleotide alone; lane 2, (TTTAGGG)5 plus nuclear extract; lanes 3–12, addition of 10 × (lanes 3, 5, 7, 9 and 11) or 100 × (lanes 4, 6, 8, 10 and 12) excess cold competitor oligonucleotides with various numbers of telomeric repeats (lanes 3–10) or with a point mutation in the repeat sequence (lanes 11 and 12).
Figure 4.
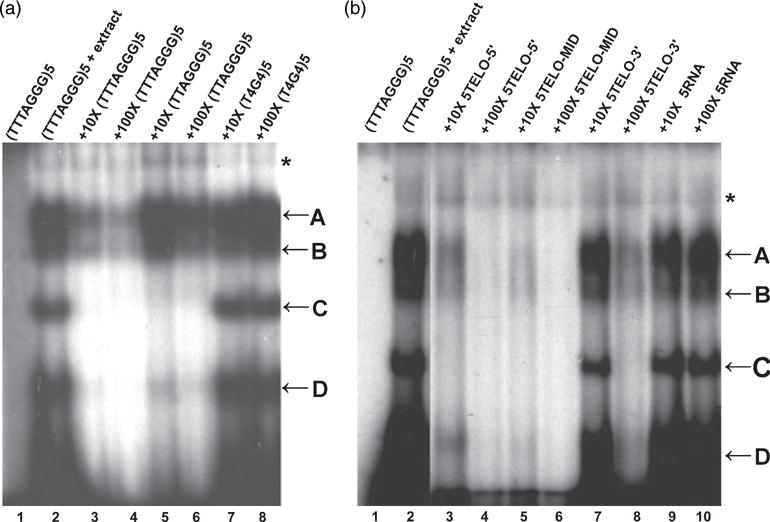
Biochemical properties of Brassica single-strand telomeric DNA binding proteins.
(a) B. oleracea proteins show a preference for plant telomeric DNA sequences. EMSAs were performed with radiolabeled (TTTAGGG)5. Lane 1, oligonucleotide alone; lane 2, oligonucleotide plus nuclear extract; lanes 3–8, competition with 10 × (lanes 3, 5 and 7) or 100 × (lanes 4, 6 and 8) excess cold competitor DNA oligonucleotides corresponding to plant (lanes 3 and 4), human (lanes 5 and 6) and ciliate (lanes 7 and 8) telomere repeat sequences.
(b) B. oleracea single-strand telomeric DNA binding proteins do not show end-binding preference. EMSAs were performed with radiolabeled (TTTAGGG)5. Lane 1, oligonucleotide alone; lane 2, oligonucleotide plus nuclear extract. Complex formation was challenged by addition of 10 × (lanes 3, 5 and 7) or 100 × (lanes 4, 6 and 8) excess cold competitor DNA with telomeric repeats positioned at the 5′ end (5TELO-5′, (TTTAGGG)5CTCTACCAAA, lanes 3 and 4), in the middle (5TELO-MID, CTCTA(TTTAGGG)5CCAAA, lanes 5 and 6) or at the 3′ end of the oligonucleotide (5TELO-3′, CTCTACCAAA(TTTAGGG)5, lanes 7 and 8). Complex formation was also challenged by addition of 10 × (lane 9) or 100 × (lane 10) excess cold 5-RNA oligonucleotide (UUUAGGG)5.
Several proteins with RNA recognition motifs display both telomeric DNA and RNA binding activities (Lin and Zakian, 1994; LaBranche et al., 1998). To determine whether the B. oleracea DNA binding proteins also interact with RNA, competition experiments were performed with the plant telomere RNA sequence. No competition was observed in the presence of up to 100-fold excess of cold (U3AG3)5 oligoribonucleotide (Figure 4b, lanes 9 and 10). Altogether, these results indicate that the B. oleracea activities we identified are highly specific for single-strand G-rich telomeric DNA substrates.
Analysis of B. oleracea single-strand telomeric DNA binding activities
In yeast and vertebrates, POT1 proteins bind to a minimum sequence of 10–12 telomeric nucleotides, roughly corresponding to two telomeric repeats (Lei et al., 2004; Wei and Price, 2004; Croy et al., 2006). Therefore, competition assays were performed with oligonucleotides of differing lengths to determine the number of telomeric repeats necessary for efficient DNA binding by B. oleracea. Similar to the situation for Arabidopsis nuclear extracts, oligonucleotides with more telomeric repeats showed progressively better competition (Figure 3, lanes 3–6). Although an oligonucleotide with five telomeric repeats was the best competitor for all complexes (Figure 3, lanes 7 and 8), some competition was detected with as few as three telomeric repeats. Notably, complex D was competed away with all of the cold telomeric oligonucleotides used in the study, suggesting that it requires the least number of telomeric repeats for binding. In contrast, complexes A, B and C preferred longer substrates for efficient binding. Surprisingly, oligonucleotides with six telomeric repeats competed less efficiently than oligonucleotides with five telomeric repeats (Figure 3, lanes 9 and 10). It is possible that the (T3AG3)6 oligonucleotide undergoes a conformational change in solution to form a secondary structure that prevents efficient protein binding. Telomeric oligonucleotides are known to form ‘G-quartet’ structures in vitro (Sundquist and Klug, 1989) that may be inhibitory for telomere protein binding. Further biochemical and biophysical analysis will be required to address this possibility.
Several single-strand telomeric DNA binding factors, including S. pombe POT1 (Sheng et al., 1995; Baumann and Cech, 2001), display a strong binding preference for the free 3′ OH. To determine whether the B. oleracea telomeric DNA binding activities exhibit a preference for 3′ ends, we performed competition experiments with oligonucleotides containing five telomeric repeats located in the middle or at the 5′ or 3′ end of the DNA. Both the 5TELO-5′ (5′ position) and 5TELO-MID oligonucleotides competed much better for binding than 5TELO-3′ (3′ position) (Figure 4b, lanes 3–8), suggesting that the B. oleracea proteins do not have a preference for the free 3′ OH. A similar result was observed for human and chicken (Gallus gallus) POT1 proteins (Loayza et al., 2004; Wei and Price, 2004), as well as for the single-strand telomeric DNA binding activities from rice (Oryza sativa, Kim et al., 1998). Overall, B. oleracea complexes A and B appear to harbor DNA binding proteins with the greatest specificity for the plant telomere sequence, and thus may represent the true G-overhang binding factors.
Purification and characterization of telomere binding proteins in B. oleracea
To further investigate the biochemical properties of the B. oleracea single-strand telomere binding proteins, we subjected B. oleracea nuclear extracts to size fractionation on a Superose 12 column. Each eluted fraction was analyzed for DNA binding activity. As shown in Figure 5, the peak of activity for complexes A and D eluted in fractions 32 and 33 (50–80 kDa), while fraction 30 (150–160 kDa) contained proteins necessary for formation of complex C. Complex B had a major peak in fraction 33 (50–80 kDa) and a minor peak in fraction 30 (150–160 kDa). An additional high-molecular-weight complex, indicated by the asterisk, was formed in fractions 28 and 29 (170–200 kDa). These data indicate that complexes A, B and D elute in the 50–80 kDa size range, which potentially overlaps with the predicted sizes of B. oleracea POT1 proteins (52.5 kDa for BoPOT1a and 51.5 kDa for BoPOT1b, based on cloned cDNA).
Figure 5.
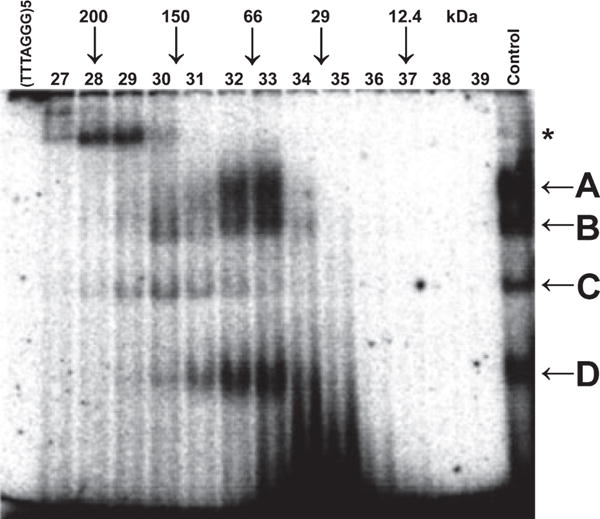
Brassica single-strand telomeric DNA binding proteins form complexes of various molecular weights.
Fractionation of B. oleracea nuclear proteins was performed on a Superose 12 column. EMSAs were performed on fractions 27–39. The positions of molecular weight protein markers are shown at the top of the gel. Unfractionated nuclear extract was used as a control in the reaction.
As discussed above, the telomeric DNA binding activities present in B. oleracea complexes A and B show the highest specificity for the plant telomere sequence, a characteristic feature of bona fide single-strand telomere binding proteins. Complex A routinely eluted as a single peak, while complex B segregated into two peaks on a number of columns (see below), suggesting that it was not homogeneous. Thus, we focused on analysis of the DNA binding component(s) within complex A. B. oleracea nuclear extracts were subjected to ammonium sulfate precipitation, preparative isoelectric focusing and size-exclusion chromatography (Figure 6a, Figures S2 and S3 and Appendix S1). This protocol allowed us to selectively purify complex A from other B. oleracea telomeric DNA binding activities.
Figure 6.
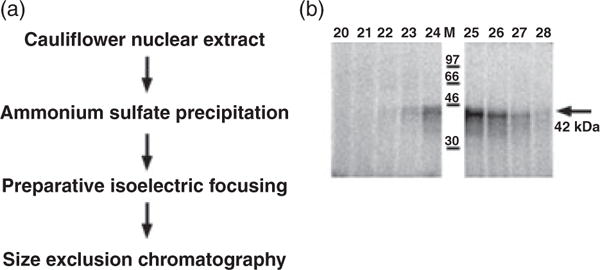
Purification and analysis of DNA binding component of B. oleracea complex A.
(a) Protein purification scheme used to partially purify single-strand telomeric DNA binding activity present in B. oleracea complex A.
(b) Autoradiograph of SDS–PAGE of proteins eluted in Superose 12 fractions 20–28 showing the highest DNA binding activity (see Figure S3). Partially purified proteins in complex A were cross-linked to the radioactively labeled telomeric DNA prior to running SDS–PAGE. A 42 kDa product is observed.
To estimate the apparent molecular weight of DNA binding components within complex A, proteins in Superose 12 fractions 20–28 (Figure S3) were allowed to interact with 32P-labeled (T3AG3)5, and then were irradiated with UV light to form covalent protein–DNA bonds. The cross-linked fractions were subjected to SDS–PAGE, and the gel was autoradiographed. A major cross-linked band was visible across fractions 22–28, peaking in fraction 25 (Figure 6b), which coincides with the peak of complex A DNA binding activity in the EMSA (Figure S3). The estimated size of this band is approximately 42 kDa, which includes both the protein and the DNA oligonucleotide cross-linked to it. Upon subtracting the weight of the DNA, the size of the putative telomeric DNA binding protein is less than 30 kDa.
As the size of this B. oleracea DNA binding protein is 2–3 times smaller than the size observed for complex A as a whole (50–80 kDa), complex A may either be formed by oligomerization of the DNA binding protein itself or by its association with additional subunits through protein–protein interactions. This DNA binding protein subunit may also be shared by complex B, as fractions 22 and 23 with the peak of complex B activity (Figure S3) appear to have the same DNA binding component as complex A (Figure 6b).
In conclusion, regardless of the overall protein subunit composition in complexes A and B, their DNA binding protein component is considerably smaller in size than the predicted molecular weights of B. oleracea POT1a and POT1b proteins (approximately 50 kDa). This finding provides further evidence that Brassicaceae POT1 proteins are unlikely to contribute to the formation of the highly specific single-strand telomeric DNA binding complexes observed in wild-type Arabidopsis and Brassica nuclear extracts.
DISCUSSION
Since the green plant lineage separated from the rest of the eukaryotes approximately 1.5 billion years ago (Hedges et al., 2004; Yoon et al., 2004), dramatic differences in the composition and functions of telomere-associated proteins may have accumulated between plants and other organisms. In support of this, at least three members of the vertebrate shelterin complex (TPP1, TIN2 and RAP1) have no obvious sequence homologs in the sequenced genomes of Arabidopsis or other plants. In contrast, POT1-like proteins appear to be conserved across eukaryotic evolution and perform essential functions in telomere biology for a number of organisms, including S. pombe, vertebrates and plants. In yeast and vertebrates, telomeric DNA binding is required for POT1 function in vivo. Genetic experiments demonstrated that the absence of functional OB folds in POT1 leads to phenotypes similar to those displayed by null mutants, and result in telomere de-protection (Bunch et al., 2005; Barrientos et al., 2008) and perturbations of telomere length regulation (Loayza and de Lange, 2003; Bunch et al., 2005). In striking contrast, the data presented here argue that single-strand telomeric DNA binding is not a function shared by the POT1 proteins in the plant kingdom.
The A. thaliana POT1a and POT1b genes were predicted to encode bona fide POT1 proteins, based on the presence of two OB folds of the Telo_bind_N type (Baumann et al., 2002; Shakirov et al., 2005). The overall structure of these plant OB folds is remarkably conserved with respect to the corresponding protein domains in the human and S. pombe POT1 proteins (J. Croy and D. Wuttke, University of Colorado, personal communication). However, the current genetic data are inconsistent with a major role for AtPOT1a and AtPOT1b in G-overhang binding and protection. First, unlike its vertebrate counterparts, AtPOT1a is not a stable component of plant telomeres. Instead, AtPOT1a physically associates with the telomerase RNP and is enriched at telomeres only in S phase (Surovtseva et al., 2007). Second, while dominant-negative experiments implicated AtPOT1b in chromosome end protection (Shakirov et al., 2005), AtPOT1b null mutants display only mild defects in telomere architecture, inconsistent with a major role in G-overhang protection (E. Shakirov, A. Nelson, D. Shippen, unpublished data).
Here we provide biochemical evidence indicating that Brassicaceae POT1 proteins do not bind single-strand telomeric DNA in vitro or in vivo. We used a combination of endogenous protein purification and recombinant protein analysis to assay for single-strand telomeric DNA binding activity. Not only did we fail to detect binding to single-strand G-rich telomeric DNA for the A. thaliana POT1a and POT1b proteins, but also for their orthologs from two other Brassicaceae species and from several more distantly related dicots that encode only a single POT1 protein. One drawback of our study is that we were unable to obtain soluble recombinant protein from E. coli or using the baculovirus expression system, limiting the amount of protein that we could analyze and thus potentially preventing the detection of low-affinity interactions between the plant POT1 proteins and telomeric DNA. However, telomeric DNA binding is readily detected by RRL-expressed POT1 proteins from yeast and mammals under the same in vitro gel-shift conditions that we employed (Figure 2) (Baumann and Cech, 2001; Wu et al., 2006). We cannot rule out the possibility that single-strand telomeric DNA binding by plant POT1 proteins requires additional post-translational modifications, oligomerization of AtPOT1a and AtPOT1b polypeptides, or protein interaction partners that are not needed for telomeric DNA binding by yeast and vertebrate POT1.
Two additional lines of evidence reinforce the conclusion that telomeric DNA binding is not a major function for POT1 proteins from Brassicaceae, and possibly other plants. First, the single-strand telomeric DNA binding activity in A. thaliana nuclear extracts is not perceptibly decreased or altered by the absence of POT1a or POT1b. Second, UV cross-linking of partially purified single-strand telomere binding proteins from B. oleracea, which display high specificity for the plant telomere repeat sequence, identified polypeptides of less than 30 kDa in size, much smaller than the predicted size of BoPOT1a or BoPOT1b (approximately 50 kDa). Furthermore, as discussed below, all previous reports of TTTAGGG repeat binding proteins from plants as evolutionarily diverse as the dicots Silene latifolia, Nicotiana tabacum, Arabidopsis thaliana and Vigna radiate, the monocots Muscari armeniacum and Scilla peruviana, and the unicellular green algae Chlamydomonas reinhardtii identified proteins in the 15–40 kDa range (Petracek et al., 1994; Fulneckova and Fajkus, 2000; Lee et al., 2000; Hirata et al., 2004; Kwon and Chung, 2004; Rotkova et al., 2007; Yoo et al., 2007), again significantly smaller than the POT1 proteins encoded in these genomes (E. Shakirov and D. Shippen, unpublished data).
While the precise function of plant POT1 proteins remains unclear, one intriguing possibility is that they have evolved a role in the recruitment of telomerase to the G-overhang in a manner similar to the Est1 protein from budding yeast (reviewed by Lundblad, 2003). Est1 interacts with Cdc13, a core component of the CST G-overhang binding complex, thereby linking the telomerase RNP to the chromosome terminus. Loss of POT1a in Arabidopsis leads to an ever-shorter telomere phenotype (Surovtseva et al., 2007), just as the name given to the yeast Est1 mutants implies.
Single-strand telomeric DNA binding proteins in plants
If POT1 is not the major G-overhang binding protein in plants, then what is responsible for protecting telomeric DNA on the chromosome terminus? To address this question, we examined telomeric DNA binding factors in B. oleracea and found four activities that display sequence-specific binding to single-strand telomeric DNA in vitro. Like G-strand binding proteins in vertebrates and rice (Kim et al., 1998; Loayza et al., 2004; Wei and Price, 2004), the B. oleracea proteins do not have a preference for a free 3′ telomeric overhang, suggesting that they may also bind to the displaced G-rich strand in the t-loop structure. Nuclear proteins in both Arabidopsis and Brassica prefer to bind to at least three consecutive telomere repeats, with the best substrate having five repeats. These findings contrast sharply with those for POT1 proteins from yeast and vertebrates, which only require two telomeric repeats or fewer for efficient binding (Lei et al., 2004; Wei and Price, 2004; Croy et al., 2006). The biochemical properties of Brassicaceae proteins resemble previously characterized single-strand telomere binding factors from other angiosperms, including soybean (Glycine max), rice (Oryza sativa) and mung bean (Vigna radiata) (Kim et al., 1998; Lee et al., 2000; Kwon et al., 2004). All of these plant proteins exhibit strong specificity for their cognate telomere repeat sequences and require three and more telomeric repeats for efficient binding.
The presence of four distinct complexes in B. oleracea versus one in A. thaliana is intriguing. A similar difference in the number of telomeric DNA-bound complexes has previously been reported for Fabaceae, with three complexes in mung bean and only one complex in soybean (Lee et al., 2000; Kwon et al., 2004). Both A. thaliana and B. oleracea are diploid species that probably originated from ancient polyploids (Town et al., 2006) through independent diploidization events. Thus, their genomes may differ in the number of retained genes that encode single-strand telomere binding proteins, although this observation alone is unlikely to explain such a striking difference in the number of shifted bands observed in EMSAs. Another interesting possibility is that this variation reflects developmental differences in the number and composition of the telomeric DNA binding complexes in A. thaliana and B. oleracea, as may be the case in mung bean (Lee et al., 2000) and senescing Arabidopsis leaves (Zentgraf et al., 2000).
Several of the previously characterized single-strand telomeric DNA binding factors from plants are classified as either proteins with RNA recognition motifs (Petracek et al., 1994; Hirata et al., 2004) or plant-specific transcription factors (Kwon and Chung, 2004; Yoo et al., 2007), and share no sequence or structural similarity with POT1 or any other OB-fold-containing proteins. Although the genetic and cytogenetic evidence to support a direct role at the telomeric G-overhang is currently lacking, the available biochemical data for at least some of these plant proteins indicate that they contribute to telomere function. Even more intriguing is the recent discovery of a Stn1 ortholog in Arabidopsis that is required for chromosome end protection (Song et al., 2008). Stn1 sequence homologs are also present in other sequenced plant genomes, from green algae to rice, poplar and probably Brassica. This finding strongly argues that components of the CST complex function at plant telomeres. Thus, despite 20 years of extensive telomere research, the full complement of G-strand binding proteins in plants and probably other eukaryotes remains to be elucidated.
EXPERIMENTAL PROCEDURES
Brassicaceae POT1 cDNA cloning
Cauliflower samples were obtained from commercial varieties. A. lyrata seeds were purchased from the Arabidopsis Biological Research Center. RNA extraction and general RT-PCR conditions were as described previously (Shakirov et al., 2005). To amplify POT1 cDNAs from Brassicaceae species, degenerate primers were designed to several consensus regions that show a high degree of amino acid similarity between Arabidopsis thaliana and Carica papaya POT1 proteins. Partial POT1 cDNA products were amplified by degenerate RT-PCR using SuperScript II reverse transcriptase (Invitrogen, http://www.invitrogen.com/). Full-length POT1 coding regions were subsequently amplified by 5′ and 3′ RACE (Ambion http://www.ambion.com) using gene-specific primers. The sequences of cDNAs encoding the following Brassicaceae POT1 proteins were submitted to GenBank: AlPOT1a (EU880293), AlPOT1b (EU880294), BoPOT1a (EU880299) and BoPOT1b (EU880300). CpPOT1 (accession number EU887728) has been described previously (Shakirov et al., 2008).
Brassica and Arabidopsis nuclear extract preparation
For the Brassica nuclear extract, up to 12 cauliflower heads were ground in a Waring blender (http://www.waringproducts.com) with 2 ml of grinding buffer (25 mM MES pH 6.0, 10 mM MgCl2, 5 mM NaCl2, 0.5 M sucrose, 40% glycerol, 14 mM β-mercaptoethanol) per gram of tissue. The resulting mass was homogenized using a Polytron (Glenmills, http://www.glenmills.com), and the extract was filtered through cheesecloth and then through Miracloth (Calbiochem, http://www.emdbiosciences.com). The suspension was spun for 10 min at 150 g at 4 °C, and then for 30 min at 1400 g in a JA-14 rotor (Beckman Coulter, http://www.beckmancoulter.com). To break chloroplasts, the pellet was resuspended several times in 10 ml wash buffer (25 mM MES pH 6.0, 10 mM MgCl2, 5 mM NaCl2, 0.5 M sucrose, 25% glycerol, 0.1% Triton X-100, 1.4 mM β-mercaptoethanol), and spun for 30 min at 4 °C in a JA-20 rotor (Beckman Coulter) at 3000 g. The final pellet was resuspended in extraction buffer (50 mM Tris/HCl pH 7.8, 10 mM MgCl2, 2M NaCl, 0.6 M KCl, 0.2 mM EDTA, 0.5 mM PMSF, 1 mM DTT), and left overnight at 4 °C for protein extraction. Nuclear extract was dialyzed against dialysis buffer (50 mM Tris/HCl pH 7.8, 5 mM MgCl2, 40 mM KCl, 0.2 mM EDTA, 0.01 mM PMSF, 1 mM DTT, 20% glycerol), and DNA was removed using DNase I. Aliquots were quick-frozen in liquid nitrogen and stored at −80 °C until needed.
To prepare Arabidopsis nuclear extract, 2–3 g of Arabidopsis 14-day-old seedlings were ground in liquid nitrogen and incubated on ice for 5–10 min in a 50 ml Falcon tube (BD Biosciences, http://www.bdbiosciences.com) with 20 ml of NIB buffer containing 5 mM EDTA, 50 mM Tris pH 8.0, 10 mM KCl, 250 mM sucrose, 1.5 mM MgCl2, 0.3% Triton X-100, 1mM PMSF, 5 mM β-mercaptoethanol, 1 mM spermine, 1 mM spermidine and protease inhibitor cocktail (Roche, http://www.roche.com). The material was filtered through one layer of Miracloth and spun at 3000 g for 20–30 min in a cold room. After removing the supernatant, the pellet was resuspended in 1 ml of Triton buffer (0.25 M sucrose, 10 mM Tris/HCl, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 1 mM PMSF, 1 mM spermine, 1 mM spermidine and protease inhibitors), and incubated on ice for 5–10 min in a 1.5 ml tube. The extract was then centrifuged in a cold room at 2000 g for 1 min, 4000 g for 1 min and 8000 g for 2 min). If the pellet was still green, indicating that not all chloroplasts are lysed, the steps above were repeated twice. The nuclei pellet was then resuspended in 500 μl of 1.5 M sucrose in NIB buffer (NIB buffer as above plus 1.5 M sucrose, 5 mM β-mercaptoethanol, 1 mM PMSF, 1 mM spermine, 1 mM spermidine and protease inhibitors), and layered on top of 500 μl of 1.5 M sucrose in NIB cushion. Following centrifugation at 14 000 g for 30 min in a cold room, nuclei were frozen in liquid nitrogen and stored in −80 °C until needed. To extract nuclear proteins, isolated nuclei were resuspended in 500 μl of extraction buffer containing 20 mM HEPES pH 8, 1 mM MgCl2, 0.2 mM EDTA, 300 mM NaCl, 10% glycerol, 1% Triton X-100, 0.1% NP-40, 1 mM DTT, 1 × complete protease inhibitors, and mixed by rotation for 30 min at 4 °C. After incubation, the nuclei suspension was spun down at 15 000 g for 15 min at 4 °C. Supernatants were collected, instantly frozen in liquid nitrogen and stored at −80 °C for further analysis.
EMSA and DNA cross-linking with Arabidopsis and Brassica nuclear extracts
Various concentrations of plant nuclear extract were mixed with 0.5 pmol of 32P-labeled (TTTAGGG)5 oligonucleotide in DNA binding buffer (20 mM Tris/HCl pH 8.0, 50 mM NaCl, 10 mM MgCl, 1mM EDTA, 1 mM DTT, 5% glycerol, 15 μg HaeIII-digested E. coli DNA), and incubated at room temperature for 15 min. The complexes were separated on 5% polyacrylamide gels (acrylamide:bisacrylamide 29:1) for 4 h at 150 V in 1 × TBE at room temperature, dried and exposed to film or PhosphorImager screens (Molecular Dynamics, http://www.gehealthcare.com). For RRL-expressed samples, reactions were performed as described previously (Wu et al., 2006) with slight modifications as described by Surovtseva et al. (2007).
For DNA cross-linking assays, partially purified protein samples from Superose 12 column (GE Healthcare, http://www.gehealthcare.com) chromatography were incubated with radioactively labeled (TTTAGGG)5 as described above for 15 min, and then cross-linked in a UV Stratalinker 1800 (Stratagene, http://www.stratagene.com/) for 15 min, followed by 10% SDS–PAGE. The gels were dried and exposed to PhosphorImager screens.
Supplementary Material
Appendix S1. Supplementary data.
Figure S1. Electrophoretic mobility shift assay with Brassicaceae POT1 proteins using (TTTAGGG)5 as a probe.
Figure S2. Purification of telomere binding proteins from B. oleracea nuclear extract.
Figure S3. Size-exclusion chromatography of B. oleracea telomere binding proteins present in complex A.
Acknowledgments
We are grateful to Sandy Chang for providing the mouse POT1a_N construct, Yulia Surovtseva and Xiangyu Song for help with EMSA experiments, and other members of the Shippen lab for insightful discussions. This work was supported by grants from the National Institutes of Health (GM065383) to D.E.S. and from the National Science Foundation (MCB 0 244 159) to T.D.M.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bailey CD, Koch MA, Mayer M, Mummenhoff K, O’Kane SL, Jr, Warwick SI, Windham MD, Al-Shehbaz IA. Toward a global phylogeny of the Brassicaceae. Mol Biol Evol. 2006;23:2142–2160. doi: 10.1093/molbev/msl087. [DOI] [PubMed] [Google Scholar]
- Barrientos KS, Kendellen MF, Freibaum BD, Armbruster BN, Etheridge KT, Counter CM. Distinct functions of POT1 at telomeres. Mol Cell Biol. 2008;28:5251–5264. doi: 10.1128/MCB.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Baumann P, Podell E, Cech TR. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol Cell Biol. 2002;22:8079–8087. doi: 10.1128/MCB.22.22.8079-8087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch JT, Bae NS, Leonardi J, Baumann P. Distinct requirements for Pot1 in limiting telomere length and maintaining chromosome stability. Mol Cell Biol. 2005;25:5567–5578. doi: 10.1128/MCB.25.13.5567-5578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11–389. J Mol Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Fajkus J, Sykorova E, Leitch AR. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005;13:469–479. doi: 10.1007/s10577-005-0997-2. [DOI] [PubMed] [Google Scholar]
- Fulneckova J, Fajkus J. Inhibition of plant telomerase by telomere-binding proteins from nuclei of telomerase-negative tissues. FEBS Lett. 2000;467:305–310. doi: 10.1016/s0014-5793(00)01178-9. [DOI] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y, Suzuki C, Sakai S. Characterization and gene cloning of telomere-binding protein from tobacco BY-2 cells. Plant Physiol Biochem. 2004;42:7–14. doi: 10.1016/j.plaphy.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Hong JP, Byun MY, Koo DH, An K, Bang JW, Chung IK, An G, Kim WT. Suppression of RICE TELOMERE BINDING PROTEIN 1 results in severe and gradual developmental defects accompanied by genome instability in rice. Plant Cell. 2007;19:1770–1781. doi: 10.1105/tpc.107.051953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. Composition and conservation of the telomeric complex. Cell Mol Life Sci. 2003;60:2295–2302. doi: 10.1007/s00018-003-3245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva ZN, Surovtseva YV, Vespa L, Shakirov EV, Shippen DE. A C-terminal Myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J Biol Chem. 2004;279:47799–47807. doi: 10.1074/jbc.M407938200. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim WT, Chung IK. Rice proteins that bind single-stranded G-rich telomere DNA. Plant Mol Biol. 1998;36:661–672. doi: 10.1023/a:1005994719175. [DOI] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Kuchar M, Fajkus J. Interactions of putative telomere-binding proteins in Arabidopsis thaliana: identification of functional TRF2 homolog in plants. FEBS Lett. 2004;578:311–315. doi: 10.1016/j.febslet.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Kwon C, Chung IK. Interaction of an Arabidopsis RNA-binding protein with plant single-stranded telomeric DNA modulates telomerase activity. J Biol Chem. 2004;279:12812–12818. doi: 10.1074/jbc.M312011200. [DOI] [PubMed] [Google Scholar]
- Kwon C, Kwon K, Chung IK, Kim SY, Cho MH, Kang BG. Characterization of single stranded telomeric DNA-binding proteins in cultured soybean (Glycine max) cells. Mol Cells. 2004;17:503–508. [PubMed] [Google Scholar]
- LaBranche H, Dupuis S, Ben-David Y, Bani MR, Wellinger RJ, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim JH, Kim WT, Kang BG, Chung IK. Characterization and developmental expression of single-stranded telomeric DNA-binding proteins from mung bean (Vigna radiata) Plant Mol Biol. 2000;42:547–557. doi: 10.1023/a:1006373917321. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1–3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 1994;22:4906–4913. doi: 10.1093/nar/22.23.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- Lundblad V. Telomere replication: an Est fest. Curr Biol. 2003;13:R439–R441. doi: 10.1016/s0960-9822(03)00365-8. [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Petracek ME, Konkel LM, Kable ML, Berman J. A Chlamydomonas protein that binds single-stranded G-strand telomere DNA. EMBO J. 1994;13:3648–3658. doi: 10.1002/j.1460-2075.1994.tb06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008;132:745–757. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Fajkus J, Vyskot B, Shippen DE. Analysis of the G-overhang structures on plant telomeres: evidence for two distinct telomere architectures. Plant J. 2000;23:633–641. doi: 10.1046/j.1365-313x.2000.00831.x. [DOI] [PubMed] [Google Scholar]
- Rotkova G, Sykorova E, Fajkus J. Characterization of nucleoprotein complexes in plants with human-type telomere motifs. Plant Physiol Biochem. 2007;45:716–721. doi: 10.1016/j.plaphy.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Shakirov EV, Shippen DE. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Salzberg SL, Alam M, Shippen DE. Analysis of Carica papaya telomeres and telomere-associated proteins: insights into the evolution of telomere maintenance in Brassicales. Trop Plant Biol. 2008;1:202–215. doi: 10.1007/s12042-008-9018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H, Hou Z, Schierer T, Dobbs DL, Henderson E. Identification and characterization of a putative telomere end-binding protein from Tetrahymena thermophila. Mol Cell Biol. 1995;15:1144–1153. doi: 10.1128/mcb.15.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Surovtseva YV, Shakirov EV, Vespa L, Osbun N, Song X, Shippen DE. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani A, Murata M. Alternative splicing of Pot1 (protection of telomere)-like genes in Arabidopsis thaliana. Genes Genet Syst. 2005;80:41–48. doi: 10.1266/ggs.80.41. [DOI] [PubMed] [Google Scholar]
- Town CD, Cheung F, Maiti R, et al. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Skopp R, Scofield M, Price C. Euplotes crassus has genes encoding telomere-binding proteins and telomere-binding protein homologs. Nucleic Acids Res. 1992;20:6621–6629. doi: 10.1093/nar/20.24.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Price CM. Cell cycle localization, dimerization, and binding domain architecture of the telomere protein cPot1. Mol Cell Biol. 2004;24:2091–2102. doi: 10.1128/MCB.24.5.2091-2102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom N, Savolainen V, Chase MW. Evolution of the angiosperms: calibrating the family tree. Proc Biol Sci. 2001;268:2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Songyang Z. The telosome/shelterin complex and its functions. Genome Biol. 2008;9:232. doi: 10.1186/gb-2008-9-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Kim SK, Kim WT. Perturbation of NgTRF1 expression induces apoptosis-like cell death in tobacco BY-2 cells and implicates NgTRF1 in the control of telomere length and stability. Plant Cell. 2004;16:3370–3385. doi: 10.1105/tpc.104.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HH, Kwon C, Lee MM, Chung IK. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 2007;49:442–451. doi: 10.1111/j.1365-313X.2006.02974.x. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- Zentgraf U, Hinderhofer K, Kolb D. Specific association of a small protein with the telomeric DNA–protein complex during the onset of leaf senescence in Arabidopsis thaliana. Plant Mol Biol. 2000;42:429–438. doi: 10.1023/a:1006324008600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supplementary data.
Figure S1. Electrophoretic mobility shift assay with Brassicaceae POT1 proteins using (TTTAGGG)5 as a probe.
Figure S2. Purification of telomere binding proteins from B. oleracea nuclear extract.
Figure S3. Size-exclusion chromatography of B. oleracea telomere binding proteins present in complex A.


