Abstract
The cancer-risk-associated rs6983267 single nucleotide polymorphism (SNP) and the accompanying long noncoding RNA CCAT2 in the highly amplified 8q24.21 region have been implicated in cancer predisposition, although causality has not been established. Here, using allele-specific CCAT2 transgenic mice, we demonstrate that CCAT2 overexpression leads to spontaneous myeloid malignancies. We further identified that CCAT2 is overexpressed in bone marrow and peripheral blood of myelodysplastic/myeloproliferative neoplasms (MDS/MPN) patients. CCAT2 induces global deregulation of gene expression by down-regulating EZH2 in vitro and in vivo in an allele-specific manner. We also identified a novel non-APOBEC, non-ADAR, RNA editing at the SNP locus in MDS/MPN patients and CCAT2-transgenic mice. The RNA transcribed from the SNP locus in malignant hematopoietic cells have different allelic composition from the corresponding genomic DNA, a phenomenon rarely observed in normal cells. Our findings provide fundamental insights into the functional role of rs6983267 SNP and CCAT2 in myeloid malignancies.
The cancer genome shows a remarkable degree of variation, as revealed by the pervasive prevalence of copy number variations, amplifications, inversions, rearrangements, and single nucleotide polymorphisms (SNPs) (Stratton et al. 2009). Most SNPs of interest are found in the noncoding regions (Freedman et al. 2011), and because three-quarters of the human genome is transcribed (Djebali et al. 2012), these SNPs could be sites of active transcription of noncoding RNAs. The rs6983267(G/T) SNP in the 8q24.21 region is one such actively transcribed SNP that confers an increased risk of colon, prostate, breast, and bladder cancers (Haiman et al. 2007; Tomlinson et al. 2007; Zanke et al. 2007; Ghoussaini et al. 2008).
Notably, the long noncoding RNA (lncRNA) Colon Cancer Associated Transcript 2 (CCAT2) maps to this SNP and is transcribed into 1.7-kb RNAs containing either the G (CCAT2-G) or T (CCAT2-T) nucleotide at base pair 662 (Ling et al. 2013). Variation in this single nucleotide results in an altered secondary structure of the RNA at the SNP locus, accounting for the distinct functional roles of the two RNAs. In colon cancer, CCAT2 alleles can bind the two subunits of the Cleavage Factor I (CFIm) complex with distinct affinities, thus regulating the alternative splicing of glutaminase (GLS) and consequently reprogramming glutamine metabolism (Redis et al. 2016). CCAT2 transcript is overexpressed in microsatellite-stable (MSS) colon cancer, breast cancer, gastric cancer, esophageal squamous cell carcinoma and non-small cell lung adenocarcinomas (Ling et al. 2013; Redis et al. 2013; Qiu et al. 2014; Cai et al. 2015; Wang et al. 2015a,b; Zhang et al. 2015) and was shown to induce chromosomal instability and metastases in colon cancer by increasing MYC expression (Ling et al. 2013). Although these studies support an oncogenic role of rs6983267 SNP and CCAT2, a deeper investigation is warranted to conclusively determine whether CCAT2 plays a causal role in vivo in tumor initiation and if the G/T alleles have functional consequences on the CCAT2-induced phenotype. In this study, we undertook a comprehensive approach using two genetically engineered in vivo models to determine (a) the tissue/organ that is most sensitive to CCAT2 overexpression, (b) if CCAT2 overexpression alone is enough to induce spontaneous tumorigenesis in vivo, and finally, (c) if the G/T SNP variation contributes to the function of CCAT2.
Results
CCAT2 mice develop spontaneous myeloid malignancies
In order to study the role of CCAT2 and its specific alleles in the regulation of cellular processes, we generated two transgenic mouse models overexpressing allele-specific CCAT2 lncRNA. A transcript of 1.7-kb human CCAT2 cDNA (CCAT2-G or CCAT2-T) was expressed under the CAG promoter in C57BL/6N background (Fig. 1A). Real-time qPCR (RT-qPCR), end-point PCR, and in situ hybridization (ISH) confirmed overexpression of human CCAT2 in these mice (using human-specific primers and probes) (Supplemental Fig. S1; Supplemental Table S1A). The ∼1.7-kb transcript has ∼82% homology between human and mice. As a result, the CCAT2 primers used to detect the ∼1.7-kb transcript could also potentially detect minimal basal murine CCAT2 transcription. However, the CCAT2 primers used in the following experiments detect a shorter transcript surrounding the SNP region that is human specific. The CCAT2 overexpression level mimicked the clinically relevant CCAT2 expression reported in several malignancies (Ling et al. 2013; Redis et al. 2013; Qiu et al. 2014; Cai et al. 2015; Wang et al. 2015a,b; Zhang et al. 2015).
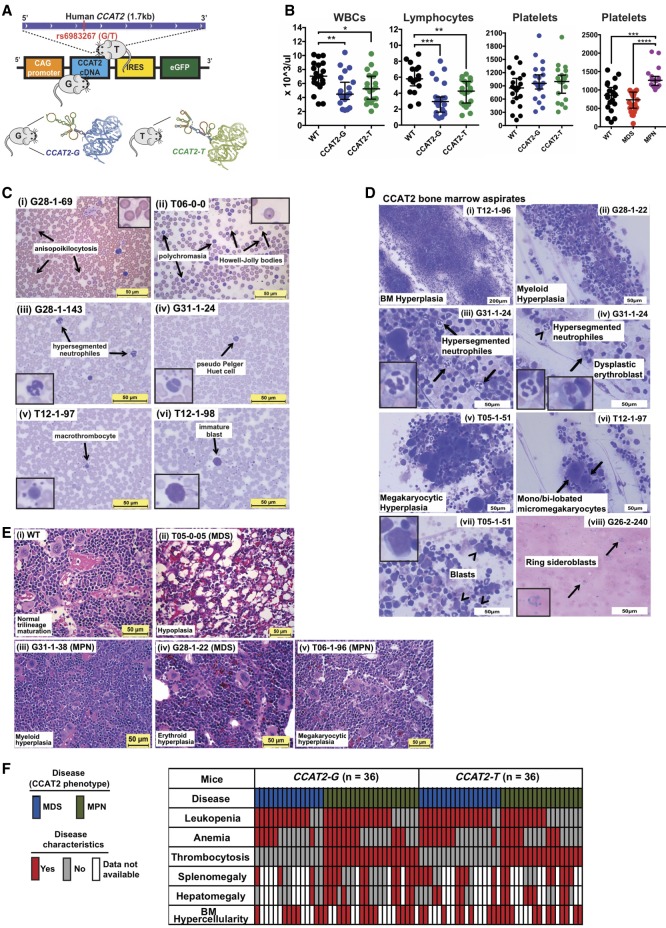
Figure 1.
CCAT2 mice display bone marrow insufficiency with clinical features of myeloid malignancies. (A) Schematic of CCAT2-plasmid cassette inserted into the mouse genome using pronuclear injection. A cartoon depicting the allele-specific secondary structure of 1.7-kb CCAT2 transcripts, which differ mostly at the ∼500 bp region surrounding the rs6983267(G/T) SNP, is shown at the bottom of panel A. (B) Peripheral blood counts of WT, CCAT2-G, and CCAT2-T mice. (C) Representative images of aberrant circulating peripheral blood cells in CCAT2-G and CCAT2-T mice. Arrows indicate the aberrations described. Images in the inset are 40× magnified aberrant cells. (D) Representative images of Hema III-stained aberrant cells present in bone marrow smears of CCAT2-G and CCAT2-T mice. Arrows and arrowheads indicate the aberrations described. Images in the inset are 40× magnified aberrant cells. (E) Histologic sections of bone marrow (from femur) with H&E staining; 40× magnified. Normal bone marrow from WT mice (i) in comparison with myeloid hyperplasic (ii), erythroid hyperplasic (iii), and megakaryocytic hyperplasic (iv) bone marrow from CCAT2-G and CCAT2-T mice. (F) The incidence of myelodysplastic and myeloproliferative clinicopathological characteristics in CCAT2-G and CCAT2-T mice are shown. Data are represented as median values ±95% confidence interval. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001.
Within 7–9 mo of age, CCAT2-G mice (n = 20 from three different founders) and CCAT2-T mice (n = 20 from two different founders) showed clinical signs of an aberrant hematological phenotype compared to age- and sex-matched WT littermates (n = 20). These mice exhibited massive leukopenia (reduction in white blood cells or WBCs) and lymphocytopenia (decreased number of lymphocytes) (Fig. 1B). Mild anemia (reduction in red blood cells or RBCs) was also displayed in 47% of mice, and 54% showed thrombocytosis (increase in platelets) (Fig. 1B; Supplemental Fig. S2A). Moreover, CCAT2-G and CCAT2-T mice showed a significant increase in the number of large unstained cells (LUCs), which are large atypical lymphocytes or blast cells, in their peripheral blood (PB) (Supplemental Fig. S2B). Consistent with complete blood counts, morphological analysis of CCAT2-G and CCAT2-T PB smears (n = 10 for each) by Hema III staining revealed the presence of several aberrant circulating blood cells, including polychromatic RBCs, Howell-Jolly bodies, hypersegmented neutrophils, pseudo Pelger Huet cells, and macrothrombocytes (Fig. 1C). Bone marrow (BM) aspirates from CCAT2-G and CCAT2-T mice (n = 10 for each) also exhibited consistent multilineage proliferative and dysplastic changes (Fig. 1D). Hematoxylin and eosin (H&E) staining of CCAT2-G and CCAT2-T mice BM sections (n = 15 for each allele) showed significantly increased BM cellularity in one or multiple cell lineages in comparison with WT mice (n = 15) (Fig. 1E; Supplemental Fig. S2C). Gross examination and histological diagnosis confirmed marked extramedullary hematopoiesis (EMH) in the spleen and liver of CCAT2 mice (P < 0.01), causing splenomegaly and hepatomegaly (Supplemental Fig. S2D,E). These mice also showed reduced iron deposition in their BM (Supplemental Fig. S2F). Periodic Acid-Schiff (PAS), myeloperoxidase (MPO), butyrate, reticulin, and trichrome stainings of the BM sections did not show any significant aberrations or myelofibrosis in these mice (Supplemental Fig. S2G–K). Collectively, because the features exhibited by CCAT2-G and CCAT2-T mice were characteristic hallmarks of myelodysplastic/myeloproliferative neoplasms (MDS/MPN) (Tefferi and Vardiman 2009; Bejar and Steensma 2014), we concluded that these mice developed a phenotype resembling the clinical manifestation of myeloid malignancies in humans (Fig. 1F). The incidence of MDS/MPN was similar in both CCAT2-G and CCAT2-T mice, implying that both alleles are equally important and single-handedly sufficient in the development of MDS/MPN (Fig. 1F; Supplemental Fig. S2L), and CCAT2 overexpression is the primary driver in MDS/MPN initiation.
CCAT2 induces genomic instability in vivo
Because CCAT2 induces genomic instability and aneuploidy in microsatellite-stable colon cancer (Ling et al. 2013) and BM cells of MDS/MPN patients are often genomically unstable, we evaluated metaphase spreads from BM cells of 8- to 9-mo-old WT (n = 4) and CCAT2-G and CCAT2-T (n = 6 each) mice for abnormal karyotypes. The BM cells of CCAT2-G and CCAT2-T mice had significantly higher frequency of structural cytogenetic aberrations, especially chromosomal breaks, than those of the WT mice (P < 0.05) (Supplemental Fig. S3A). These results showed that widespread global genomic instability and consequent dysregulation of hematopoiesis by CCAT2 might be a key event in the initiation of MDS/MPN. Ki67 proliferation analysis demonstrated that BM of CCAT2-G and CCAT2-T mice presented significantly more Ki67-positive cells than WT BM (n = 3 for each group, P < 0.001) (Supplemental Fig. S3B). Similarly, the rate of apoptosis was also significantly higher in these BM (n = 3 for each group, P < 0.01) (Supplemental Fig. S3C). These data suggested a prominent dysregulation of hematopoietic cell maturation and maintenance in CCAT2 mice, which might play an important role in BM failure.
CCAT2 mice display hematopoietic stem cell exhaustion
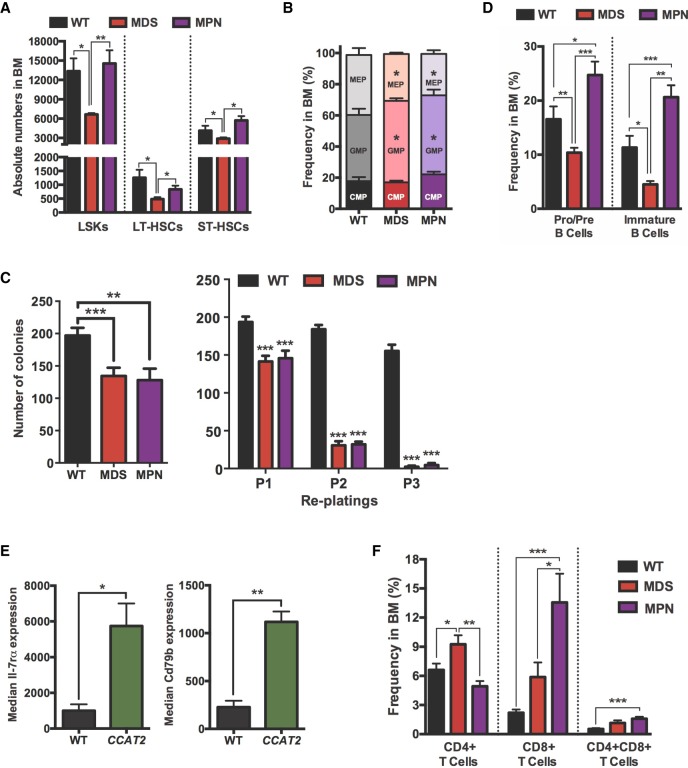
In order to characterize the hematopoietic stem cell (HSC) population in these mice, we performed flow cytometry on BM cells from CCAT2-G, CCAT2-T, and WT mice (n = 8 for each group). The MDS-like and MPN-like mice had significantly different HSC populations in their BM. The proportion of LSK (Lin−Sca1+cKit+) cells was significantly lower in MDS-like CCAT2 mice than in MPN-like CCAT2 and WT mice (Fig. 2A). Additionally, the percentages of long-term HSCs (LT-HSCs) and short-term HSCs (ST-HSCs) were also significantly lower only in MDS-like CCAT2 mice (Fig. 2A). Further analysis of myeloid-committed progenitor cells revealed a significant expansion of the granulocytic–monocytic progenitor (GMP) cell compartment, whereas the megakaryocytic–erythroid progenitor (MEP) population was reduced (P < 0.05) (Fig. 2B). In vitro HSC colony formation assay demonstrated a significant decrease in colony-forming capabilities of HSCs from both MDS-like and MPN-like CCAT2 mice (P < 0.01 and P < 0.001, respectively, in Fig. 2C, left). Serial replating assay of HSCs from MDS-like and MPN-like CCAT2 mice revealed a remarkable decrease in their repopulating efficiency in vitro compared to WT HSCs (P < 0.001) (Fig. 2C, right). These data confirmed that the self-renewal and differentiation efficiency of HSCs was significantly diminished in CCAT2 mice.
Figure 2.
Bone marrow cells of CCAT2 mice display exhaustion of hematopoietic stem cells. (A) Flow cytometry analysis of hematopoietic stem cells in MDS- and MPN-like CCAT2 mice compared to WT mice. Cells analyzed include LSK (Lin−Sca1+cKit+) cells; long-term HSCs (LT-HSCs, defined by Lin−cKit+Sca1+CD34loCD135lo population); and short-term HSCs (ST-HSCs, defined by Lin−cKit+Sca1+CD34hiCD135lo population). (B) Flow cytometric analysis of hematopoietic progenitor cells in MDS- and MPN-like CCAT2 mice compared to WT mice. Cells analyzed include common myeloid progenitors (CMPs, Lin−cKit+Sca1−/loCD34+FcγRlo population); granulocyte–macrophage progenitors (GMPs, Lin−cKit+Sca1−CD34+FcγR+ population) and megakaryocyte–erythroid progenitors (MEPs, Lin−cKit+Sca1−CD34−FcγR− population). (C) In vitro colony formation assay (left) and in vitro serial replating analysis (right) using bone marrow cells of WT, MDS-, and MPN-like CCAT2 mice. Data are average of three independent experiments done in triplicates. (D,E) Flow cytometry analysis of B cells in different developmental phases (D) and expression of developmental markers Il-7rα and CD79b in pro–pre B cells (E) of CCAT2 mice compared to WT mice. (F) Flow cytometric analysis of T cells in different developmental phases in CCAT2-G and CCAT2-T mice compared to WT mice. Data are represented as mean values ±SEM. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Among lineage-positive cells, the percentage of early-stage (pro–pre) and immature B cells was significantly reduced in MDS-like CCAT2 mice compared to WT and MPN-like mice (Fig. 2D). This correlated with HSC exhaustion and lower peripheral WBC levels in MDS-like CCAT2 mice. Conversely, MPN-like CCAT2 mice displayed a significant increase in early-stage and immature B cells, suggesting a block in B-cell differentiation (Fig. 2D). Further analysis of early-stage (pro–pre) and immature B cells revealed an increase in the expression of Il-7 receptor alpha (Il-7rα) and CD79b markers that play an important role in B-cell development (Fig. 2E). Dysregulation of these genes might help explain the B-cell aberrations exhibited by CCAT2 mice. Additionally, we noted a distinct infiltration of activated CD8+ T cells in the BM of these mice (Fig. 2F). However, no distinct dysregulation of developmental markers (including CD44, CD62l, CD25, CD49b, CD223, CD357, and Folate Receptor 4) was identified in immature T cells. Collectively, the flow cytometry data highlighted two distinct phenotypes (MDS-like and MPN-like) induced in CCAT2 mice, suggesting that CCAT2 plays an important role in regulating the HSC pool.
CCAT2-induced myeloid malignancies show age-dependent progression
The blood counts, BM histology, and cytology analysis of CCAT2-G and CCAT2-T mice at different ages (young: 4–6 mo; old: 15–18 mo) detected an increased incidence of MPN with age. Early signs of mild hyperplasia were noted in the BM of young mice, which corroborated with mild cytopenias observed in their PB (n = 8 for each group) (Supplemental Fig. S4A,B). Conversely, PB counts, H&E staining, and histology showed widespread hyperplasia, cytopenia, and splenic EMH in old mice (n = 8 for each group) (Supplemental Fig. S4C,D). CyTOF mass cytometry on BM cells of young mice (n = 3 for each group) showed mild expansion of myeloid lineage cells and reduction of erythroid cells in CCAT2 mice compared to age-matched WT mice (Supplemental Fig. S4E). In addition, a threefold increase in CD34+ blast cells was also observed in the BM of these mice (Supplemental Fig. S4F), suggesting that CCAT2 induced significant HSC dysfunction at an early age in these mice. Similar incidence of myeloid expansion and blast cells was also observed in old CCAT2 mice compared to age-matched WT mice (n = 3 for each group, P < 0.05). A significant age-dependent increase in immature/progenitor cells was observed in the BM of CCAT2 mice compared to WT mice (Supplemental Fig. S4G), indicating inhibition of HSC maturation. In addition, we observed a gradual progression to MPN-like phenotype with age, with 95% of old mice displaying MPN-like symptoms (Supplemental Fig. S4H). However, no difference was observed between the overall survival of WT, CCAT2-G, and CCAT2-T mice (n = 65 until the age of 24 mo, P = 0.424). Taken together, these results indicated that CCAT2 induced gradual HSC exhaustion and dysfunction, resulting in an age-dependent progression to MPN, but no progression to acute leukemia, characteristics consistent with low-risk MDS/MPN in humans.
CCAT2-induced myeloid malignancies are transplantable
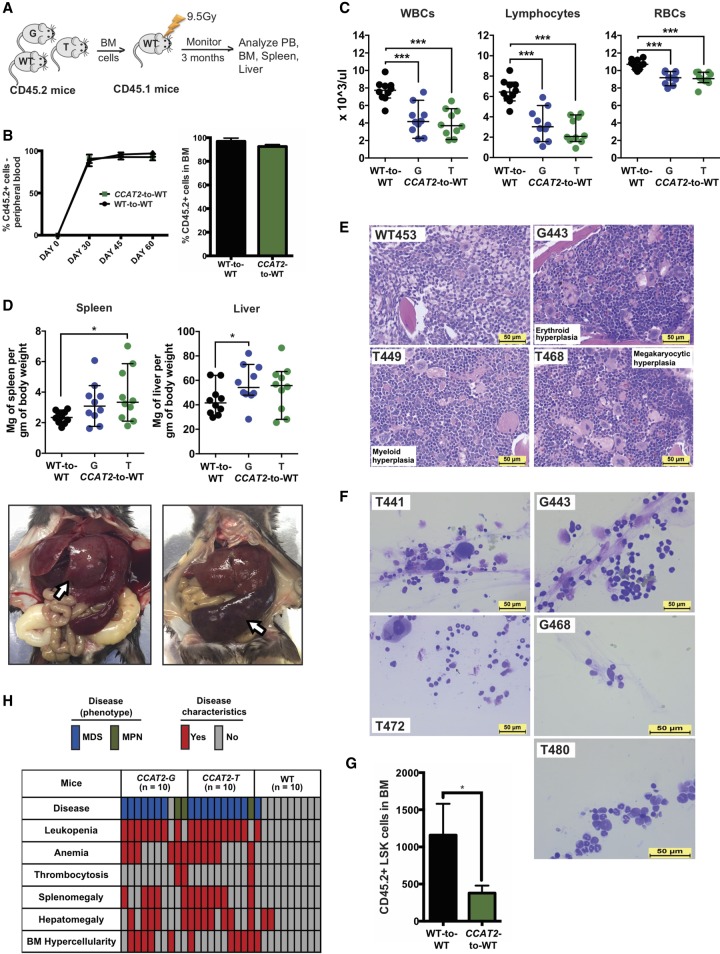
To test if the CCAT2-induced myeloid malignancies were caused by a primary defect in the BM cells or by a dysregulated microenvironment, we performed transplantation experiments. Lethally irradiated 3-mo-old CD45.1 WT mice (congenic to CD45.2 C57Bl6 mice used to generate CCAT2 transgenic mice) were reconstituted with BM cells from 9-mo-old CCAT2-G and CCAT2-T mice with MDS or MPN (CCAT2-to-WT transplantation) or WT mice (WT-to-WT transplantation) (n = 10 for each group) (Fig. 3A). The CD45.1 recipient mice showed full reconstitution of the donor CD45.2 BM cells 1 mo post-transplantation (Fig. 3B). Within 3 mo of transplantation, the CCAT2-to-WT group mice developed MDS/MPN features. PB showed progressive leukopenia, lymphocytopenia, and anemia (Fig. 3C). Necropsies demonstrated splenomegaly and hepatomegaly in the CCAT2-to-WT group, but not in the WT-to-WT group (Fig. 3D). CCAT2-to-WT group BM was dysplastic and hyperproliferative for one or multiple cell lineages in comparison with WT-to-WT mice (Fig. 3E). Additionally, BM aspirates from CCAT2-to-WT group exhibited consistent multilineage dysplastic changes (n = 5 mice for each group) (Fig. 3F). Only one of the WT-to-WT controls showed signs of early-stage MDS. Finally, flow cytometry analysis confirmed exhaustion of HSCs in CCAT2-to-WT mice compared to WT-to-WT mice (Fig. 3G). Collectively, 16/20 (80%) CCAT2-to-WT mice developed MDS, whereas 3/20 (15%) developed MPN (Fig. 3H). These findings confirmed that the CCAT2-to-WT group mice presented dysplastic changes identical to CCAT2-G and CCAT2-T mice and indicated that CCAT2 overexpressing myelodysplastic BM cells are sufficient to initiate MDS/MPN in radiation conditioned WT mice.
Figure 3.
CCAT2-induced myeloid malignancies are transplantable. (A) Schematic of the transplantation experiments. (B) Flow cytometric analysis to identify proportion of CD45.2+ cells in peripheral blood and bone marrow of CCAT2-to-WT and WT-to-WT groups. (C) Peripheral blood counts (CD45.2+) of WT-to-WT and CCAT2-to-WT groups mice. Data are represented as mean values ± SEM. (D) Spleen-to-body-weight ratio (left) and liver-to-body-weight ratio (right) of WT-to-WT and CCAT2-to-WT groups (upper). Representative images of spleen and liver from CCAT2-to-WT mice are shown below. (E) Histologic sections of bone marrow (from femur) with H&E staining of WT-to-WT and CCAT2-to-WT groups; 40× magnified. (F) Representative images of Hema III-stained aberrant cells present in bone marrow smears of CCAT2-to-WT mice. (G) Flow cytometric analysis of hematopoietic stem cells in CCAT2-to-WT mice compared to WT-to-WT mice. Cells analyzed include LSK (Lin-Sca1+cKit+) cells. (H) Incidence of myelodysplastic and myeloproliferative clinicopathological characteristics in CCAT2-to-WT and WT-to-WT mice are shown. Data are represented as median values ± 95% confidence interval. (*) P < 0.05; (***) P < 0.001.
CCAT2 is overexpressed in MDS patients
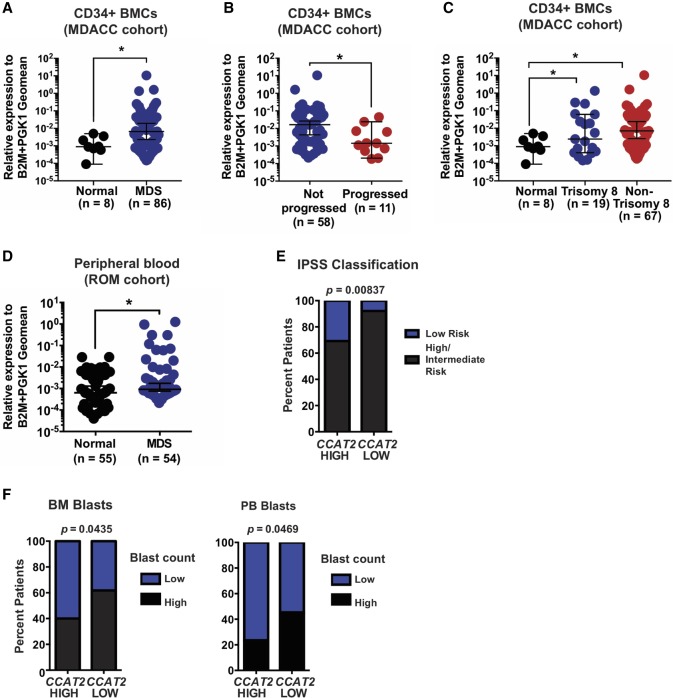
To assess the relevance of these findings in humans, we examined CCAT2 expression in CD34+ BM cells (MDACC cohort, n = 86) and PB mononuclear cells (ROM cohort, n = 54) of MDS patients and age- and sex-matched healthy individuals (n = 8 CD34+ BM samples and n = 55 PB mononuclear cell samples). We identified significantly higher CCAT2 expression in MDS patients (P < 0.05) (Fig. 4A). CCAT2 expression was also significantly higher in patients with stable MDS compared to patients that presented with MDS at diagnosis but eventually succumbed to AML (P < 0.05) (Fig. 4B). Further, CCAT2 expression in patients with Trisomy 8, who have an extra copy of Chromosome 8 (Paulsson and Johansson 2007), was similar to CCAT2 expression in patients with other genetic abnormalities (P < 0.05) (Fig. 4C), implying that an extra Chromosome 8 is not a major factor promoting CCAT2 overexpression in MDS patients. CCAT2 expression levels were also significantly higher in the PB mononuclear cells (MNCs) of MDS patients when compared to normal healthy individuals (ROM cohort, P < 0.05) (Fig. 4D). In addition, high CCAT2 expression levels correlated with low risk (P < 0.01) (Fig. 4E) and low percentage of BM and PB blasts in MDS patients (P < 0.05) (Fig. 4F). Collectively, MDS patient results validated our transgenic mice data that CCAT2 is important in MDS, and further study with more patient data sets will be required to understand the relationship between CCAT2 expression and MDS progression.
Figure 4.
CCAT2 is overexpressed in MDS patients. (A,B) CCAT2 expression in CD34+ bone marrow cells of MDS patients and healthy individuals. (A) Comparison between healthy individuals and MDS patients. (B) Comparison between MDS patients who progressed and those who did not progress to secondary AML. (C) CCAT2 expression in CD34+ bone marrow cells of MDS patients with Trisomy 8, other genetic abnormalities, and healthy individuals. (D) CCAT2 expression in peripheral blood of MDS patients and healthy individuals. (E) Correlation between CCAT2 expression in MDS patients and their risk category (classified according to IPSS risk classification). (F) Correlation between CCAT2 expression and percent blasts in bone marrow and peripheral blood of MDS patients. Data are represented as median values ±95% confidence interval. (*) P < 0.05.
CCAT2 exhibits startling DNA-to-RNA allelic imbalances at the SNP locus in MDS
Because rs6983267 SNP has been linked to cancer risk, we next sought to determine the correlation between CCAT2 overexpression and allelic composition at the SNP locus in MDS patients. For this purpose, we performed two independent assays, Sanger sequencing and allele-specific restriction enzyme digestion (using Tsp45I enzyme that cuts only CCAT2-T) (Supplemental Fig. S5A,B) on PCR-amplified region surrounding the rs6983267 SNP (using primers CCAT2_2F+2R in Supplemental Table S1A) in both genomic DNA (gDNA) and cDNA from CD34+ BM cells and PB MNCs of these patients. We identified a normal distribution of allelic frequencies in the gDNA of this population (GG = 0.30, GT = 0.55, and TT = 0.15, n = 161), which matches the natural allelic distribution of this SNP in normal population (NCBI SNP database). We found that the gDNA and cDNA sequences from the same patient were not an exact match at the SNP locus (Fig. 5A,i–ii). Although the gDNA of only 52% patients in the MDACC cohort (CD34+ BM cells) was heterozygous at the SNP locus, 74% of these patients expressed both CCAT2 alleles at the RNA level (CCAT2-GT, P = 0.003) (Fig. 5A,i; Supplemental Fig. S5C). We also observed this phenomenon in the PB MNCs of MDS patients (ROM cohort) (Fig. 5A,ii; Supplemental Fig. S5D). In this cohort, cDNA of 87% patients was heterozygous at the SNP locus compared to only 60% patients that were heterozygous at the gDNA (P = 0.002) (Fig. 5A,ii). These discrepancies between DNA and RNA were only observed at this specific locus in CCAT2. In addition, we never identified nonphysiological alleles (C or A) at this locus. We concluded that this phenomenon of DNA-to-RNA allelic imbalances represented a novel non-APOBEC, non-ADAR, SNP rs6983267-specific RNA editing event. Only samples confirmed by both Sanger sequencing and restriction digestion were labeled as rs6983267-specific RNA editing (RE) positive (RE+) (Supplemental Fig. S5A). In addition, we used various types of reverse transcriptases and polymerases to confirm this was not a technique-introduced artifact (Methods).
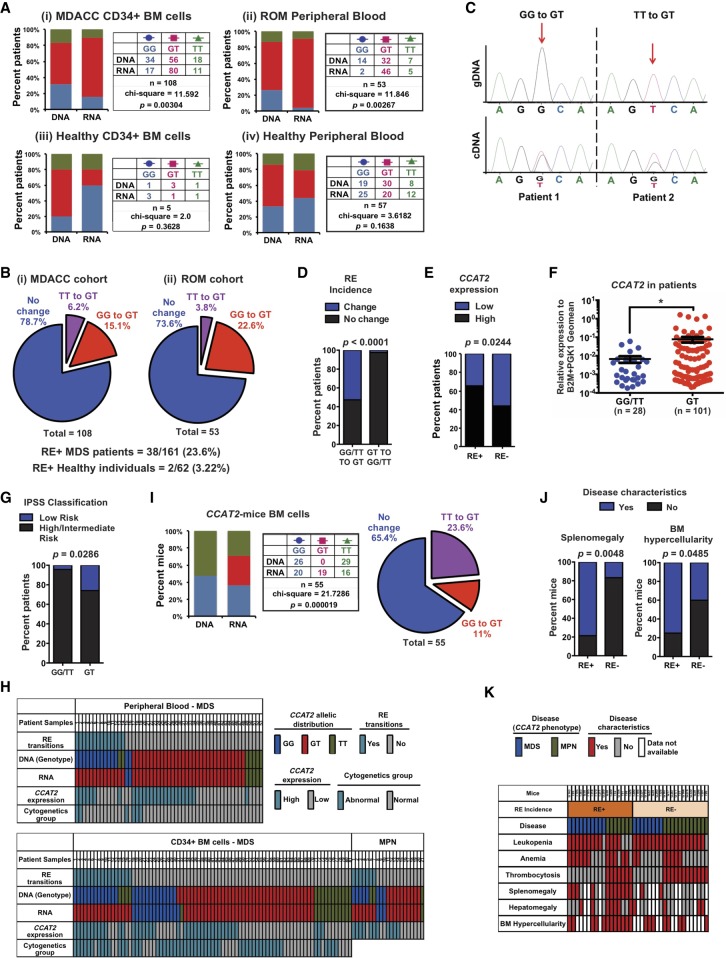
Figure 5.
MDS patients display a novel non-APOBEC, non-ADAR, SNP-specific RNA editing at rs6983267 SNP. (A) Allelic imbalance between genomic DNA and CCAT2 cDNA at the SNP locus in CD34+ bone marrow cells (MDACC cohort) (i) or peripheral blood mononuclear cells (ROM cohort) of MDS patients (ii), and in CD34+ bone marrow cells (iii) or peripheral blood (iv) of healthy individuals. (B) Pie charts depicting the rate and types of rs6983267-RE observed in CD34+ bone marrow cells or peripheral blood mononuclear cells of MDS patients compared to healthy individuals. (C) Representative examples of two rs6983267-RE+ patients by Sanger sequencing of the genomic DNA and CCAT2 cDNA at the SNP locus. (D) Correlation between incidence of rs6983267-RE according to the genotype of MDS/MPN patients. (E) Correlation between CCAT2 expression levels and incidence of rs6983267-RE in MDS/MPN patients. (F) CCAT2 expression in patients that express CCAT2-GT compared to patients that express CCAT2-GG or CCAT2-TT. (G) Correlation between patients that express CCAT2-GT, CCAT2-GG, or CCAT2-TT and their risk category (classified according to IPSS risk classification). (H) CCAT2 expression levels, incidence of rs6983267-RE, and clinical features for each MDS/MPN patient analyzed are shown. (I,J) Rate of rs6983267-RE occurrence in the bone marrow cells of CCAT2 mice. (K) The incidence of rs6983267-RE and MDS/MPN clinicopathological characteristics displayed by CCAT2 mice are shown. Data are represented as mean values ± SEM. (*) P < 0.05.
Next, to determine if rs6983267-RE was disease specific, we sequenced the gDNA and cDNA at the SNP locus in BM (n = 5) and PB (n = 57) of age- and sex-matched healthy individuals. No significant differences between gDNA and cDNA were detected in these individuals (Fig. 5A,iii–iv). The rate of rs6983267-RE incidence was much higher in MDS/MPN patients compared to healthy individuals (23.6% versus 3.2%, respectively) (Fig. 5B), suggesting that CCAT2-associated rs6983267-RE was significantly enriched in MDS/MPN patients. We then analyzed paired BM and PB samples from six MDS patients, and detected that two (33%) patients had the exact same editing in their BM and PB, suggesting that rs6983267-RE could be potentially induced in the HSC compartment first. Figure 5C shows the Sanger sequencing data for two RE+ MDS/MPN patients. rs6983267-RE predominantly induced conversion of homozygous DNA to heterozygous RNA (P < 0.0001) (Fig. 5D). Further, rs6983267-RE+ patient samples have significantly higher CCAT2 expression levels compared to rs6983267-RE-negative patients (RE−, P = 0.024) (Fig. 5E). Because the functional output of rs6983267-RE is expression of CCAT2-GT, we compared patients that expressed CCAT2-GG and CCAT2-TT to patients that expressed CCAT2-GT. CCAT2-GT patients had higher CCAT2 levels in their BM and PB compared to CCAT2-GG or CCAT2-TT patients (P < 0.05) (Fig. 5F). As expected, CCAT2-GT also correlated with low-risk MDS compared to patients with CCAT2-GG or CCAT2-TT (P = 0.028) (Fig. 5G). Figure 5H describes the rs6983267-RE profile for all patients evaluated in this study, along with their clinical characteristics. Collectively, our data identified a novel form of RNA modification prevalent in MDS/MPN patients.
CCAT2 induces rs6983267-RE in vivo
Based on our patient data, we believe CCAT2 expression was a prerequisite for rs6983267-RE; however, CCAT2 expression alone might not be enough to induce this editing. To determine if CCAT2 was important for rs6983267-RE occurrence, we analyzed the gDNA and cDNA of CCAT2 transgenic mice. We reasoned that if the human CCAT2 transcript played an active role in induction of rs6983267-RE, then the transgenic mice would also display rs6983267-RE. Analysis of the gDNA and cDNA sequences from BM and PB of CCAT2-G and CCAT2-T mice using human-specific CCAT2 primers revealed a similar incidence of rs6983267-RE in these mice (P < 0.0001) (Fig. 5I; Supplemental Fig. S5E,F), and 34% of the mice were heterozygous for CCAT2 transcript, expressing both G- and T- nucleotides. We confirmed our sequencing and restriction digestion data using digital droplet PCR (ddPCR) on two rs6983267-RE− and one rs6983267-RE+ mice samples with enough DNA and RNA to be tested by all three methods (Supplemental Fig. S5G). This was remarkable considering that the genomic cDNA inserted into these mice contained only a single nucleotide (either G or T) (Supplemental Fig. S1C). Use of human-specific CCAT2 primers eliminated the possibility that endogenous mouse CCAT2 might be confounding the data. To understand the role of editing in CCAT2-induced phenotype, we compared RE+ and RE− CCAT2-mice. Phenotypic comparison between these mice revealed that rs6983267-RE+ mice were more prone to have splenomegaly and BM hypercellularity compared to rs6983267-RE− mice (P = 0.004 and 0.048, respectively) (Fig. 5J). Figure 5K shows the detailed phenotypic comparison between rs6983267-RE+ and rs6983267-RE− mice. Spontaneous occurrence of RNA editing in CCAT2 mice implied that human CCAT2 sequence played an important role in the incidence of rs6983267-RE.
rs6983267-RE causes significant immune dysregulation in CCAT2-induced myeloid malignancies
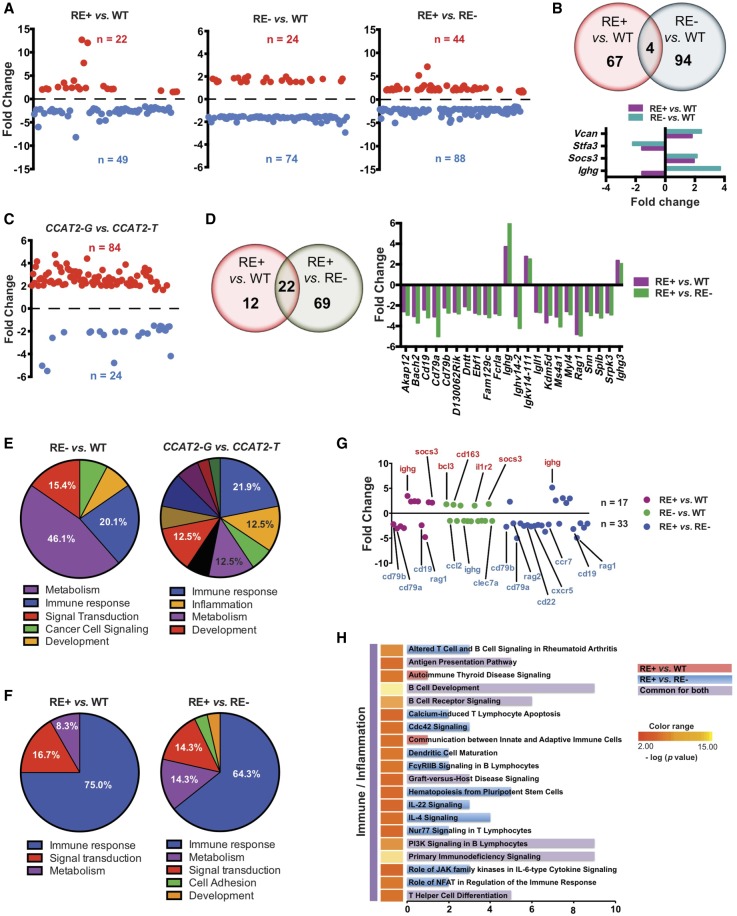
To study the global alterations induced by rs6983267-RE, we performed a genome-wide expression profiling (Affymetrix microarray) on BM cells of WT mice (n = 4), CCAT2-G and CCAT2-T mice (RE− group, n = 8), and CCAT2-GT mice (RE+ group, n = 6). We found significant expression variations in RE+ or RE− mice compared to WT mice, as well as RE+ mice compared to RE− mice (Fig. 6A). We identified Socs3, an inhibitor of JAK/STAT pathway and Ighg, a critical immune regulator, to be significantly dysregulated in both rs6983267-RE+ and rs6983267-RE− mice compared to WT mice (Fig. 6B). Significant changes were observed in RE− CCAT2-G versus RE− CCAT2-T mice (Fig. 6C), indicating that both alleles might function via different mechanisms to initiate myeloid malignancies. Genome-wide comparison of RE+ mice with RE− and WT mice revealed 22 genes that were significantly altered by rs6983267-RE (Fig. 6D). These included important immune regulators and B-cell receptor signaling pathway genes CD16, CD79a, CD79b, Igll1, and Ighg (Fig. 6D).
Figure 6.
CCAT2 induces global gene expression dysregulation in vivo. (A) Scatter plots representing genes that were significantly up-regulated (in red) and down-regulated (in blue) in bone marrow cells of rs6983267-RE+ versus WT mice, rs6983267-RE− versus WT mice, and rs6983267-RE+ versus rs6983267-RE− mice (P ≤ 0.05, fold change ≥1.5). (B) Venn diagram showing genes dysregulated in rs6983267-RE+ versus WT mice and rs6983267-RE− versus WT mice. Expression level fold changes of commonly regulated genes are shown below. (C) Scatter plots representing genes that were significantly up-regulated (in red) and down-regulated (in blue) in bone marrow cells of CCAT2-G mice compared to CCAT2-T mice. (D) Venn diagram showing genes dysregulated in rs6983267-RE+ versus WT mice and rs6983267-RE+ versus rs6983267-RE− mice. Expression level fold changes of commonly regulated genes are shown to the right. (E,F) Pie diagrams depicting the top differentially regulated pathways, grouped according to their molecular function, in bone marrow cells of rs6983267-RE− (E) or rs6983267-RE+ (F) mice as described. (G) Scatter plot of significantly aberrantly regulated immune genes in the bone marrow cells of rs6983267-RE+ or rs6983267-RE− mice as described. Most important genes are listed. (H) Top canonical pathways altered in bone marrow cells of rs6983267-RE+ mice compared to WT or rs6983267-RE− mice, grouped according to their molecular function as determined by Ingenuity Pathway Analysis.
Pathway analysis revealed that although RE− mice displayed genome-wide changes associated with diverse cellular processes (Fig. 6E), RE+ mice displayed significant dysregulation of genes involved primarily in immune regulation (Fig. 6F). We identified a significant down-regulation of important immune-related genes in these mice (Fig. 6G). This implied that although CCAT2-G and CCAT2-T independently regulated a large cohort of genes, the presence of both RNAs in rs6983267-RE+ mice led to substantial immune impairment in vivo (Fig. 6F,G). We identified pathways involved in antigen presentation, autoimmunity, immunodeficiency, B-cell receptor signaling, B-cell development, T cell differentiation, CDC42 signaling, IL22 signaling, and IL4 signaling (Fig. 6H). Several of these genes and pathways, such as immunodeficiency signaling, B-cell receptor signaling, and IL4 signaling, were previously implicated as functionally altered in MDS, MPN, and other myeloid malignancies (Pellagatti et al. 2010). In order to confirm immune dysregulation by rs6983267-RE, we performed nCounter PanCancer Immune Profiling Panel (NanoString Technologies) analysis on hematopoietic stem and progenitor cells (HSPCs) from WT (n = 4), RE+ (n = 5), and RE− (n = 3) mice. We identified significant dysregulation of important immune genes only in RE+ mice compared to WT and RE− mice (Supplemental Fig. S6A). CD22, CD19 and H2-Ob (Histocompatibility 2, O region beta locus) were overlapping immune genes that were dysregulated in both total BM and enriched HSPCs. Because we observed immune dysfunction in complete BM as well as enriched HSPCs of rs6983267-RE+ mice, we concluded that immune dysregulation could potentially be a function of RNA editing and not due to differences in the cellular composition of the BM of these mice.
At the same time, commonalities and differences in gene expression patterns were also apparent between the MDS-like and MPN-like CCAT2 mice (Supplemental Fig. S6B,C). We observed a significant increase in the number of dysregulated genes and consequent enrichment of deregulated pathways in MPN-like CCAT2 mice, which possibly reflected increased disease severity. MPN-like mice showed a significant enrichment of immune-related genes and associated pathways (Supplemental Fig. S6D), suggesting that immune deregulation is an important phenomenon in CCAT2-induced myeloid malignancies. We also analyzed the microarray data to identify dysregulated processes that might provide a hint for the rs6983267-RE mechanism. We identified several genes that play an important role in DNA–RNA binding, nucleotide biosynthesis, and transcription to be deregulated in rs6983267-RE+ mice (Supplemental Fig. S6E). These data suggested that the altered function of the transcription machinery by CCAT2 might be a potential mechanism of rs6983267-RE that needs to be further evaluated.
CCAT2 regulates EZH2 in vitro and in vivo
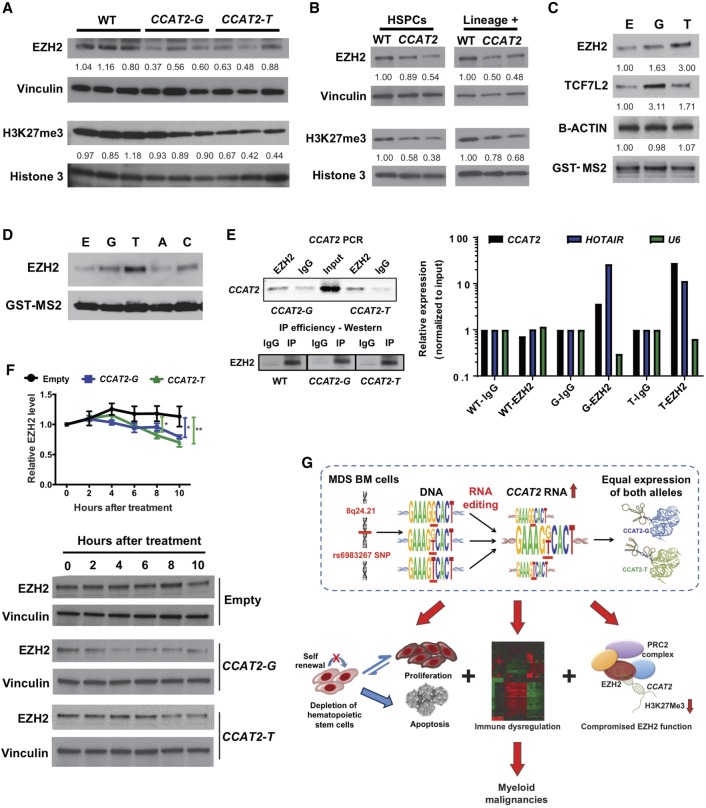
We further identified Ezh2 to be down-regulated in the BM cells of both CCAT2-G and CCAT2-T mice compared to WT littermates (Fig. 7A). A corresponding decrease in global H3K27me3 levels, an epigenetic marker for transcriptional silencing by polycomb repressive complex 2 (PRC2) was observed (Fig. 7A). Clinically, 10% of MDS patients show loss-of-function mutations in EZH2, and these mutations are associated with shorter survival (Ernst et al. 2010). EZH2 loss has also been reported to induce MDS/MPN in vivo, but mitigate its transformation to leukemia (Tanaka et al. 2012; Muto et al. 2013; Sashida et al. 2014; Mochizuki-Kashio et al. 2015). Compromised Ezh2 function in our model might potentially explain why CCAT2-G and CCAT2-T mice do not progress to AML, even after 24 mo. Based on this, we decided to further study the regulation of EZH2 by CCAT2.
Figure 7.
CCAT2 regulates EZH2 in vitro and in vivo. (A,B) Western blot analysis on complete BM cells from CCAT2-G, CCAT2-T, and WT mice (A), and hematopoietic stem and progenitor cells (HSPCs) and lineage-positive cells from CCAT2-G, CCAT2-T, and WT mice (B). (C,D) Western blot analysis for EZH2 in cell lysates from GST-tagged allele-specific CCAT2-overexpressing HEK293 cells following RNA pulldown experiment. (C) TCF7L2 was used as a positive control and B-Action as a negative control for CCAT2 interaction. (D) Pulldown analysis after overexpressing nonphysiological A and C alleles of the SNP. (E) RT-qPCR for CCAT2 expression in BM cells of CCAT2-G, CCAT2-T, and WT mice following RNA Immuno-Precipitation by EZH2 antibody (right). HOTAIR was used as a positive control and U6 as a negative control for EZH2 interaction. All expression levels were normalized to input. End-point PCR for CCAT2 and Western blot analysis showing efficiency of EZH2 RIP (left). (F) Western blot analysis to detect EZH2 levels following treatment of CCAT2-overexpressing-HEK293 cells with cycloheximide for 0, 2, 4, 6, 8, and 10 h. Upper panel shows quantification of three independent Western blot experiments performed in triplicates. (G) Schematic model of proposed mechanism of CCAT2-induced MDS/MPN phenotypes. Data are represented as mean values ±SEM. (*) P < 0.05; (**) P < 0.01.
We confirmed EZH2 down-regulation in human HEK293 and mouse MEF cell lines (Supplemental Fig. S7A). Ezh2 and H3K27me3 levels are reduced in hematopoietic stem and progenitor cells (HSPCs) as well as lineage-positive BM cells of CCAT2 mice (Fig. 7B). Gene enrichment analysis of microarray data identified a positive enrichment of EZH2-regulated targets defined in human cancers (obtained from the Molecular Signature database [MSigDB]) in the BM cells of CCAT2-G and CCAT2-T mice (hypergeometric probability distribution analysis, common CCAT2:EZH2 targets = 25, representation factor: 2.0, P < 3.471 × 10−4). Several EZH2 downstream targets were identified to be dysregulated in these mice (Supplemental Fig. S8B). Because Ezh2 mRNA levels were not altered in BM cells of CCAT2 mice (Supplemental Fig. S7C), we sought to determine if CCAT2 altered EZH2 levels post-transcriptionally. EZH2 has been reported to interact with CCAT2 as well as other lncRNAs (Tsai et al. 2010; Kotake et al. 2011; Hirata et al. 2015; Wang et al. 2016; Deng et al. 2017); we thus studied CCAT2:EZH2 direct interaction. Indeed, using RNA pulldown analysis, we discovered that EZH2 binds to both CCAT2-G and CCAT2-T in HEK293 cells (stronger interaction with CCAT2-T) (Fig. 7C). Artificial in vitro mutation of the SNP to A or C nucleotides (that never occur in the human population) further confirmed the specificity of EZH2:CCAT2 interaction by RNA pulldown analysis (Fig. 7D). RNA immunoprecipitation (RIP) analysis on the BM cells from WT, CCAT2-G, and CCAT2-T mice (n = 3) detected a significant enrichment of both CCAT2-G and CCAT2-T transcripts by real-time qPCR, validating the direct interaction of EZH2 with CCAT2 (Fig. 7E). RIP analysis after overexpressing CCAT2 and EZH2 showed a significant increase in the strength of the EZH2:CCAT2 interaction (Supplemental Fig. S7D). In addition, we confirmed EZH2:CCAT2 interaction in the SET2 MPN cell line, which is a more relevant disease model (Supplemental Fig. S7E).
To understand how CCAT2 regulated EZH2 expression, we performed cycloheximide chase assay for protein stability on HEK293 cells overexpressing CCAT2-G, CCAT2-T, or Empty (pcDNA3 control) vectors. After 10 h of cycloheximide treatment, EZH2 levels were reduced to 70% and 62% compared to the 0-h time point in the CCAT2-G and CCAT2-T clones, respectively, but no significant reduction was noted in the Empty clones (P < 0.05) (Fig. 7F). We detected a strong inverse correlation between CCAT2 expression and EZH2 protein levels in CD34+ BM of MDS patients (MDACC cohort, n = 24, r = −0.435, P = 0.033) (Supplemental Fig. S7F). Finally, we compared EZH2 protein levels in BM cells of rs6983267-RE+ and rs6983267-RE− CCAT2 mice. rs6983267-RE+ mice exhibited markedly lower levels of Ezh2 protein as well as mRNA in their BM cells (Supplemental Fig. S7G). Collectively, these data indicated that CCAT2 regulated the expression of EZH2, and the simultaneous presence of both CCAT2 RNA alleles by rs6983267-RE achieved a stronger down-regulation of EZH2, thus providing a combined advantage to the oncogenic function of CCAT2. Taken together, our data indicate that CCAT2 overexpression leads to genomic instability and rs6983267-RE, which in turn results in a genome-wide gene expression dysregulation, resulting in compromised EZH2 function and impairment of immune processes, and thus inducing MDS/MPN (Fig. 7G).
Discussion
One of the greatest challenges in the post-genome-wide association studies (GWAS) era is to understand the functional and biological significance of SNPs and derive insights that can be translated to clinical benefits (Freedman et al. 2011). Through an integrated approach, we successfully deciphered the clinical importance of cancer-risk associated rs6983267 and the accompanying CCAT2 lncRNA. Constitutive overexpression of allele-specific CCAT2 altered the global gene expression landscape in the hematopoietic cells, which ultimately initiated de novo myeloid malignancies, indicating that both alleles are important. We also identified a novel SNP-associated RNA mutation that is highly prevalent in cancer cells: the rs6983267-RE. Via this phenomenon, the aberrant malignant cells can potentially control the transcription of rs6983267 SNP locus (and potentially several other loci to be identified) and preferentially produce heterozygous transcripts from homozygous DNA. This study reveals that actively transcribed SNPs might have a more fundamental role in genetic regulation than previously understood.
We propose that rs6983267-RE is a purposeful, systematic, and precisely regulated event in human MDS/MPN.
rs6983267-RE is purposeful: We found this to be significantly more frequent in premalignant MDS/MPN syndromes compared to healthy individuals, suggesting it might be an important mechanism adopted by aberrant cells, which gives rs6983267-RE a cellular purpose. The importance of this event can be emphasized by the fact that although only 54% of the MDS patients in our cohorts were heterozygous for the SNP at the DNA level (similar to the normal distribution of the SNP alleles in the healthy human population), 78% of these patients actually expressed heterozygous CCAT2 RNA. Additionally, the presence of rs6983267-RE at the cancer-predisposing rs6983267 SNP, embedded in an oncogenic lncRNA (CCAT2), and located within the most commonly amplified 8q24 region, suggest rs6983267-RE could have important biological consequences.
rs6983267-RE are systematic and regulated: If the rs6983267-RE detected in MDS/MPN patients was random cellular noise, then the nucleotide variations would be arbitrary; namely, the nonphysiological A and C alleles (that never occur in human population) would also be identified. However, the fact that the variations always involved only the physiologically relevant G and T alleles of the SNP in 100% of studied cases suggest that rs6983267-RE is systematically regulated. This is further supported by the human–mouse conservation that we observed. We identified the RE in MDS/MPN patients as well as in the CCAT2-induced MDS/MPN mice (where the human CCAT2 cDNA was inserted randomly in the mouse genome). This implied that the CCAT2 genomic locus might act in cis to induce the rs6983267-RE and thus control the transcription at its SNP site, further emphasizing that it is a carefully regulated phenomenon.
Another important implication is the significant accumulation of both CCAT2 alleles (heterozygosity at the SNP locus), implying that both alleles are important in MDS/MPN initiation. We previously reported the allele-specific functionality of CCAT2 in colon cancer. The CCAT2 G and T alleles differentially modulate cellular metabolism by regulating the alternative splicing of GLS to preferentially induce the expression of GAC, the oncogenic isoform of GLS (Redis et al. 2016). This is further supported by our data that both the G and T alleles can spontaneously initiate stable MDS/MPN phenotype in mice. Although the rs6983267 “G” allele has been touted to be the cancer-predisposing allele, our data illustrate that both alleles are functional and, in fact, equally essential and independently sufficient for MDS/MPN. The microarray data demonstrated that both alleles function via independent pathways to induce oncogenesis. However, when present concurrently (in rs6983267-RE+ cases), it initiates a global dysregulation of immune-associated genes as well as their specific targets. Although the BM microenvironment and the inherent heterogeneity of the BM composition might also play an important role in the dysregulation of immune system, our data indicate that it can, at least partially, be attributed to CCAT2 overexpression. According to our data, while the constitutive overexpression of CCAT2-G and CCAT2-T independently induce EZH2 repression, the concurrent presence of both alleles further augments the repression of EZH2. Thus, we argue that although the CCAT2 transcript is the “active” functional entity, the specific alleles contribute to its oncogenicity by independently regulating common and distinct pathways, affording it to have multiple far-reaching targets. Additionally, because rs6983267-RE also existed in CCAT2 mice, we propose the presence of a positive feedback loop, wherein CCAT2 transcript could act in cis to regulate the occurrence of rs6983267-RE at its locus, while rs6983267-RE could control the allelic distribution of CCAT2 transcripts.
For years, scientists endorsed the central dogma of biology, which holds that the RNA transcripts used as templates for protein assembly are a perfect match to the original DNA. However, with the advent of the Human Genome Project and the discovery of noncoding RNAs, this simplistic concept has been repeatedly challenged. DNA–RNA discrepancies have been previously reported: peripheral blood mononuclear cells (Ju et al. 2011), immortalized B cells (Peng et al. 2012), at the interleukin-12 receptor β1 (IL-12Rβ1) locus in peripheral blood mononuclear cells (Turner et al. 2015), and at three specific loci in the human mitochondrial DNA from different cell types (Bar-Yaacov et al. 2013). Notably, these DNA–RNA discrepancies have only been reported in normal tissues and their incidence in human diseases is still unknown. The primary difference between these DNA–RNA discrepancies and rs6983267-RE is that to date, DNA–RNA discrepancies are identified exclusively in non-SNP regions of the human genome, whereas we discovered rs6983267-RE in one of the well-characterized SNP region in the human genome. Further studies are required to understand whether SNP-specific RE exists at other actively transcribed SNP loci and in other malignancies.
One question that arises from this study is the mechanism of rs6983267-RE occurrence. Although the classical A-to-I RNA editing relies on chemical modification of the nucleotides, this cannot be the case with rs6983267-RE because it involves variation between a purine (G) and a pyrimidine (T) nucleotide. We believe that “nucleotide switching” or “nucleotide insertion” mechanisms are at play in the rs6983267-RE-positive cases; however, further work is needed to elucidate the intricate mechanisms. Because the RNA modifications introduced by rs6983267-RE are distinct from the classic A-to-I RNA editing mechanism, we propose two potential explanations for this specific RE occurrence. First, the complexity of the SNP genomic locus might be the catalyst in rs6983267-RE occurrence: The 8q24 region is one of the most commonly amplified regions in the human genome (Beroukhim et al. 2010), the rs6983267 SNP has been reported to predispose individuals to several cancers (Haiman et al. 2007; Tomlinson et al. 2007; Zanke et al. 2007), the gDNA itself forms a large molecular loop and acts as an enhancer to Myc located about 500 kb upstream (Pomerantz et al. 2009), and the oncogenic lncRNA CCAT2 transcribed from this region also acts as an enhancer to Myc (Ling et al. 2013). The instability of the genomic region might lead to active alterations in the RNA transcription mechanism, resulting in rs6983267-RE. Second, the widely described clonal heterogeneity of MDS/MPN BM cells might cause a small population of CD34+ BM cells to be genomically diverse at the SNP locus from the majority of CD34+ BM cells. The signal generated by this small subpopulation of cells will not be detected in gDNA using Sanger sequencing and restriction digestions, resulting in rs6983267-RE. One caveat is that these proposed explanations are based on the observation of rs6983267-RE in MDS patients and might not stand true in mouse model, since the genomic complexity does not occur in the mouse model.
In conclusion, this study elucidates the biological importance of actively transcribed cancer-associated rs6983267 SNP and its accompanying lncRNA CCAT2 in MDS/MPN, a disease not yet associated with this SNP. The CCAT2 transgenic mice can serve as a robust model to study stable de novo MDS/MPN that does not progress to secondary AML and as a preclinical model for evaluating new therapies for premalignant MDS/MPN. Our mouse model also provides a unique opportunity to further explore the intricate mechanisms of transcriptional regulation by nonexonic SNPs and lncRNAs. Finally, the presence of SNP-specific RE at the cancer-predisposing rs6983267 SNP located within the most commonly amplified 8q24 region is indicative of an enigmatic genomic puzzle that warrants further investigation.
Methods
Generation of CCAT2 transgenic mice
CCAT2 transgenic mice were generated using random integration approach. A 1.7-kb human cDNA of CCAT2 expressing either the G or T allele was cloned into a vector backbone containing the CAG promoter, along with the eGFP reporter gene followed by IRES site. Pronuclear injection of the entire 4.5-kb linearized insert and generation of founder mice was performed by the MDACC Genetically Engineered Mouse Facility. The founders were mated with C57BL/6N mice. Pups were screened for presence of the transgene by PCR and Southern blot analysis on tail-extracted DNA according to standard protocols. For PCR screening, three different primer pairs were used. Pups showing positive detection by both PCR and Southern blot were identified as founders. All primers and probes used for this study are given in Supplemental Table S1A. All the protocols and experiments were conducted according to the guidelines of the MDACC Institutional Animal Care and Use Committee (IACUC).
Transplantation experiment
B6.SJL-Ptprca Pepcb/BoyJ mice (Jackson Laboratories) expressing the CD45.1 marker were used for the transplantation experiments. BM cells were extracted from 9-mo-old CCAT2 (confirmed to have MDS/MPN) or WT mice, crushed in cold 1× PBS with 2% FBS, and passed through a 40 µm filter. B6.SJL-Ptprca Pepcb/BoyJ mice 6–12 wk old were lethally irradiated (9.5 Gy) and injected with 1 × 106 BM cells from CCAT2 (CCAT2-to-WT group) or WT (WT-to-WT group) mice. Irradiated recipients were maintained on sterile water containing 0.5 g/L of enrofloxacin for 2 wk after irradiation. Blood levels were monitored for 3 mo.
Clinical samples
CD34+ cells from BM of 86 myelodysplastic syndrome (MDS) patients and eight healthy individuals were obtained from MD Anderson Cancer Center tissue bank (MDACC cohort). A second set of peripheral blood mononuclear cells from 54 MDS patients and 55 healthy volunteer samples were obtained from University of Medicine and Pharmacy Iuliu Hatieganu, Romania (ROM cohort). All samples were collected according to the institutional policies and obtained following patient's informed consent. Tissue samples were obtained from fresh surgical specimens frozen in liquid nitrogen and stored at −80°C. Samples were de-identified prior to any analyses using standard procedures. Clinical information of the patients used in this study is provided in Supplemental Table S2.
Detection of rs6983267-RE
gDNA and RNA were extracted from CD34+ BM cells and PB mononuclear cells using standard protocols as described above. Reverse transcription was performed using random hexamers with SuperScript III Reverse Transcriptase or MultiScribe Reverse Transcriptase according to the manufacturer's protocol (Thermo Fischer Scientific). End-point PCR was performed on the gDNA and cDNA from the patients with high fidelity Advantage HF PCR 2 kit (Clontech) or high fidelity Taq Polymerase (Thermo Fischer Scientific) according the manufacturer's protocol to amplify the 500-bp region encompassing the SNP. For the reaction, 50 ng DNA template was used. Primer sequences are available in Supplemental Table S1A. Products were run on 2% agarose gel to verify the amplicon size and purified using QIAquick PCR purification kit (Qiagen). The purified PCR product was used for Sanger sequencing and restriction digestion as described below. Only samples identified to be rs6983267-RE+ by both Sanger sequencing and restriction digestion were considered as rs6983267-RE+ for this study.
Sanger sequencing
Purified PCR product (20 ng/µL) was submitted for Sanger sequencing to the MD Anderson Sequencing and Microarray Core Facility. Each rs6983267-RE+ sample was sequenced twice, using forward and reverse primers. The sequencing results from gDNA and cDNA from each patient were matched individually and analyzed using SeqMan Pro (DNASTAR). Only clean sequences with no background peaks were used for rs6983267-RE analysis.
Restriction digestion
Purified PCR product (200 ng) was used for restriction digestion using Tsp45I enzyme for 1 h. The products were then run on 2% agarose gel to identify the allele present.
Gene expression profiling analyses
Total RNA was extracted from BM cells of WT, CCAT2-G, and CCAT2-T mice as described above. RNA quality was assessed using RNA 6000 Nano assay (Bioanalyzer, Agilent). The labeling and the hybridization of mRNAs were performed according to Affymetrix standard protocols. Briefly, 5 µg of total RNA was reverse transcribed with an oligo(dT) primer that has a T7 RNA polymerase promoter at the 5′ end. Second-strand synthesis was followed by cRNA production with incorporation of biotinylated ribonucleotides using the BioArray High Yield RNA Transcript Labeling Kit T3 from Enzo Life Sciences. The labeled cRNA was fragmented and hybridized to Affymetrix GeneChip Mouse Genome 230 4.0 arrays. GeneSpring GX software v.13 (Agilent Technologies) was used for probe set summarization and robust multiarray average (RMA) normalization procedures. The differentially expressed genes were selected to have a >1.5-fold change difference between the compared groups (average value), a <10% FDR using Benjamini and Hochberg corrected moderated t-test and P < 0.05. Gene lists were uploaded to Ingenuity Pathway Analysis (Qiagen) for Enrichment Analysis. EZH2 target gene list was obtained from the Molecular Signatures Database (http://software.broadinstitute. org/gsea/msigdb). The significance of overlap between CCAT2-regulated genes and EZH2 targets was performed using hypergeometric probability distribution analysis.
EZH2 expression analysis
Western blot staining for EZH2, H3K27me3, Histone 3, Vinculin and GAPDH, and immunohistochemistry for EZH2 were performed on mouse tissues, cell lines, and stable clones as previously described (Ling et al. 2013). The antibodies used for the analyses are listed in Supplemental Table S1A. RNA pulldown and RNA immunoprecipitation experiments were performed on CCAT2-overexpressing cell lines as previously described (Redis et al. 2016). For EZH2 protein stability analysis, cellular protein degradation was monitored after blockage of de novo protein synthesis via cycloheximide treatment. For this purpose, HEK293 cells transfected with CCAT2-G, CCAT2-T, or Empty vector (E) were treated with 50 µg/mL final concentration of cycloheximide. Cells were harvested after cycloheximide treatment at indicated time points followed by protein extraction, SDS-PAGE, and Western blot analysis to visualize protein degradation of EZH2.
Statistical analysis
The statistical analyses were performed using GraphPad Prism. The Shapiro–Wilk test was applied to determine whether data followed a normal distribution. Accordingly, the t-test or the nonparametric Mann-Whitney–Wilcoxon test was applied to assess the relationship between CCAT2 expression levels and clinical parameters, or the experimental variables. CCAT2 RNA levels were analyzed as a log-transformed continuous variable or as a dichotomized variable based on the median level of CCAT2 RNA, and the log-rank test was used to evaluate differences. All data are presented as the mean values ± the standard error of the mean or median value with 95% confidence interval (as specified) from at least three independent experiments. Two-sided t-tests were used to test the relationships between the means of data sets, and P-values indicate the probability of the means compared, being equal with *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Additional information on hematological measurements, peripheral blood morphology, peripheral blood and bone marrow histology, in situ hybridization, genomic instability analysis, flow cytometry and mass cytometry analysis, RNA extraction, reverse transcription, and real-time qPCR performed on these mice are presented in Supplemental Materials.
Data access
Microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE106581. The Sanger sequencing data have been submitted to NCBI Trace Archive (https://trace.ncbi.nlm.nih.gov/Traces/trace.cgi) under Trace Identifier (TI) numbers 2344290315–2344290801.
Supplementary Material
Acknowledgments
Dr. Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr. Calin's laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and 1 R01 CA182905-01, the UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), Aim at Melanoma Foundation and the Miriam and Jim Mulva research funds, the Brain SPORE (2P50CA127001), the Center for Radiation Oncology Research Project, the Center for Cancer Epigenetics Pilot project, a 2014 Knowledge GAP MDACC grant, a CLL Moonshot project, the UT MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment, a SINF grant in colon cancer, the Laura and John Arnold Foundation, the RGK Foundation and the Estate of C.G. Johnson, Jr. M.J.Y. is supported in part by NIH/NCI R01 CA164346, 1R01 CA200703-01A1 and Leukemia SPORE Grant P50 CA100632, CPRIT RP140402, and IRG and Sister Institution Network fund of UT MD Anderson Cancer Center. Research support for Dr. Girnita's laboratory is received from the Swedish Research Council, Swedish Cancer Society, The Swedish Childhood Cancer Foundation, Crown Princess Margareta's Foundation for the Visually Impaired, SINF StraCan, King Gustaf V Jubilee Foundation, Stockholm Cancer Society, Stockholm County and Karolinska Institute. M.D. and I.B.N. were supported in part by a POC grant nr.35/01.09.2016, ID 37_796, entitled “Clinical and economical impact of personalized targeted anti-microRNA therapies in reconverting lung cancer chemoresistance”—CANTEMIR. We thank Dawn Chalaire of the Department of Scientific Publications at MD Anderson Cancer Center for editing the manuscript.
Author contributions: M.Y.S. and G.A.C. designed the overall project and directed the experimental studies. M.Y.S., R.R., L.F., and J.P.T. designed and executed the xenograft experiments. M.Y.S., V.P., B.C., R.R., L.F., X.Z., C.R.A., M.D., M.C., D.N., As.M., M.R.E., and H.L. did in vitro experiments and the phenotype analysis in mice. M.Y.S., M.A.C., and J.W.W. performed the transplantation experiments. M.J.Y., C.B.R., and M.G. reviewed the pathologic slides. Ma.F., C.I., and K.V.R. performed bioinformatics analysis. M.Y.S. and M.M. performed and analyzed CyTOF experiments. P.P.B., M.H.B., E.S., and K.R. performed and analyzed the immune panel analysis in mice. M.Y.S., V.P., B.C., M.S., M.D., M.I.A., M.C., T.M., S.C., C.T., D.D., H.Y., H.A., and An.M. analyzed patient samples and performed rs6983267-RE analysis. M.Y.S., M.J.Y., M.R., Mu.F., K.R., L.G., I.B.N., An.M., S.V., R.F., C.B.R., M.G., G.G.M., and G.A.C. interpreted the data. M.Y.S. and G.A.C. wrote the paper, and all authors contributed to the final format of the writing.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.225128.117.
References
- Bar-Yaacov D, Avital G, Levin L, Richards AL, Hachen N, Rebolledo Jaramillo B, Nekrutenko A, Zarivach R, Mishmar D. 2013. RNA–DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA. Genome Res 23: 1789–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Steensma DP. 2014. Recent developments in myelodysplastic syndromes. Blood 124: 2793–2803. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, He J, Zhang D. 2015. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. Onco Targets Ther 8: 2657–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Zhao Y, Wu X, Song G. 2017. Upregulation of CCAT2 promotes cell proliferation by repressing the P15 in breast cancer. Biomed Pharmacother 91: 1160–1166. [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. 2010. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42: 722–726. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, Casey G, De Biasi M, Carlson C, Duggan D, et al. 2011. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet 43: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, et al. 2008. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst 100: 962–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. 2007. A common genetic risk factor for colorectal and prostate cancer. Nat Genet 39: 954–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. 2015. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res 75: 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Kim JI, Kim S, Hong D, Park H, Shin JY, Lee S, Lee WC, Kim S, Yu SB, et al. 2011. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat Genet 43: 745–752. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. 2011. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 30: 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al. 2013. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 23: 1446–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki-Kashio M, Aoyama K, Sashida G, Oshima M, Tomioka T, Muto T, Wang C, Iwama A. 2015. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood 126: 1172–1183. [DOI] [PubMed] [Google Scholar]
- Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, Sanada M, Miyagi S, Saraya A, Kamio A, et al. 2013. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med 210: 2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson K, Johansson B. 2007. Trisomy 8 as the sole chromosomal aberration in acute myeloid leukemia and myelodysplastic syndromes. Pathol Biol (Paris) 55: 37–48. [DOI] [PubMed] [Google Scholar]
- Pellagatti A, Cazzola M, Giagounidis A, Perry J, Malcovati L, Della Porta MG, Jadersten M, Killick S, Verma A, Norbury CJ, et al. 2010. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 24: 756–764. [DOI] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. 2012. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 30: 253–260. [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. 2009. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41: 882–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, Yin R. 2014. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol 35: 5375–5380. [DOI] [PubMed] [Google Scholar]
- Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, et al. 2013. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget 4: 1748–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B, Taguchi A, Chen Y, et al. 2016. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell 61: 520–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashida G, Harada H, Matsui H, Oshima M, Yui M, Harada Y, Tanaka S, Mochizuki-Kashio M, Wang C, Saraya A, et al. 2014. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat Commun 5: 4177. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. 2009. The cancer genome. Nature 458: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Miyagi S, Sashida G, Chiba T, Yuan J, Mochizuki-Kashio M, Suzuki Y, Sugano S, Nakaseko C, Yokote K, et al. 2012. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood 120: 1107–1117. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Vardiman JW. 2009. Myelodysplastic syndromes. N Engl J Med 361: 1872–1885. [DOI] [PubMed] [Google Scholar]
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, et al. 2007. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 39: 984–988. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Aggarwal P, Miller HE, Waukau J, Routes JM, Broeckel U, Robinson RT. 2015. The introduction of RNA-DNA differences underlies interindividual variation in the human IL12RB1 mRNA repertoire. Proc Natl Acad Sci 112: 15414–15419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ, Hu JH. 2015a. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol 8: 779–785. [PMC free article] [PubMed] [Google Scholar]
- Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q, Yin R, Xu L. 2015b. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol 36: 5523–5528. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Liu JZ, Lv P, Dang Y, Gao JY, Wang Y. 2016. Long non-coding RNA CCAT2 promotes gastric cancer proliferation and invasion by regulating the E-cadherin and LATS2. Am J Cancer Res 6: 2651–2660. [PMC free article] [PubMed] [Google Scholar]
- Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, et al. 2007. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 39: 989–994. [DOI] [PubMed] [Google Scholar]
- Zhang X, Xu Y, He C, Guo X, Zhang J, He C, Zhang L, Kong M, Chen B, Zhu C. 2015. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol 111: 834–839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.