Abstract
Background
Pharmacogenomic variability can contribute to differences in pharmacokinetics and clinical responses. Pediatric patients with cerebral palsy (CP) with genetic variations have not been studied for these potential differences.
Objective
To determine the genetic sources of variation in oral baclofen clearance and clinical responses.
Design
Pharmacogenomic add-on study to determine variability in oral baclofen clearance and clinical responses.
Setting
Multicenter study based in academic pediatric cerebral palsy clinics.
Participants
49 patients with CP who had participated in an oral baclofen Pharmacokinetic/Pharmacodynamic (PK/PD) study.
Methods or Interventions
From 53 participants in a PK/PD trial, 49 underwent genetic analysis of 307 key genes and 4,535 single-nucleotide polymorphisms (SNPs) involved in drug absorption, distribution, metabolism and excretion. Associations between genotypes and phenotypes of baclofen disposition (weight-corrected and allometrically scaled clearance) and clinical endpoints (improvement from baseline in mean hamstring Modified Tardieu Scale (MTS) scores from baseline for improvement of R1 spastic catch) were determined by univariate analysis with correction for multiple testing by false discovery rate (FDR).
Main Outcome Measurements
Primary outcome measures were the genotypic and phenotypic variability of oral baclofen in allometrically scaled clearance and change in the MTS angle compared to baseline.
Results
After univariate analysis of the data, the SNP of ABCC9 (rs11046232, heterozygous AT vs. the reference TT genotype) was associated with a 2-fold increase in oral baclofen clearance (Mean 0.51 ± Standard Deviation 0.05 L/hr/kg for the AT genotype vs. 0.25 ± 0.07 L/hr/kg for the TT genotype, adjusted p<.001). Clinical responses were associated with decreased spasticity by MTS in allelic variants with SNPs ABCC12, SLC28A1, and PPARD.
Conclusions
Genetic variation in ABCC9 impacting oral baclofen clearance highlights the need for continued studies of genetic polymorphisms to better characterize variable drug response in children with CP. Single nucleotide polymorphisms in ABCC12, SLC28A1, and PPARD were associated with varied responses, which warrants further investigation to determine their effect on spasticity.
Introduction
Oral baclofen, (RS)-4-amino-3-(4-chlorophenyl) butanoic acid, is a GABAB receptor agonist that has been used to treat spasticity in adults for many years, and several studies have been conducted to characterize its disposition and dose-response relationships in this age group.1-3 Similarly, oral baclofen has been used to treat spasticity associated with cerebral palsy in children for about the same length of time, but the absence of approved guidelines for its safe and effective use in young patients has resulted in a broad range of dosing strategies by clinicians managing their care.4 There are a myriad of developmental factors that may impact drug absorption, distribution, metabolism and excretion throughout the developmental continuum, from birth to adolescence.5 Thus, it is essential for studies be conducted in an appropriately aged patient population, such as children with cerebral palsy, to generate sufficient data to establish dose ranges, dosing schedules, dose escalation strategies and adverse event profiles. Furthermore, it is important to characterize the extent of variability to be anticipated in a pediatric population as well as the primary sources of variability, such as the relative contributions of ontogeny and genetic variation to the observed variability in drug disposition and response. The ultimate goal is to minimize the risks of therapeutic failure and excessive toxicity in treated patients.
Unfortunately, pediatric pharmacokinetic (PK) data for baclofen are scarce, and there are few studies examining the population variability or factors contributing to variability in disposition and response in children. Establishment of safe and effective dosing strategies for children with cerebral palsy requires an understanding of the PK and pharmacodynamic (PD) properties of baclofen in children, and recognition of individual differences that may contribute to divergent clinical responses to baclofen among children with cerebral palsy. The Best Pharmaceuticals for Children Act (BPCA) of 2002 addressed this knowledge in a study entitled “Oral Baclofen Pharmacokinetics and Pharmacodynamics in Children with Spasticity (Best PK/PD)”, registered as clinical trial NCT00607542 at ClinicalTrials.gov. A population pharmacokinetic analysis was conducted on concentration vs. time data obtained from 49 pediatric patients with spastic cerebral palsy, ranging in age from 2 to 17 years, enrolled at 11 participating clinical sites. This analysis revealed that the oral baclofen concentrations were adequately characterized with a two-compartment PK model with linear elimination and a delayed absorption process.6 Furthermore, body weight was identified as a covariate that contributed to variability in the estimate of apparent oral clearance. The study also found a single nucleotide polymorphism (SNP) in the ABCC9 gene to be associated with an increase in oral baclofen clearance. In this report, we present the results of an exploratory pharmacogenomic study to assess the contribution of other genetic variants to determine various pharmacogenomic sources of oral baclofen clearance and pharmacodynamic outcomes in this cohort of children with cerebral palsy.
Methods
Patients and Samples
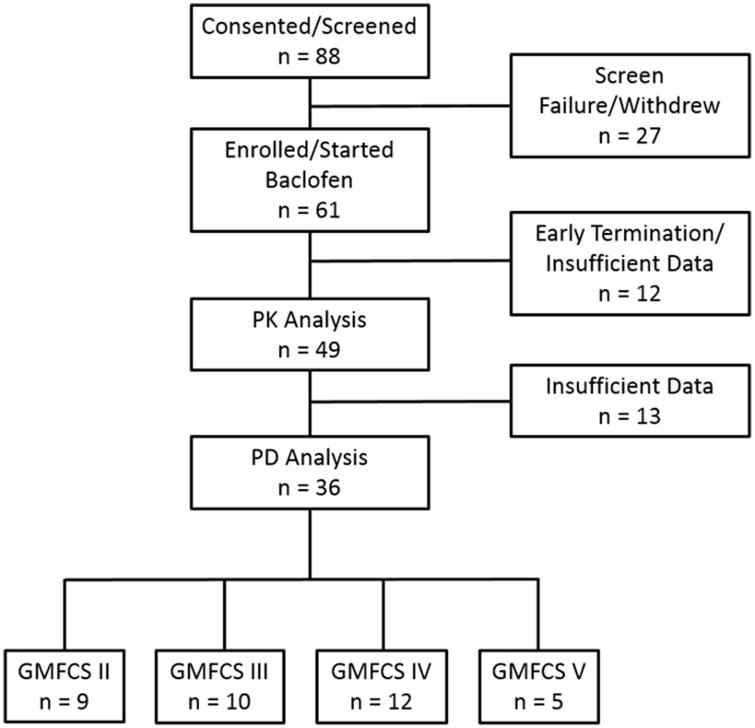
After parental permission was obtained, DNA isolated from whole blood or saliva was available for 53 children participating in a pharmacokinetic study for pharmacogenetic analysis; adequate post-dose PK data at the PK visit were available for 49 of the subjects (19 female, 30 male) all of which were included in the population PK analysis (Figure 1). DNA was isolated using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) in the Division of Clinical Pharmacology, Toxicology and Therapeutic Innovation at Children's Mercy Kansas City. Quality was assessed by agarose gel electrophoresis, and DNA concentration was determined spectrophotometrically with a NanoDrop instrument at the time of isolation.
Figure 1.
Demographics of patients included in the clinical response scores when shown as an overall composite score. Pharmacokinetic (PK) analysis and pharmacodynamic (PD) analysis were performed from the same group. Gross Motor Function Classification System (GMFCS) scores ranged from II-V.
Genotyping
DNA samples were genotyped at the Canadian Pharmacogenomics Network for Drug Safety (CPNDS), British Columbia Children's Hospital Research Institute, Vancouver, BC, Canada) for 4,535 SNPs using a customized Illumina GoldenGate SNP genotyping assay (Illumina, San Diego, CA) designed to capture the genetic variation of 307 key genes involved in drug absorption, distribution, metabolism and excretion (ADME), including phase I and II drug metabolism enzymes, drug transporters, drug targets, drug receptors, transcription factors, ion channels and other disease-specific genes. All SNP genotype data were manually clustered using GenomeStudio software (Illumina). The 384 SNPs that could not be clustered were excluded from further analyses, leaving a total of 4,151 SNPs. The ADME panel consisted of 488 coding SNPs, identified by literature review and database queries that cause nonsynonymous or synonymous amino acid changes or have been associated with changes in enzyme activity or function. The panel also consisted of 4,048 tag SNPs identified using the ldSelect algorithm to select a maximally informative set of tag SNPs to assay in the candidate genes. The tag SNP selection was performed using data from the International HapMap project that included all four populations (CEU, CHB, JPT and YRI) with a threshold for the LD statistic r2 of 0.8 and a minor allele frequency of more than 0.05.7 The concordance of genotype calls between replicate genotyped samples was greater than 99.9% (n = 8). If the call rate for any sample was below 95%, the sample was excluded from further analysis; genotyping was repeated in 5 samples failing this threshold on the first attempt. The average genotyping call rate for all samples was 98.7%.
Of the 4,151 SNPs passing quality control, 233 deviated from Hardy-Weinberg equilibrium (p<0.05) and were excluded from the analysis, leaving a total of 3,918 for analysis of statistical associations with PK and drug response outcome variables.
Pharmacokinetic Outcome Measures
Estimates of uncorrected apparent oral baclofen clearance (CL/F) and apparent oral baclofen clearance corrected for body weight (L/h/kg) from the population PK analysis were provided as the primary outcome measures for the exploratory pharmacogenetic investigation. For this cohort (n=49) the mean ± SD uncorrected CL/F was 6.6 ± 2.3 L/h (median 6.1 L/h) with a range of 3.1 to 11.4 L/h, and the weight corrected CL/F varied approximately 5-fold, from 0.113 to 0.562 L/hr/kg with a mean ± SD of 0.261 ± 0.090 L/h/kg (median 0.247 L/h/kg). Allometrically-scaled apparent oral clearance data were also provided, with a mean ± SD of 1.455 ± 0.364 L/h/kg0.529 (median 1.388 L/h/kg0.529; range 0.821 to 2.760 L/h/kg0.529).6
Clinical Outcome Measures
Spasticity was primarily assessed at baseline and a follow-up PK visit using the Modified Tardieu Scale (MTS). The severity of resistance to stretch was evaluated using the 0 to 2 Tardieu scale score (TSS), and the angle of spastic catch (R1) was evaluated using manual and electronic goniometers when a clear catch (TSS=2) was found. Improvement in the Ashworth Scale was also assessed. Hamstring measurements were available in 36 subjects. Assessment of genetic variants associated with clinical response was determined using the following clinical endpoints: 1) improvement in the mean hamstring MTS score from the baseline visit to the PK study visit and 2) improvement in the mean hamstring MTS angle of spastic catch (R1) from the baseline visit to the PK study visit, measured in degrees using a manual and an electronic goniometer. To calculate improvement, the original visit mean change scores were rescaled so that positive improvements indicate a reduction in spasticity from baseline.
Statistical Analysis
A univariate analysis was conducted to determine the association between each outcome variable, corrected oral clearance and drug response, and each SNP. Continuous outcome variables were analyzed by an analysis of variance testing to compare the means of these variables by genotype. Categorical variables were analyzed by Fisher's exact test to compare the percentage (risk) of outcomes by genotype. Multiple tests were adjusted by false discovery rate (FDR). Tests with adjusted p-value (FDR) less than .05 were considered statistically significant. Statistical analyses were conducted with SAS software version 9.2 and JMP version 10 (SAS Institute Inc., Cary, NC).
Results
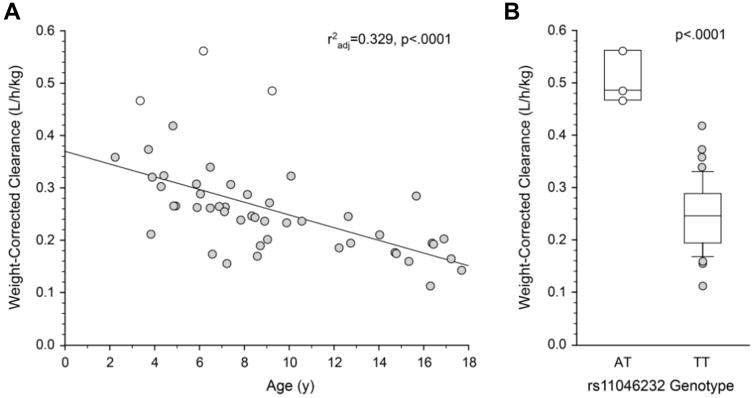
A total of 3,918 SNPs were available for analysis in 53 subjects, and clearance estimates were available for 49 of these subjects. The univariate analysis on all 3,918 SNPs for body-weight corrected clearance revealed many SNPs with nominal p-values <.05. The top 20 SNPs with the smallest p-values are presented in Table 1. Consistent with primary elimination of unchanged baclofen in the urine, 10 of the top 20 genes/SNPs were associated with transporter genes; no cytochrome P450 genes were found in the top 20 genes/SNPs. However, only one SNP, rs11046232 in ABCC9, remained significantly associated with clearance adjusted for body weight after correction of the p-values for FDR (0.51 ± 0.05 L/hr/kg for the AT genotype vs 0.25 ± 0.07 for the TT genotype, adjusted p<.001). The homozygous reference TT genotype was observed in 48 subjects while the heterozygous AT genotype was observed in 3 subjects (one subject failed genotyping). The three subjects heterozygous for rs11046232T>A had the highest values for weight-corrected clearance (Figure 2A, 2B). No statistically significant associations were observed for uncorrected apparent oral clearance after FDR adjustment.
Table 1. Top 20 single-nucleotide polymorphisms (SNPs) and their corresponding genes associated with weight-corrected apparent oral clearance of baclofen (L/h/kg).
| SNP | RS Number | Var/Var | Var/Ref | Ref/Ref | P Value | Adjusted P |
|---|---|---|---|---|---|---|
| ABCC9 | rs11046232 | 0.51 ± 0.05 | 0.25 ± 0.07 | <.001 | <.001 | |
| ALDH1A2 | rs11858606 | 0.56 | 0.18 ± 0.04 | 0.26 ± 0.07 | <.001 | .12 |
| NNMT | rs2244175 | 0.41 ± 0.12 | 0.24 ± 0.07 | 0.24 ± 0.06 | <.001 | .15 |
| SULT1C1 | rs752273 | 0.27 ± 0.08 | 0.21 ± 0.06 | 0.4 ± 0.1 | <.001 | .41 |
| CES2 | rs11568314 | 0.43 ± 0.08 | 0.25 ± 0.08 | <.001 | .33 | |
| FMO1/2 | rs2307492 | 0.5 | 0.21 ± 0.04 | 0.26 ± 0.08 | <.001 | .31 |
| ABCA4 | rs6658767 | 0.4 ± 0.14 | 0.25 ± 0.08 | <.001 | .29 | |
| SLC31A1 | rs2233914 | 0.44 ± 0.03 | 0.32 ± 0.12 | 0.24 ± 0.07 | <.001 | .33 |
| SLCO3A1 | rs960440 | 0.56 | 0.23 ± .07 | 0.26 ± 0.08 | <.001 | .30 |
| ALDH5A1 | rs807515 | 0.24 ± 0.08 | 0.27 ± 0.08 | 0.48 ± 0.01 | <.001 | .28 |
| ALDH3B1 | rs479763 | 0.56 | 0.24 ± 0.06 | 0.26 ± 0.09 | <.001 | .39 |
| SLC22A1/2 | rs3798156 | 0.56 | 0.27 ± 0.07 | 0.25 ± 0.08 | <.001 | .39 |
| CYP1B1 | rs1800440 | 0.56 | 0.23 ± 0.05 | 0.26 ± 0.09 | <.001 | .38 |
| SLCO3A1 | rs2283458 | 0.43 ± 0.12 | 0.25 ± 0.08 | 0.25 ± 0.08 | .002 | .41 |
| SLC22A15 | rs12023924 | 0.23 ± 0.05 | 0.26 ± 0.08 | 0.38 ± 0.17 | .002 | .39 |
| SLC13A1 | rs4288315 | 0.56 | 0.24 ± 0.06 | 0.26 ± 0.08 | .002 | .39 |
| FMO1/2 | rs2421707 | 0.56 | 0.26 ± 0.09 | 0.25 ± 0.06 | .002 | .38 |
| SLCO3A1 | rs8025658 | 0.56 | 0.26 ± 0.09 | 0.25 ± 0.08 | .002 | .40 |
| DPYD | rs11587873 | 0.56 | 0.26 ± 0.08 | 0.25 ± 0.08 | .002 | .39 |
| SLC22A3 | rs2292334 | 0.56 | 0.26 ± 0.06 | 0.25 ± 0.09 | .002 | .38 |
Var/Var denotes a genotype that is homozygous for the minor allele at that position; Var/Ref indicates heterozygosity; Ref/Ref is a homozygous reference (or “wild-type”) genotype. SNPs were selected from 3918 candidate SNPs that passed quality control and were in Hardy-Weinberg equilibrium. All SNPs except ABCC9 rs11046232 had false discovery rate–adjusted P values >.1; several SNPs in this top 20 list are characterized by a single Var/Var genotype with relatively high weight-corrected clearance.
Figure 2.
(A) Relationship between weight-corrected apparent oral baclofen clearance and age. Subjects with the ABCC9 rs11046232 AT genotypes had the highest values for oral baclofen clearance at their respective ages. (B) Significant effect (P < .0001) of the ABCC9 genotype (rs11046232) on the weight-corrected apparent oral clearance of baclofen (L/h/kg) after false discovery rate adjustment. Boxes represent the 25th and 75th percentiles; the solid line within the interquartile range represents the median. Whiskers represent the 10th and 90th percentiles. All subjects are presented for the heterozygous AT genotype (open symbols), whereas only subjects with data lying beyond the 10th and 90th percentiles are presented for the homozygous TT genotype.
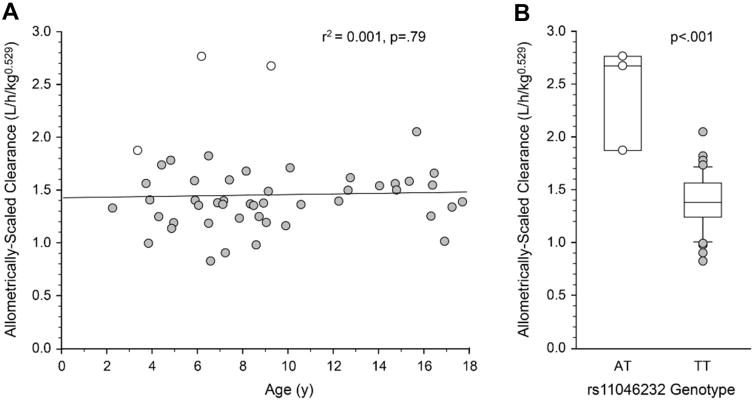
Allometric scaling is required when comparing children across different ages as the size of a child and their organs alters the clearance of medications. When an allometric-scaling factor of 0.529 is applied to weight (kg0.529) and apparent oral clearance is corrected for allometrically-scaled weight (L/hr/kg0.529), age-related changes in oral baclofen clearance are no longer apparent (Figure 3A). However, allometrically-scaled apparent oral clearance remained statistically significantly higher in subjects heterozygous for rs11046232 (2.43 ± 0.49 L/hr/kg0.529) compared to subjects with the reference TT genotype (1.39 ± 0.25 L/hr/kg0.529, adjusted p=.002; Figure 3B). Interestingly, a second SNP, rs11858606 in ALDH1A2, was marginally associated with allometrically-scaled apparent oral clearance (p=0.04 after adjustment for multiple testing, Table 2), but this effect is limited to a single subject with a homozygous variant genotype.
Figure 3.
(A) Allometric scaling of apparent oral clearance of baclofen (L/h/kg0.529) removes age-associated changes in age. (B) A significant relationship (P < .001) between the ABCC9 genotype (rs11046232) and allometrically scaled clearance is present after false discovery rate adjustment. Boxes represent the 25th and 75th percentiles; the solid line within the interquartile range represents the median. Whiskers represent the 10th and 90th percentiles. All 3 subjects with heterozygous genotypes are presented (open symbols), whereas only subjects with data lying beyond the 10th and 90th percentiles are presented for the homozygous genotype.
Table 2. Single-nucleotide polymorphisms (SNPs) and their corresponding genes associated with allometrically scaled, apparent oral clearance of baclofen (L/h/kg0.529) with adjusted P values <.1.
| SNP | RS Number | Var/Var | Var/Ref | Ref/Ref | P Value | Adjusted P |
|---|---|---|---|---|---|---|
| ABCC9 | rs11046232 | 2.43 ± 0.49 | 1.39 ± 0.25 | <.001 | <.001 | |
| ALDH1A2 | rs11858606 | 2.76 | 1.36 ± 0.23 | 1.41 ± 0.26 | <.001 | .04 |
Var/Var denotes a genotype that is homozygous for the minor allele at that position; Var/Ref indicates heterozygosity; Ref/Ref is a homozygous reference (or “wild-type”) genotype. SNPs were selected from 3918 candidate SNPs that passed quality control and were in Hardy-Weinberg equilibrium.
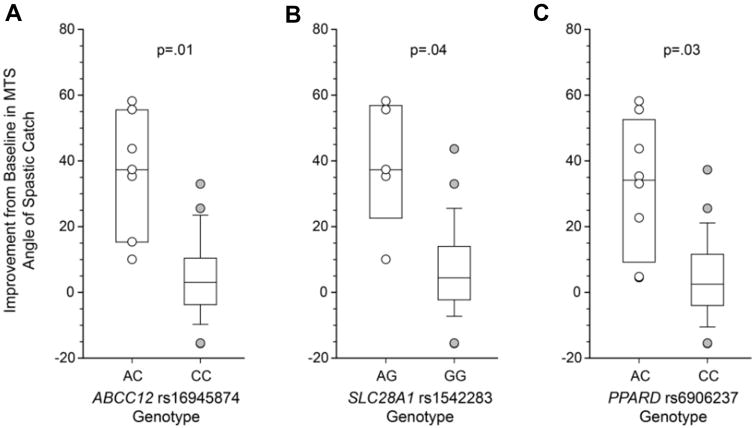
Genotype associations with the three clinical improvement measures are presented in Table 3. Statistically significant associations (p-values <.05 after correction for FDR) were apparent only when response was measured as improvement in the mean MTS R1 hamstring angle measured without an electronic device. Carriers of the variant “A” allele for rs16945874 in ABCC12 (CA genotype; n=7) had significantly greater mean MTS R1 relative to subjects with subjects homozygous for the wildtype “C” (36.4 ± 18.4 vs 4.4 ± 11.6, adjusted p=0.01; Figure 4A). Similar differences in response were observed between carriers of variant alleles and homozygous reference genotypes for two SNPs in SLC28A1 (rs1542283 and rs16974641) and in PPARD (rs7770619 and rs6906237) (Table 3). The data for SLC28A1 rs1542283 and PPARD rs6906237 are also presented in Figure 4B and Figure 4C, respectively.
Table 3. Top single-nucleotide polymorphisms (SNPs) and genes associated with response to baclofen.
| SNP | RS Number | Var/Var | Var/Ref | Ref/Ref | P Value | Adjusted P |
|---|---|---|---|---|---|---|
| PON2/3 | rs43037* | 1.38 ± 0.44 | 0.23 ± 0.38 | 0.13 ± 0.41 | <.001 | .06 |
| ABCC12 | rs16945874† | 36.4 ± 18.4 | 4.4 ± 11.6 | <.001 | .01 | |
| SLC28A1 | rs1542283† | 39.2 ± 19.4 | 6.2 ± 13.3 | <.001 | .04 | |
| SLC28A1 | rs16974641† | 39.2 ± 19.4 | 6.2 ± 13.3 | <.001 | .04 | |
| PPARD | rs7770619† | 33.7 ± 21.6 | 4.7 ± 12.0 | <.001 | .04 | |
| PPARD | rs6906237† | 32.2 ± 20.6 | 4.5 ± 12.0 | <.001 | .04 | |
| EPHX1 | rs868966† | 30.8 ± 20.29 | 3.4 ± 11.9 | 4.8 ± 10.5 | <.001 | .09 |
| ABCC12 | rs16945874‡ | 34.4 ± 19.1 | 3.1 ± 13.7 | <.001 | .08 |
Var/Var denotes a genotype that is homozygous for the minor allele at that position; Var/Ref indicates heterozygosity; Ref/Ref is a homozygous reference (or “wild-type”) genotype. Adjusted P values reflect correction for false discovery rate.
Response measure is improvement in the mean Modified Tardieu Scale (MTS) Tardieu scale score (TSS) from the baseline visit to the pharmacokinetics study visit (hamstring).
Response measure is improvement in the mean MTS angle of spastic catch (R1) from the baseline visit to the pharmacokinetics study visit, measured in degrees using a manual goniometer (hamstring).
Response measure is improvement in the mean MTS R1 score from the baseline visit to the pharmacokinetics study visit, measured by electronic goniometer (hamstring).
Figure 4.
Genetic associations with clinical response to baclofen as indicated by the improvement in the mean Modified Tardieu Scale angle of spastic catch from the baseline visit to the pharmacokinetic study visit, measured in degrees. Positive values indicate a reduction in spasticity relative to baseline. Boxes represent the 25th and 75th percentiles; the solid line within the interquartile range represents the median. Whiskers represent the 10th and 90th percentiles. All subjects are presented for heterozygous genotypes (open symbols), whereas only subjects with data lying beyond the 10th and 90th percentiles are presented for homozygous genotypes. Data are presented for rs16945874 in ABCC12 (A), rs1542283 in SLC28A1 (B), and rs6906237 in PPARD (C). Statistical comparisons were corrected for false discovery rate.
Discussion
The purpose of this add-on pharmacogenetic investigation to ClinicalTrials.gov study NCT00607542, “Oral Baclofen Pharmacokinetics and Pharmacodynamics in Children with Spasticity (Best PK/PD)” was to explore potential genetic contributions to observed variability in pharmacokinetic factors determined in the course of the primary study. As essentially no information is available concerning the specific pathways involved in baclofen disposition, a global ADME approach was taken that utilized a genotyping platform comprising 307 ADME genes that was developed by the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) at the British Columbia Children's Hospital Research Institute (Vancouver, BC). This platform has been successfully applied to identify genetic variants associated with an increased risk of cisplatin ototoxicity and anthracycline-associated cardiotoxicity in pediatric cancer patients.8,9,10,11
Finding ten transporter genes, but no Cytochrome P450 genes among the top 20 genes associated with weight-corrected apparent oral baclofen clearance were consistent with available information indicating that baclofen is primarily excreted unchanged by the kidney. Approximately 15% of a dose undergoes deamination to form 3-(p-chlorophenyl)-4-hydroxybutyric acid, and some drug is excreted unchanged in the feces. After correction for FDR, however, only one SNP, rs11046232 in the ABCC9 transporter gene, was significantly associated with apparent baclofen clearance. This association was not an artifact of age effects on clearance as the association was present regardless of whether allometrically-scaled apparent oral clearance or clearance corrected for body weight was the phenotype of interest.
ABCC9 is a member of the multi-drug resistance protein (MRP) group of transporters with ATP binding and hydrolysis domains, that includes the CFTR gene (ABCC7) and transporters involved in the transport of many endogenous molecules, such as glucuronide and glutathione conjugates, and resistance to anti-cancer drugs.12 The gene is located on chromosome 12p12.1, and undergoes alternative splicing of mutually exclusive terminal exons to form ATP-sensitive potassium channels known as the sulfonylurea receptors SUR2A, which utilizes exon 38A, and SUR2B (exon 38B). ABCC9 is located immediately adjacent to KCNJ8, which encodes the channel-forming inward rectifier subunit Kir6.1. The proteins Kir6.1 and SUR2B are key components of the smooth muscle ATP-sensitive potassium channel (KATP channel), an octameric structure of Kir6 and SUR subunits in which structural predictions indicate the presence of one ABCC9 (SUR2B) ATP binding site at each of the Kir6 interfaces.13 Non-synonymous (amino acid-changing) mutations in ABCC9 have been associated with cardiomyopathy dilated type 10 and Cantu syndrome.14,15
ABCC9 rs11046232 is an intronic SNP with a minor allele frequency of 2.0% in the 1000 Genomes dataset, consistent with the frequency of 3/96 = 3.1% in the current study. It is reported to belong to a 4-SNP haplotype that includes ABCC9 intronic SNPs rs2900492, rs4148649 and rs4762865. This haplotype occurs at a higher frequency (approximately 4%) in individuals with a more pronounced decrease in blood pressure upon standing compared to subjects who demonstrate a much lower postural change in blood pressure (frequency ∼1%). However, the authors of this study urged cautious interpretation of the genotype-phenotype association due to the limited power of the study and the fact that rare haplotypes were inferred, rather than phased.16 The only other SNP in this haplotype that was assessed in the current study was rs4148649, and this SNP had no effect on weight-corrected apparent clearance (p=0.509).
Although many MRP transporters encoded by ABCC genes transport a variety of endogenous and xenobiotic substrates, the substrate profile of ABCC9 is not well characterized.17 Therefore, the relevance of genetic variation in ABCC9 on drug transport and observed variability remains unknown.
Transporters were also prominent among the limited number of genes associated with clinical response to baclofen in this cohort. ABCC12 (also known as MRP9) is a member of the same gene family as ABCC9, but it remains unclear whether the ABCC12 gene encodes a functional transporter.12 Nevertheless, the rs16945874 C>A variant is a non-synonymous SNP that results in an amino acid change from alanine to glutamic acid, with the potential for functional implications. ABCC12 is of particular interest, as it is primarily expressed in the brain and central nervous system which is where baclofen is thought to act.18 SLC28A1 is also known as the concentrative nucleoside transporter-1 (CNT1), and is involved in the cellular uptake of nucleoside analogs, such as the anti-cancer compound gemcitabine (2′,2′-difluorodeoxycytidine) and the reverse transcriptase inhibitor, zidovudine (azidothymidine, or AZT). PPARD, peroxisomal proliferative activated receptor delta, is a member of the peroxisomal proliferative activated receptor (PPAR) family of transcription factors whose ligands have peroxisomal proliferating properties and control the number and size of peroxisomes. Other members of the family are involved in the pathogenesis of chronic diseases such as diabetes, obesity, atherosclerosis, and cancer; genetic variation in PPARD has been associated with the presence of bipolar disorder.19 As in the case of ABCC9 and baclofen clearance, the relevance of SNPs in ABCC12, SLC28A1 and PPARD to observed variability in clinical response to baclofen is unknown, but is sufficiently intriguing to warrant future prospective validation studies in a larger cohort of patients.
No other SNPs were significantly associated with the various drug disposition and response phenotypes, and therefore it is important to avoid over-interpretation of the results. However, one cannot exclude the possibility that some of the (statistically) non-significant findings might nevertheless be functionally relevant even though the study was not sufficiently powered to detect associations that may be real. In this context, some of the top 20 genes in Table 1 may be of interest for future replication studies. For example, genotypes with at least one variant allele of rs807515 in ALDH5A1 were associated with a weight-corrected apparent oral baclofen clearance that is approximately half that of the Ref/Ref genotype (Table 1). ALDH5A1 encodes, a mitochondrial enzyme that is highly expressed in the brain and commonly referred to as succinic semialdehyde dehydrogenase, which is an enzyme is involved in the catabolism of the gamma-aminobutyric acid (GABA). Although nominally significant (p<.001), significance was not retained after FDR adjustment (p=0.28). Rare coding and non-coding mutations in ALDH5A1 result in an autosomal recessive metabolic disorder that is characterized by the accumulation of GABA and 4-hydroxybutryic acid leading to several moderate to severe phenotypic neurological disorders including mental retardation, ataxia and seizures.20
Alterations in drug exposure lead to increased variability of drug response. As oral baclofen has been reported to have up to 90% inter-individual variability in oral absorption and elimination processes2, the level of drug exposure affects the pharmacodynamic portion of the dose-exposure-response paradigm. In the case of oral baclofen, the ultimate response of decreased spasticity is dependent on both the drug dosage and the amount of drug exposure at the site of action. No current studies have evaluated the pharmacodynamic outcomes when controlling for the same level of drug exposure. A recent systematic review of the literature to determine a practice parameter for oral baclofen concluded there was not sufficient evidence for or against the use of oral baclofen.21 Previous trials have controlled for the oral baclofen dosage to evaluate for response, but no studies have incorporated the exposure of oral baclofen to the response. By controlling the level of exposure, future studies could better evaluate the pharmacodynamic parameters of interest, such as change in spasticity or prevention of adverse events. A significant need exists to evaluate this relationship.
Conclusion
This exploratory pharmacogenetic add-on study has identified rs11046232 in the ABC transporter gene ABCC9 as being associated with increased clearance of baclofen in children with cerebral palsy, and SNPs in ABCC12, SLC28A1 and PPARD with the clinical response to the drug in this same population. Given the relatively small number of subjects in this study, especially those with clinical response data, these associations should be considered preliminary and requires validation in a prospective study to determine if they can serve as biomarkers of clinical response in this underserved patient population. A significant need exists to examine the relationships and genetic determinants of oral baclofen clearance and outcomes with more targeted studies for these specific genotypes.
Acknowledgments
Funding Source: Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) NICHD-2005-13-2; Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) NICHD 5T32HD069038-07
Abbreviations
- CP
Cerebral Palsy
- PK
Pharmacokinetics
- PD
Pharmacodynamics
- SNP
Single-nucleotide polymorphism
- FDR
False-discovery rate
Footnotes
Financial Disclosures: None.
Conflict of Interest: None of the authors declare a conflict of interest in this manuscript.
Clinical Trial Registration: NCT00607542
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wuis EW, Dirks MJ, Termond EF, Vree TB, Van der Kleijn E. Plasma and urinary excretion kinetics of oral baclofen in healthy subjects. Eur J Clin Pharmacol. 1989;37:181–184. doi: 10.1007/BF00558228. [DOI] [PubMed] [Google Scholar]
- 2.Shellenberger MK, Groves L, Shah J, Novack G. A controlled pharmacokinetic evaluation of tizanidine and baclofen at steady state. Drug Metab Dispos. 1999;27:201–204. [PubMed] [Google Scholar]
- 3.Chou R, Peterson K, Helfand M. Comparative efficacy and safety of skeletal muscle relaxants for spasticity and musculoskeletal conditions: a systematic review. J Pain Symptom Manage. 2004;28:140–175. doi: 10.1016/j.jpainsymman.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Milla PJ, Jackson AD. A controlled trial of baclofen in children with cerebral palsy. The Journal of international medical research. 1977;5(6):398–404. doi: 10.1177/030006057300100203. [DOI] [PubMed] [Google Scholar]
- 5.Kearns GL, Abdel-Rahman SM, Alander SW, Blowey DL, Leeder JS, Kauffman RE. The impact of ontogeny on drug disposition and action. New Engl J Med. 2003;349:1157–1167. doi: 10.1056/NEJMra035092. [DOI] [PubMed] [Google Scholar]
- 6.He Y, Brunstrom-Hernandez JE, Thio LL, et al. Population pharmacokinetics of oral baclofen in pediatric patients with cerebral palsy. The Journal of pediatrics. 2014 May;164(5):1181–1188.e1188. doi: 10.1016/j.jpeds.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International HapMap C. The International HapMap Project. Nature. 2003 Dec 18;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 8.Ross CJ, Katzov-Eckert H, Dube MP, et al. Genetic variants in TPMT and COMT are associated with hearing loss in children receiving cisplatin chemotherapy. Nat Genet. 2009 Dec;41(12):1345–1349. doi: 10.1038/ng.478. [DOI] [PubMed] [Google Scholar]
- 9.Visscher H, Ross CJ, Rassekh SR, et al. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol. 2012 May 1;30(13):1422–1428. doi: 10.1200/JCO.2010.34.3467. [DOI] [PubMed] [Google Scholar]
- 10.Aminkeng F, Bhavsar AP, Visscher H, et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nature genetics. 2015 Sep;47(9):1079–1084. doi: 10.1038/ng.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pussegoda K, Ross CJ, Visscher H, et al. Replication of TPMT and ABCC3 genetic variants highly associated with cisplatin-induced hearing loss in children. Clinical pharmacology and therapeutics. 2013 Aug;94(2):243–251. doi: 10.1038/clpt.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006 Jul;86(3):849–899. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- 13.Flagg TP, Enkvetchakul D, Koster JC, Nichols CG. Muscle KATP channels: recent insights to energy sensing and myoprotection. Physiol Rev. 2010 Jul;90(3):799–829. doi: 10.1152/physrev.00027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bienengraeber M, Olson TM, Selivanov VA, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004 Apr;36(4):382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Bon BW, Gilissen C, Grange DK, et al. Cantu syndrome is caused by mutations in ABCC9. Am J Hum Genet. 2012 Jun 8;90(6):1094–1101. doi: 10.1016/j.ajhg.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis JA, Lamantia A, Chavez R, Scurrah KJ, Nichols CG, Harrap SB. Genes controlling postural changes in blood pressure: comprehensive association analysis of ATP-sensitive potassium channel genes KCNJ8 and ABCC9. Physiol Genomics. 2010 Feb 4;40(3):184–188. doi: 10.1152/physiolgenomics.00173.2009. [DOI] [PubMed] [Google Scholar]
- 17.Zhou SF, Wang LL, Di YM, et al. Substrates and inhibitors of human multidrug resistance associated proteins and the implications in drug development. Curr Med Chem. 2008;15(20):1981–2039. doi: 10.2174/092986708785132870. [DOI] [PubMed] [Google Scholar]
- 18.Hartz P. ATP-Binding Cassette, Subfamily C, Member 12: ABCC12. [Accessed February 22, 2017];2017 https://www.omim.org/entry/607041.
- 19.Zandi PP, Belmonte PL, Willour VL, et al. Association Study of Wnt Signaling Pathway Genes in Bipolar Disorder. Arch Gen Psychiatry. 2008;65:785–793. doi: 10.1001/archpsyc.65.7.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson KM, Christensen E, Jakobs C, et al. The clinical phenotype of succinic semialdehyde dehydrogenase deficiency (4-hydroxybutyric aciduria): case reports of 23 new patients. Pediatrics. 1997 Apr;99(4):567–574. doi: 10.1542/peds.99.4.567. [DOI] [PubMed] [Google Scholar]
- 21.Quality Standards Subcommittee of the American Academy of N, the Practice Committee of the Child Neurology S. Delgado MR, et al. Practice parameter: pharmacologic treatment of spasticity in children and adolescents with cerebral palsy (an evidence-based review): report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology. 2010 Jan 26;74(4):336–343. doi: 10.1212/WNL.0b013e3181cbcd2f. [DOI] [PMC free article] [PubMed] [Google Scholar]