Abstract
During development, transcription factors select distinct gene programs, providing the necessary regulatory complexity for temporal and tissue-specific gene expression. How related factors retain specificity, especially when they recognize the same DNA motifs, is not understood. We address this paradox using basic helix-loop-helix (bHLH) transcription factors ASCL1, ASCL2, and MYOD1, crucial mediators of lineage specification. In vivo, these factors recognize the same DNA motifs, yet bind largely different genomic sites and regulate distinct transcriptional programs. This suggests that their ability to identify regulatory targets is defined either by the cellular environment of the partially defined lineages in which they are endogenously expressed, or by intrinsic properties of the factors themselves. To distinguish between these mechanisms, we directly compared the chromatin binding properties of this subset of bHLH factors when ectopically expressed in embryonic stem cells, presenting them with a common chromatin landscape and cellular components. We find that these factors retain distinct binding sites; thus, specificity of binding is an intrinsic property not requiring a restricted landscape or lineage-specific cofactors. Although the ASCL factors and MYOD1 have some distinct DNA motif preference, it is not sufficient to explain the extent of the differential binding. All three factors can bind inaccessible chromatin and induce changes in chromatin accessibility and H3K27ac. A reiterated pattern of DNA binding motifs is uniquely enriched in inaccessible chromatin at sites bound by these bHLH factors. These combined properties define a subclass of lineage-specific bHLH factors and provide context for their central roles in development and disease.
Developmental lineage specification is orchestrated by the complex interaction of transcription factors (TFs). These factors identify, bind, and regulate relevant gene targets to establish and maintain cell fate. Multiple factors influence how TFs recognize specific regulatory sequences in the genome including chromatin accessibility, protein–DNA interactions, and cofactor recruitment of other TFs, noncoding RNAs, and chromatin modifying enzymes (Massari and Murre 2000; Spitz and Furlong 2012; Calo and Wysocka 2013; Morris 2016; Zhao et al. 2016). Despite decades of study, fundamental gaps remain in understanding the regulatory mechanisms underlying tissue-specific gene expression. In particular, how TFs of the same class retain specificity of activity, especially when they bind the same DNA motifs, remains unresolved. One clear example of this conundrum involves a subset of the class II basic helix-loop-helix (bHLH) tissue-specific TFs that are considered to be master regulators of discrete cell fate: ASCL1, ASCL2, and MYOD1.
The functional capacity of DNA binding class II bHLH factors is dependent on their ability to recognize and activate transcription of specific gene targets across the genome. In vitro assays have shown that bHLH factors activate transcription through specific enhancer regions, and in vivo binding comparisons have demonstrated that despite their class-defining bHLH domain, they bind to different sites throughout the genome (Cao et al. 2010; Castro et al. 2011; Fong et al. 2012, 2015; Meredith et al. 2013; Borromeo et al. 2014; Schuijers et al. 2015), thereby activating distinct transcriptional targets for expression. Factor-specific preference in DNA binding motifs can account for some of this specificity between class II bHLH factors. Genome-wide binding studies demonstrate that the neural-specific factor NEUROD1 and muscle-specific factor MYOD1 can bind both a shared Ebox motif, as well as factor-specific Ebox motifs (Fong et al. 2012). However, the neural bHLH ASCL1, and the related factor ASCL2, share the same preferred Ebox motif as the muscle-specific bHLH factors MYOD1, MYF5, and MYOG when examined in their respective lineages (Wright et al. 1991; Castro et al. 2011; Soleimani et al. 2012; Borromeo et al. 2014; Liu et al. 2014; Schuijers et al. 2015). Thus, additional mechanisms must be involved to supplement TF preference in DNA binding motifs.
ASCL1 and ASCL2 are nearly identical in their bHLH domains, but they are present in distinct lineages and drive distinct gene expression programs; ASCL1 directs neural lineages, whereas ASCL2 is important in multiple lineages including trophectoderm (Guillemot et al. 1994), T-helper cells (Liu et al. 2014), and intestinal stem cells (van der Flier et al. 2009). MYOD1, a master regulator of skeletal muscle, shares bHLH domain-defining amino acids with the ASCL factors but is more divergent in protein sequence. This specific subset of bHLH factors recognizes the same Ebox motif, CAGSTG (Cao et al. 2006; Borromeo et al. 2014; Schuijers et al. 2015), but paradoxically, these factors bind distinct genomic sites in their respective lineages in vivo. Lineage-specific differences in chromatin accessibility and the presence of cell-type–specific cofactors likely play a role in determining where these factors bind. However, given that ASCL1, ASCL2, and MYOD1 induce disparate tissue-specific gene expression programs when ectopically expressed, additional mechanisms must exist to account for the distinct activities (Nakada et al. 2004; Nishiyama et al. 2009; Fong et al. 2012). In this study, we seek to distinguish between different models of TF specificity, and we ask whether binding differences depend solely on chromatin state or whether factor-specific differences intrinsic to these factors result in distinct interactions with DNA to enact discrete transcriptional programs.
Results
Differences in class II bHLH factor binding in the genome are a consequence of intrinsic properties of each factor
The related class II bHLH factors ASCL1, ASCL2, and MYOD1 heterodimerize with class I bHLH factors and selectively bind Ebox motifs (Fig. 1A,B). Previous studies showed these factors bind to distinct genomic sites in the lineages where they are expressed; however, they recognize the same CAGSTG motif (Cao et al. 2006; Borromeo et al. 2014; Schuijers et al. 2015). To remove the variability in the chromatin landscape experienced by ASCL1, ASCL2, and MYOD1 in vivo, we directly compared their DNA binding properties when ectopically expressed in embryonic stem cells (Fig. 1C). Although the genomic binding pattern of this subset of bHLH factors has previously been compared in their respective lineages, their DNA binding behavior in a single, common cell type mitigates the influence of cell-type–specific influences such as chromatin accessibility and differentially expressed proteins. Embryonic stem cells (ESCs) demonstrate embryonic totipotency, the capacity to develop into any tissue in the embryo (Evans and Kaufman 1981; Martin 1981). ASCL1, ASCL2, and MYOD1, which serve as crucial lineage specifiers, are essentially absent from the transcriptional profile of ESCs (Fig. 1), yet when ectopically expressed, they activate lineage-specific gene transcription programs (ASCL1, neural; ASCL2, trophectoderm; MYOD1, muscle) (Nishiyama et al. 2009). To decipher fundamental mechanisms by which the bHLH factors specifically identify targets and regulate transcription across a complex genome, modified ESCs were used, and represent a tabula rasa on which the activity of these bHLH factors can be compared, minimizing the potential confounding influence of developmental cues present in partially or fully defined lineages.
Figure 1.
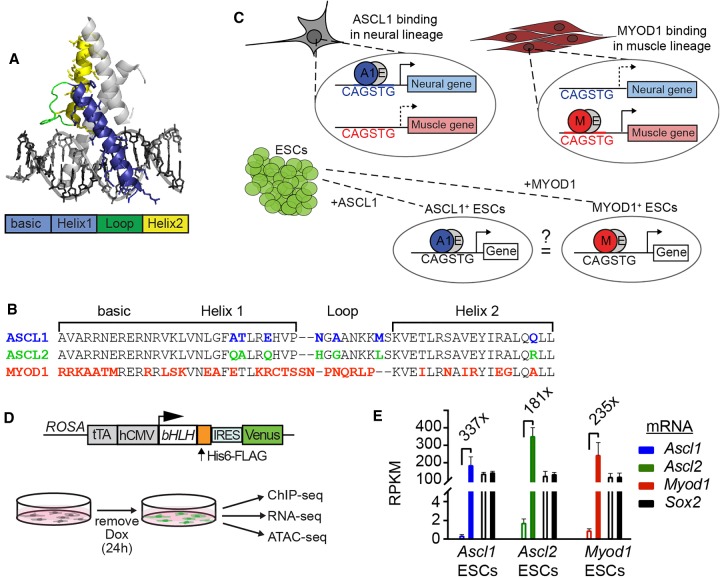
ASCL1, ASCL2, and MYOD1 are related class II bHLH transcription factors. Class II bHLH factors heterodimerize with class I bHLH factors and selectively bind CANNTG Eboxes in vitro (Johnson et al. 1992; Murre et al. 1994). (A) Illustration showing conserved basic helix-loop-helix structural domains modeled on the NEUROD1:E47 crystal structure (Longo et al. 2008). Colored bar shows defined domains and corresponds to the colors on the structure. (B) Comparison of the amino acid sequence of ASCL1, ASCL2, and MYOD1 bHLH domains. The colors represent distinctions within the bHLH domain unique to only one of the proteins compared. (C) Illustration showing the concept tested in this study. Previous work shows that ASCL1 and MYOD1 bind to distinct sites in the lineages where they are expressed, neural and muscle, respectively; however, they recognize the same CAGSTG motif. We ask whether they will bind the same sites or distinct sites when presented with the same chromatin landscape. (D) Diagram of the transgene present in the ESC lines that express a single bHLH factor when cultured in the absence of doxycycline (DOX) (Gossen and Bujard 1992; Guturu et al. 2013). (E) RNA-seq RPKM values from ESCs before and after induction of the individual bHLH factors (Supplemental Table S2). The mean fold change across replicates is shown for each. mRNA levels for an endogenous TF in ESCs, Sox2, are shown for comparison.
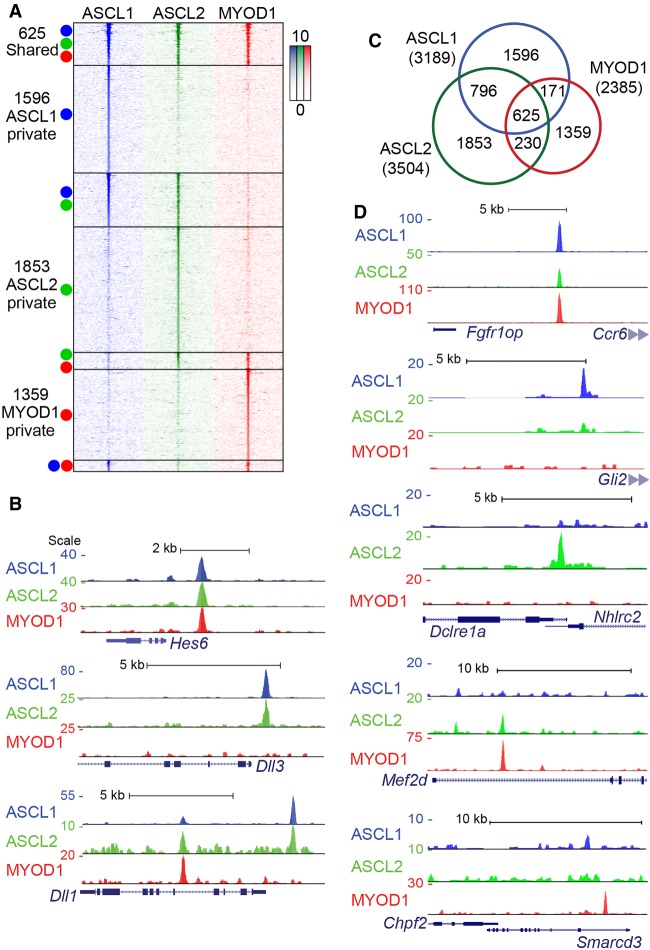
We determined the binding sites for ASCL1, ASCL2, and MYOD1 in ESCs engineered to ectopically express each factor in a knock-in tet-OFF system (Fig. 1D; Nishiyama et al. 2009). Removal of doxycycline (DOX) (Guturu et al. 2013) from the culture medium leads to expression of the transgenic construct (Gossen and Bujard 1992), here featuring a single FLAG-tagged bHLH transgene. The level of each bHLH mRNA after induction was similar to the mRNA level for an endogenous transcription factor, Sox2 (Fig. 1E). To directly compare the binding of the bHLH factors in an unbiased cellular context, ChIP-seq for ASCL1, ASCL2, and MYOD1 was performed on cells 24 h after removal of DOX (Fig. 1D,E). Antibodies against the FLAG moiety were used for ASCL2 and MYOD1 ChIP; however, for ASCL1, the FLAG is not efficiently detected (Nishiyama et al. 2009), necessitating the use of antibodies directed against ASCL1. ASCL1, ASCL2, and MYOD1 were each found to bind a few thousand sites genome-wide (Fig. 2A; Supplemental Table S1). Strong enrichment was detected near previously identified targets of bHLH function, including binding sites near Notch pathway genes Dll1, Dll3, and Hes6 (Fig. 2B). Some sites, such as a site ∼1 kb upstream of the Hes6 locus, show clear evidence of shared binding by these factors. Crucially, not all sites are bound by all factors, and many sites showing strong enrichment for a single factor show no enrichment for the others. Sites such as those located near Dll3 show no enrichment for MYOD1, but are bound by both ASCL1 and ASCL2. Dll1 features two binding sites: one located upstream of the TSS that is bound by both ASCL1 and ASCL2, but not MYOD1; and one located in the fourth intron that is bound robustly by MYOD1, but shows little enrichment for ASCL1. Notably, expression of these, and other known targets, is detected at 24 h by RNA-seq, suggesting that the binding observed for these factors is sufficient for gene regulatory activity (Supplemental Table S2). A second independent experiment validated the finding that ASCL1, ASCL2, and MYOD1 retain distinct genomic binding sites even when assayed in a common cell context (Supplemental Fig. S1).
Figure 2.
Tissue-specific bHLH factors maintain distinct binding in the ESC genome revealing intrinsic binding specificity properties. (A) Heatmap of ChIP-seq data sets from ESCs at 24 h post-induction of bHLH expression. Each column shows the ChIP-seq signal for the factor indicated. Each row indicates a single interval ±3 kb centered on the peak apex, with the ChIP-seq signal indicated by color density. Colored circles indicate which ChIP-seq data set(s) identified a significant peak. (B,D) UCSC Genome Browser tracks comparing bHLH ChIP-seq enrichment near known targets including the Notch pathway genes Hes6, Dll3, and Dll1 (B), and novel targets (D) with either shared or private peaks. Each track shown represents ChIP-seq data from the indicated bHLH factor, normalized to 10 million reads and aligned to the mm10 genome. Track scale represents the number of reads at a given position. (C) Proportional area diagram of bHLH binding sites shown in the heatmap (A) with numbers representing distinct peaks identified in each subset. Peaks were considered overlapping if the peak apex was within 150 bp.
Genome-wide, 625 sites were bound by all three factors, reflecting their shared recognition for a CAGSTG Ebox. However, >50% of the sites bound by each factor are factor specific (Fig. 2C; Supplemental Table S1). Comparison of the binding sites reveals no difference in distribution with respect to genic features such as transcription start site or introns, and all three factors show similar preference for distal enhancer regions, as previously described (Supplemental Fig. S2). This demonstrates that ASCL1, ASCL2, and MYOD1 maintain distinct patterns of binding even when compared in a common cell type with the same chromatin landscape and cellular components. Genome browser tracks for additional examples for shared binding sites (i.e., Fgfr1op/Ccr6), and distinct binding sites (i.e., Gli2, Dclre1a/Nhlrc2, Mef2d, Smarcd3/Chpf2) are shown (Fig. 2D). Examples such as these demonstrate that overall, ASCL1, ASCL2, and MYOD1 do not simply bind the same set of sites when assayed in a common cellular context. As ChIP-seq data from all three cell lines reveal robust enrichment for specific binding sites, and many peaks are clearly absent from one or more data sets, these differences cannot be attributed to differences in sequencing depth or technical bias and represent clear, veritable distinction between the factors tested. Thus, the distinct pattern of binding observed in differentiated cell or tissue types is not simply due to differential chromatin accessibility. This finding is particularly striking for ASCL1 and ASCL2, which have virtually identical bHLH domains (Fig. 1).
bHLH factors ASCL1, ASCL2, and MYOD1 preferentially bind similar motifs
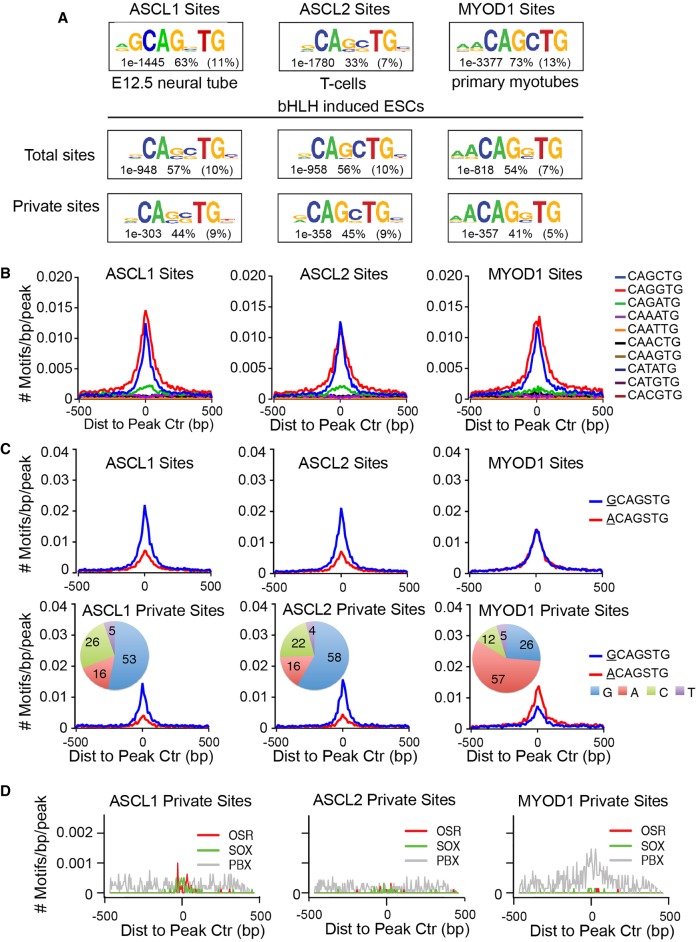
The mouse genome contains approximately 14 million Ebox (CANNTG) sites genome-wide. These features have long been identified as the preferred DNA binding motifs for bHLH factors (Ephrussi et al. 1985; Murre et al. 1989), which bind to the major groove of DNA as homodimeric or heterodimeric complexes (Ferré-D'Amaré et al. 1993; Ellenberger et al. 1994; Ma et al. 1994). The in vivo motif preferences of ASCL1, ASCL2, and MYOD1 are similar when compared in their respective lineages such as neural tube, T-helper cell, and myoblast (Fig. 3A; Cao et al. 2010; Castro et al. 2011; Liu et al. 2014). To test whether the specificity observed in ESCs might reveal additional motif preference in the form of primary (Ebox), we performed de novo motif analysis to identify the preferred motif of each factor (Heinz et al. 2010). We targeted our analysis to the 50 bp surrounding the peak apex. This analysis identified a strong Ebox motif, as expected for class II bHLH factors (Fig. 3A).
Figure 3.
ASCL1, ASCL2, and MYOD1 bind similar motifs. (A) Comparison of primary Ebox motifs identified by de novo motif discovery in ChIP-seq for ASCL1, ASCL2, and MYOD1 from differentiated tissues, as indicated, and the ESCs with the ectopic factors using a 50 bp interval centered on the peak apex. Comparative motifs in differentiated cell types were generated from previously published data sets for ASCL1 (Borromeo et al. 2014), ASCL2 (Liu et al. 2014), and MYOD1 (Cao et al. 2010). Numbers reflect the P-value significance of the specified motif, the percent incidence of the specific motif shown in ChIP-seq peak regions, and the percent incidence in a normalized random background set. (B,C) Frequency and distribution of Ebox motifs (B), or expanded Eboxes (C), across a 1-kb interval surrounding peak centers. Each colored plot represents the rate of Ebox occurrence for one Ebox permutation as indicated. (D) Frequency and distribution of primary and secondary motifs identified by de novo motif discovery within a 1-kb interval centered on peak apex from ASCL1 and MYOD1 binding sites. Text indicates the motif specified and the factor family predicted to bind the motif.
To compare Ebox selectivity, we determined the distribution of each possible permutation of the canonical Ebox, relative to the peak centers identified from ChIP-seq for each factor. This comparison supports the findings from de novo motif analysis and reveals strong enrichment for features matching a CAGSTG motif near the peak centers (Fig. 3B, red and blue), as well as modest enrichment for GA/TC core dinucleotide Eboxes (Fig. 3B, green). This is in striking contrast to other Ebox permutations, which show no enrichment across the interval surrounding the peaks identified. The genomic incidence of each permutation suggests that Ebox motif preference is not dictated by the abundance of each motif (Supplemental Fig. S3). Thus, even in pluripotent ESCs lacking lineage-restricted chromatin features or lineage-specific cellular properties, ASCL1, ASCL2, and MYOD1 preferentially bind the same Ebox motif, CAGSTG.
In addition to comparison of the canonical hexameric Ebox sequence identified, we tested whether an expanded motif provides additional binding site specificity. This analysis revealed the presence of additional enriched nodes in the position-weight-matrix (PWM) adjacent to the Ebox. ASCL1/ASCL2-preferred motifs show enrichment for an adjacent guanine nucleotide, compared to MYOD1, which shows enrichment for an adenine residue at this position. To explore whether the observed binding specificity was due to the presence of these additional flanking nucleotides, we compared their distribution at binding sites identified for each bHLH factor (Fig. 3C). Sites bound by ASCL1 or ASCL2 demonstrate greater association with GCAGSTG motifs, whereas sites bound by MYOD1 are equally likely to associate with motifs with a flanking adenosine, ACAGSTG. This result holds even after deconvolution to force flanking site analysis for a single orientation of the site by comparing the distribution of each permutation of flanking motifs (Supplemental Fig. S4). Motifs featuring these additional flanking nucleotides are overrepresented by 25%–30% at the binding sites of these factors (Fig. 3C). Thus, the flanking nucleotide appears to influence binding specificity and may represent an important mechanism in site selection in some contexts. Nevertheless, the differences observed cannot be solely attributed to these flanking motifs, as ∼65% of the differential binding occurs at sites lacking such flanking nucleotide positions, and sites featuring such positions are present in shared as well as private sites.
Secondary cofactor motifs are not a primary contributor to the distinct genomic binding
Another possible mechanism for selective and differential binding would be through factor-co-factor interactions providing additional specificity through the presence of secondary motifs proximal to the primary Ebox motif. To test this possibility, de novo motif discovery was again performed with a broader window (150 bp) for motif analysis to allow for identification of motifs neighboring the primary Ebox binding motif located at the apex of the ChIP-seq peaks (Fig. 3D; Supplemental Fig. S5). A homeobox motif resembling that bound by a PBX factor was enriched in MYOD1-bound sites (7%). PBX and SWI/SNF complexes are well-known components of MYOD1 regulatory function in the muscle lineage, including at important myogenic genes (de la Serna et al. 2001, 2005; Berkes et al. 2004). Pbx2 and Pbx3 are present in the ESCs, and thus, may facilitate selective binding by MYOD1 at sites not bound by ASCL1 or ASCL2. However, these sites are numerically insufficient (7%) to be a primary determinant of differential binding. Similarly, secondary motifs identified in the ASCL1 or ASCL2 ChIP-seq data, SOX and OSR factors, were found in <3% of sites. Thus, although cofactors may play a major role in specific binding of bHLH factors in differentiated tissues, the rarity of these motifs near bHLH bound sites in the ESCs suggests cofactors are not the primary mechanism for the differential binding detected in this paradigm.
ASCL1, ASCL2, and MYOD1 bind open and closed chromatin and satisfy multiple criteria as pioneering transcription factors
TFs are one component of the regulatory schema underlying lineage specification. However, TF binding has long been observed to be highly restricted, and all known TFs have been observed to bind only a small subset of the total set of motifs present in the genome. Thus, TF binding is itself regulated by mechanisms other than simple motif recognition. The specific mechanisms underlying selection of appropriate binding sites from the many potential motif loci remains incompletely understood. One previously identified mechanism of lineage-specific transcriptional regulation is the influence of higher-order chromatin structure on binding site accessibility, wherein nucleosomal DNA is rendered inaccessible to transcription factor binding (Vierstra et al. 2014).
One possible distinction in ASCL1, ASCL2, and MYOD1 binding preference could be differential ability to bind inaccessible (closed) sites within the chromatin landscape. The ability to bind inaccessible chromatin is known as “pioneering,” and distinctions in pioneering capability were previously described for the archetypical pioneering TFs of the FOXA and GATA families (Gualdi et al. 1996). Such distinctions were further observed by ChIP-seq between different classes of TFs (Cirillo et al. 2002; Soufi et al. 2012) and suggested for class II bHLHs (Wapinski et al. 2013; Soufi et al. 2015). The pioneering ability of ASCL1 was suggested as a mechanism for the distinct binding and activity of the master regulators ASCL1 and MYOD1, based on structural differences in the bHLH domains of these factors (Soufi et al. 2015). To test the possibility that a differential ability to bind closed chromatin might provide a primary mechanism underlying the distinct binding observed for these factors, we compared global chromatin accessibility in ESCs at ASCL1, ASCL2, and MYOD1 binding sites using ATAC-seq, a method for characterizing the distribution of open chromatin genome-wide (Buenrostro et al. 2013, 2015). Comparison of ATAC-seq signal in the three engineered ESC lines shows similar distribution of open chromatin prior to bHLH induction (Supplemental Fig. S6). Consistent with previous findings demonstrating that ASCL1 exhibits pioneering binding activity in fibroblasts (Wapinski et al. 2013), both ASCL1 and its closely related factor, ASCL2, bind ATAC-seq defined inaccessible (closed) and accessible (open) chromatin (Fig. 4A). However, in contrast to what was modeled for MYOD1 in Soufi et al. (2015), MYOD1 also binds both closed and open chromatin in this assay and does so to a similar extent as ASCL1 (Fig. 4A). ASCL2 also binds both closed and open chromatin, but the aggregate accessibility of ASCL2 binding sites appears higher compared to the other two factors (Fig. 4A; Supplemental Fig. S6). Comparison of these data to previously published genome-wide MNase-seq from ESCs (GEO accession number GSE58101) (Carone et al. 2014) supports the ability of these factors to bind closed chromatin. Similar to what was seen in the ATAC-seq, ASCL2 does this to a lesser extent than ASCL1 and MYOD1 (Supplemental Fig. S7; Carone et al. 2014). Thus, by these criteria, ASCL1 and MYOD1 appear to exhibit similar chromatin binding properties. However, more stringent criteria for pioneering activity, such as nucleosomal binding with nucleosome positioning assays, would be necessary to define these factors as pioneer factors.
Figure 4.
ASCL1, ASCL2, and MYOD1 bind open and closed chromatin and satisfy multiple criteria for pioneering transcription factors. (A) Heatmap of ChIP-seq and ATAC-seq from ESCs. Each plot compares the total set of binding sites identified for the factor indicated and shows the ChIP-seq (left) and ATAC-seq (right) signal for each peak region in a 6-kb interval centered on the peak apex. Rows are ordered based on the ATAC-seq signal. Dashed red line approximates the upper and lower quartiles used to define “open” versus “closed” binding sites. (B) Histogram of mean bHLH binding and chromatin accessibility (left) and H3K27ac enrichment (right) at binding sites identified by ChIP-seq for each factor. For each, the bHLH ChIP-seq signal (blue, left axis) is enriched at the peak center. At these sites, there is an increase in ATAC-seq and H3K27ac signal post bHLH induction (green versus red, right axis). (C) Heatmap of ASCL1 ChIP-seq and the ATAC-seq signal for regions showing a significant increase (left) or decrease (right) in chromatin accessibility upon ASCL1 induction. Each row represents a single peak interval with ASCL1 ChIP-seq (left), ATAC-seq from uninduced control ESCs (The ENCODE Project Consortium et al. 2007), or ESCs post-induction (right). (D) Frequency and distribution of Ebox motif distribution at bHLH binding sites identified in nucleosome-occupied (left) and nucleosome-depleted (right) chromatin. Each plot shows the frequency and distribution of a single Ebox permutation across the intervals surrounding the peak apex for the transcription factor, as shown. Motif shown represents the most significantly enriched de novo motif as compared to sequence-normalized background regions.
Transactivation activity of the class II bHLH factors has long been observed (Lassar et al. 1986; Davis et al. 1987; Johnson et al. 1990, 1992; Weintraub et al. 1990; Guillemot et al. 1993, 1994), but the exact mechanisms mediating the progression from binding to transcriptional regulation remain unclear. To test whether bHLH binding might lead to changes in chromatin accessibility in cis, facilitating binding of transcriptional machinery, we compared the chromatin accessibility (by ATAC-seq) at bHLH binding sites in both uninduced ESCs and at 24 h post-induction of each bHLH factor. We also performed ChIP-seq for H3K27ac to assess if the bHLH TFs initiate changes in this active enhancer mark. Both ATAC-seq and H3K27ac ChIP-seq showed a significant aggregate increase in signal at bHLH bound sites (Fig. 4B; Supplemental Fig. S8). Both of these properties support the characterization of these bHLH factors as pioneer TFs (Gualdi et al. 1996; Cirillo et al. 2002; Soufi et al. 2012). However, not all bHLH binding sites were associated with increases in open chromatin and the active enhancer mark. To compare the degree to which bHLH factor binding is associated with these local increases or decreases, we directly compared the ASCL1 ChIP-seq signal on intervals surrounding these regions in accessible chromatin as assessed by ATAC-seq (Fig. 4C). Sites showing increases in local accessibility were more closely associated with ChIP enrichment for bHLH factors than those showing a decrease, further supporting a role for these factors in the regulation of chromatin structure.
To test whether chromatin accessibility might reveal distinct characteristics suggesting a mechanism for the distinct binding observed for each bHLH factor, we performed motif analysis at the regions bound by each factor found within open versus closed chromatin. We ranked the binding sites identified for each factor based on the mean ATAC-seq signal surrounding the peak apex in uninduced ESCs. We then performed de novo motif discovery on the upper and lower quartile of these peaks. This approach revealed an unexpected distinction in motif distribution, with closed sites enriched for CAGGTG Eboxes, and more open sites showing a greater association with CAGCTG Eboxes (Fig. 4D). A distinction in motif preference between nucleosome-occupied and nucleosome-depleted sites was previously reported for MYC (Soufi et al. 2015). Sites undergoing an increase in chromatin accessibility after bHLH induction were more associated with CAGGTG motifs, similar to findings in Fong et al. (2012) (Supplemental Fig. S9). Although Ebox motifs were enriched at these dynamic sites, no secondary motifs were identified as significantly enriched (Supplemental Fig. S9).
Histone modification signatures at ASCL1-, ASCL2-, and MYOD1-bound sites are similar
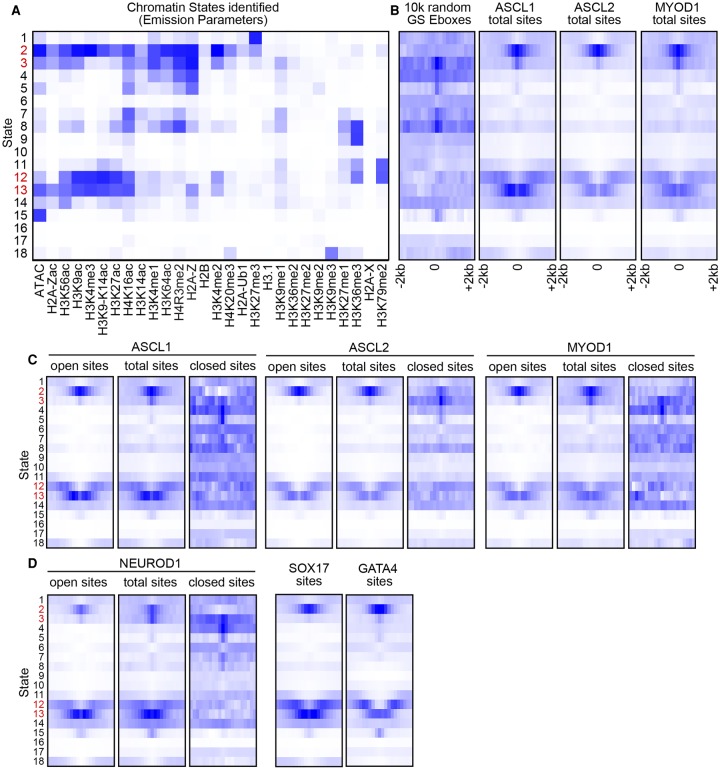
In addition to nucleosomal occupancy, post-translational modifications of the histone octamer were observed, and demonstrated to both correlate with transcriptional regulation and consistently vary between cells of distinct lineages (The ENCODE Project Consortium et al. 2012; Kundaje et al. 2012; Wang et al. 2012). Such patterns suggest that a “histone code” is also an integral component of the mechanisms defining lineage specification and cell fate. Thus, another potential mechanism to guide the bHLH factors to distinct sites would be if they bind specific histone modifications. Such a model has been proposed for ASCL1 in the context of a fibroblast-to-neuron transdifferentiation assay, wherein a conflicting trivalent signature of active marks H3K4me1 and H3K27ac, along with the repressive mark H3K9me3, were found to be predictive of bHLH binding (Wapinski et al. 2013). To test whether these or other histone decoration signatures might provide a mechanism for differential binding, we compared the genomic distribution of multiple modifications to the binding sites identified for these factors. Using 28 distinct chromatin features obtained from published ChIP-seq data sets, and our H3K27ac ChIP-seq and ATAC-seq, we performed undirected hidden Markov modeling to define chromatin states genome-wide (Fig. 5A; Supplemental Table S3). These states were then used to compare the chromatin features present at bHLH binding sites identified from the ESCs when each bHLH TF was induced (Fig. 5B).
Figure 5.
ASCL1, ASCL2, and MYOD1 bind similar chromatin states enriched with active features. (A) Chromatin states (Ernst and Kellis 2012) identified by hidden Markov modeling of chromatin features identified by ChIP-seq and ATAC-seq in ESCs from published data sets and data generated in this study. The strength of the state association with each chromatin feature (columns) is indicated by increasing color intensity. States indicated in red represent those highlighted in text. (B–D) State enrichment diagrams of genomic sites identified by random CAGSTG Ebox sites and ASCL1, ASCL2, and MYOD1 ChIP-seq (B), by bHLH ChIP-seq binned into those sites in open versus closed chromatin from Figure 4A (C), and by additional TF binding sites for comparison (D). Each plot represents the degree of association of the sites (color intensity) versus the states indicated in A in the 4-kb interval centered on the ChIP-seq peak apex. See Methods for a list of GEO accession numbers for all data sets used.
Hidden Markov model analysis with this set of 28 chromatin features at the ASCL1-, ASCL2-, and MYOD1-bound sites reveals that the bound sites for each bHLH are similarly enriched for markers of active enhancers and promoters, characterized by H3K9ac, and H3K27ac, and H3K4me1 (enhancers and promoters), or H3K4me3 (promoters) (Creyghton et al. 2010). Sites associated with these marks are also enriched for open chromatin as measured by ATAC-seq, further supporting this definition. In contrast to ASCL1-bound sites in mouse fibroblasts, no enrichment for H3K9me3 was detected in ASCL1-bound sites in ESCs. Additionally, no mark, or combination of marks, demonstrates clear bHLH factor-specific association even when assessed in the total subset and open/closed subsets of sites bound by the three bHLH factors (Fig. 5C). As such, differential preference for the histone modifications included in this analysis does not appear to be a primary factor in binding site selection and specificity in ESCs, suggesting that the mechanism supporting their distinct binding and function lies elsewhere.
There was, however, a distinction noted between open and closed binding sites. Open sites closely mirrored the overall pattern of state enrichments for these factors. In contrast, closed sites did not demonstrate enrichment of specific states, and partially resembled the randomized control set (Fig. 5C). To test whether these results were unique to this set of bHLH factors when ectopically expressed in ESCs, we performed similar analysis on sites bound by the neural class II bHLH factor, NEUROD1, and other TF classes including SOX17 and GATA4. In each case, when these TFs were ectopically expressed in ESCs, they also show binding to chromatin with a similar histone modification signature as ASCL1, ASCL2, and MYOD1, and the distinction between open and closed chromatin is detected (Fig. 5D). These results demonstrate that the ESC chromatin environment at ASCL1, ASCL2, and MYOD1 binding sites is not directing factor-specific binding, but rather the chromatin environment is characteristic for TF binding in general.
ASCL1, ASCL2, and MYOD1 bind a novel pattern of DNA motifs identified specifically in closed chromatin
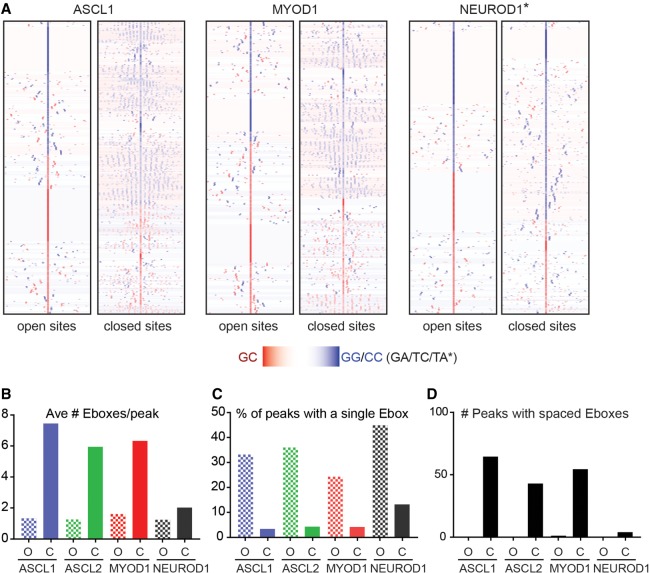
The ability of bHLH factors to bind both accessible and inaccessible chromatin regions suggests that these factors either do not distinguish between disparate chromatin states, or that specific features of these binding sites may facilitate or allow their binding and function. Comparison of the de novo binding motifs between open and closed chromatin suggests that the number and distribution of these motifs may be distinct between these chromatin states (Fig. 4D; Supplemental Table S4). To uncover novel features in motif distribution, we compared Ebox distribution at the bHLH bound genomic intervals identified by ChIP-seq that fell within open versus closed chromatin using an in-house software pipeline (Fig. 6A; Supplemental Table S5; Supplemental Methods). A striking pattern in Ebox distribution was revealed specifically in closed chromatin that was not detected in open chromatin. This Ebox pattern in closed chromatin has two notable features. First, bHLH binding sites located in closed chromatin have an overall higher number of Eboxes present relative to that seen in open chromatin (three- to fourfold difference) (Fig. 6B). bHLH bound sites in open chromatin often have only a single, centrally located Ebox at the peak center (Fig. 6C). What is striking, however, is that the Ebox distribution in the closed chromatin reveals the presence of spatially reiterated patterns of Ebox distribution, roughly 10–15 bp apart, in peaks associated with closed chromatin; this spacing is equivalent to 1–1.5 turns of the DNA helix (Fig. 6A). Such patterns of Ebox spacing is detected in 40%–60% of closed binding sites for ASCL1, ASCL2, and MYOD1, but not in peaks associated with regions of open chromatin (Fig. 6D).
Figure 6.
Unique stereotypical patterning of Ebox motifs in ASCL1-, ASCL2-, and MYOD1-bound sites specifically in closed chromatin. (A) Heatmap comparison of CAGSTG Ebox motifs identified in the upper and lower quartile of binding sites identified by ChIP-seq for each factor, as indicated, reflecting the same peaks as shown in Figure 4A. NEUROD1 quartiles were determined based on chromatin accessibility in uninduced ESCs, as assayed by ATAC-seq. Plots reflect cluster analysis of the distribution of each motif, with peak coordinates computationally centered on the CAGSTG Ebox motif closest to the peak apex, within ±25 bp of peak apex, and visualized across 300 bp, centered on the motif specified. Peak intervals lacking a CAGSTG motif within this interval are not shown in this comparison. (B) The average number of CAGSTG Ebox motifs per peak (within 300 bp) around bHLH bound regions identified in open (O) chromatin or closed (C) as determined by ATAC-seq on uninduced ESCs. (C) The percentage of total bHLH bound regions featuring a single Ebox motif in chromatin defined as open versus closed. (D) Number of peaks featuring the spatially patterned CAGSTG Ebox motifs as shown in A, demonstrating that this feature is restricted to closed chromatin and is not a feature of all class II bHLH TF.
The patterned Ebox features are similarly found in ASCL1-, ASCL2-, and MYOD1-bound closed chromatin. To determine if these features are specific to the class II bHLH TFs or generally represent a property of TF binding to closed chromatin, we analyzed additional TF ChIP-seq data sets in which the TFs were also ectopically expressed in ESCs or were constitutively present. In ESCs with ectopically induced NEUROD1, another neurogenic class II bHLH TF, there were fewer Eboxes in its bound sites relative to that seen for ASCL1, ASCL2 and MYOD1, and no evidence of the reiterated Ebox patterning in open or closed chromatin was detected (Fig. 6A–D). Thus, this property of ASCL1, ASCL2, and MYOD1 binding in closed chromatin defines a distinct subclass of class II bHLH TFs. We analyzed ChIP-seq data from other classes of TFs induced in ESCs including SOX17, GATA4, and FOXA2, and found no evidence for patterned motifs in open or closed chromatin. TFs constitutively present in ESCs, including TCF12 (Brookes et al. 2012), a class I bHLH, and SOX2 also do not show this patterned distribution of motifs at their bound intervals (Supplemental Fig. S10). Taken together, the presence of increased numbers of Eboxes and the stereotypical patterning of these motifs in ASCL1-, ASCL2-, and MYOD1-bound sites in closed chromatin are unique features specific to this subclass of TF regulators.
Discussion
Class II bHLH factors maintain distinct binding in a common chromatin landscape
The ability of class II bHLH TFs to establish and maintain appropriate cell lineages is dependent on their ability to regulate gene expression, which is a function of their ability to recognize their cognate binding sites within the genome. In this study, we tested the ability of ASCL1, ASCL2, and MYOD1 to identify and bind to distinct genomic loci when ectopically expressed in ESCs, which lack the lineage-specific chromatin features and gene expression of their respective cell lineages. We demonstrated that these factors maintain distinct binding in this reduced system, and thus possess intrinsic specificity in binding that exists beyond their endogenous cellular environments.
We explored whether we could attribute the intrinsic binding specificity between these factors to differences in DNA motif specificity, cofactor interactions, or other chromatin features. The bound sites are enriched for Eboxes resembling those previously reported for these factors in differentiated tissues and in vitro (CAGSTG) (Blackwell and Weintraub 1990; Cao et al. 2010; Castro et al. 2011; Liu et al. 2014). There is additional specificity between the ASCL factors and MYOD1 conferred by the Ebox flanking nucleotide; ASCL factors prefer GCAGSTG and MYOD1 has a preference for ACAGSTG. Although this feature has the greatest impact on the differential binding, it can account for only a third of the private sites between MYOD1 and ASCL2. The MYOD1/ASCL2 comparison is particularly key given the technical advantage of using the anti-FLAG antibodies to determine binding by ChIP. The Ebox flanking nucleotide provides no distinction between ASCL1 and ASCL2 as both are enriched for GCAGSTG.
Factor-specific secondary motifs were identified that point to PBX-like factors as influencing MYOD1 binding in the ESCs. PBX factors are components of MYOD1 regulatory function in the muscle lineage (de la Serna et al. 2001, 2005; Berkes et al. 2004), and Pbx2 and Pbx3 mRNA are present in undifferentiated ESCs. The contribution of this cofactor motif, however, only accounts for <10% of the MYOD1 private sites. Cofactor motifs for the ASCL factors such as SOX and OSR are present in <3% of sites and thus are predicted to contribute even less to ASCL1 and ASCL2 differential binding. Thus, a combinatorial model of factor–cofactor binding is likely too simplistic to adequately describe the distinct binding activity of these factors. However, this analysis does not rule out a role for regulatory cofactors, as such factors may not function through motif-specific DNA interactions. Examples include the Id (Inhibition of Binding) family that possesses an HLH domain and can interact with MYOD1 but not TAL1, another bHLH, to repress its activity (Langlands et al. 1997).
Chromatin features such as accessibility and histone modifications also contribute little to the differential binding of ASCL1, ASCL2, and MYOD1. All three factors can access closed chromatin, although in aggregate ASCL2 sites, they have higher accessibility based on ATAC-seq and MNase-seq than the other two factors. Comparing MYOD1 and ASCL2 specifically, this difference could contribute up to ∼20% of the differential binding. Thus, even combining contributions from DNA motif specificity, cofactor interactions, and ability to bind closed chromatin, we can account for only 50%–60% of the differential binding between MYOD1 and ASCL2. Additional mechanisms, still to be discovered, are needed to explain the differential binding and function of these important lineage-specifying transcription factors.
Class II bHLH factors are pioneering factors and chromatin interactors
Although transcription factors are largely characterized based on their ability to recognize and interact with specific DNA binding motifs against the broader context of the genome, it has long been understood that both broad and local epigenetic features are an integral component of gene regulation. Key among these is chromatin accessibility, which is known to affect genomic engagement and transcriptional regulation by transcription factors (Gross and Garrard 1988; Clark et al. 1993; Merika and Orkin 1993; Gerber et al. 1997). A subset of factors, known as “pioneer” factors, possesses the ability to engage the genome despite apparently unfavorable access to the DNA helix (Zaret and Carroll 2011). This ability has been suggested as a crucial mechanism in cellular reprogramming assays. However, of the four canonical reprogramming factors, only POU5F1, SOX2, and KLF4 but not MYC have demonstrated this ability, suggesting that differential pioneering activity represents a potential mechanism defining transcription factor function (Soufi et al. 2012).
Recently, the binding of ASCL1 has been characterized in a fibroblast-to-neuron reprogramming assay and found to bind to inaccessible chromatin (Wapinski et al. 2013). MYOD1, however, was predicted to have diminished, or absent pioneering activity, based on structural modeling of its bHLH domain (Soufi et al. 2015). Here, we demonstrate that despite the fact that ASCL1, ASCL2, and MYOD1 bind to separate sites, they can all access closed chromatin based on ATAC-seq and MNase-seq. Nevertheless, this raises the possibility that the ability to bind closed chromatin is a feature of class II bHLH proteins as a class and suggests that the structural differences present within the bHLH domain of ASCL1 and MYOD1 are not sufficient to dramatically alter this ability in the context of ESCs. Because a number of bHLH factors act as crucial regulators of lineage and cell-fate specification (Massari and Murre 2000), the ability to access closed chromatin may be central to this capacity, especially with regard to their roles in “reprogramming cocktails” of transcriptional regulators, which can transition cells to alternative lineages.
Although the epigenetic landscape is increasingly identified as important in controlling gene expression, our findings demonstrate that it is not sufficient to account for the distinct binding, and thus, specific function of the lineage-specific bHLH TFs. It is likely the case that favorably bound sites may regulate expression of genes that facilitate additional TF binding and chromatin modification. The changes in chromatin accessibility observed upon TF induction suggest that these factors recruit chromatin remodelers to their respective binding sites. Given the ability of MYOD1 and ASCL1 to identify specific binding targets in the lineage-inappropriate environment of embryonic fibroblasts (Vierbuchen et al. 2010; Wapinski et al. 2013), it is possible that some previously unidentified cofactor may be partially responsible for directing these bHLH factors to the specific sites necessary for lineage-relevant function in development, disease, and reprogramming. The observation that binding sites for ASCL1 in ESCs are not characterized by the same epigenetic profile as those identified in differentiated cell types, i.e., H3K27ac, H3K4me1, and H3K9me3 (Wapinski et al. 2013), could suggest that the binding of these factors is governed by alternative rules in different cellular contexts. More likely, however, is the possibility that the pluripotent ESCs lack some of the repressive elements of a differentiated cell type that are present for stable repression of lineage-inappropriate gene targets. If bHLH factors exhibit preference for specific loci, the bound sites likely feature different, lineage-dependent epigenetic states.
One surprising feature identified in the bHLH bound regions was a clear distinction in motif density and distribution in open versus closed chromatin. This property is specific to the ASCL1, ASCL2, and MYOD1 subset of class II bHLH factors and does not extend to the other bHLH factors NEUROD1 or TCF12. The canonical model of bHLH function is predicated on a single dimeric complex interacting with a single Ebox motif. An increase in Ebox motif density may provide a means by which transcription factors can access closed DNA, via increased recruitment due to the presence of multiple Eboxes. The striking spacing of the Eboxes identified suggests helical patterning, which may be of particular significance when viewed in the quaternary complex of an intact nucleosome. This suggests that a series of Ebox motifs may be accessible on the surface of a nucleosome, potentially serving as a beacon for bHLH binding, or as a pathfinder, directing bHLH factors to a specific binding site necessary for enhancer function. Additionally, although the results of our de novo motif discovery are not suggestive of bHLH-cofactor interactions as a primary driver of factor-specific binding, the presence of multiple Eboxes on adjacent helical turns may be evidence of tetrameric bHLH:bHLH complexes, as recently shown for the TWIST family of transcription factors (Chang et al. 2015).
In summary, ASCL1, ASCL2, and MYOD1 bind to distinct sites and direct lineage-specific transcriptional programs in their respective tissues. Here, we demonstrated that these factors maintain distinct binding when ectopically expressed in undifferentiated cells, a property that cannot be fully explained by dependence on distinct motifs. They possess intrinsic specificity that goes beyond simply binding their specific Ebox motif in accessible genomic sites. These results suggest that additional regulatory complexity is needed to direct bHLH transcription factor binding selectivity. Indeed, the extent of factor-specific binding suggests that these factors can identify their many cognate binding sites even when the loci are present in closed chromatin. DNA sequence at the bHLH bound sites has not identified cofactors that would explain distinctions in binding specificity. However, this does not preclude the possibility that cofactors influence the binding of these factors. Additional mechanisms must be explored going forward, including factor-specific interactions with distinct bHLH complex binding partners (such as E-proteins), with non-DNA binding proteins, and/or with noncoding RNA (ncRNA). The role of an ncRNA as a site selective cofactor was previously demonstrated (Zhao et al. 2008), and ncRNAs are known to interact with MYOD1 in skeletal muscles (Yu et al. 2017). Future efforts to uncover cofactors, whether protein or ncRNA, that modulate specificity of the class-related bHLH factors are necessary.
Methods
Cell culture
Murine ESCs engineered to express a specific TF upon removal of DOX from the growth media were generated by Dr. M. Ko at the National Institute of Aging and are available from the Coriell Institute for Medical Research (Nishiyama et al. 2009). Cells were grown essentially as previously described (Nishiyama et al. 2009) on Mitomycin-C treated SNLP (puromycin resistant murine embryonic fibroblast feeder cells), passaged every 48 h to fresh plates and were fibroblast-depleted prior to experiments by serial passage on gelatinized plates without feeder cells. Induction of TF expression by removal of DOX was accomplished by three serial washes in phosphate buffered saline (PBS) every 3 h followed by replacement of the media with no DOX. Cells were collected 24 h post-induction for all experiments. VENUS fluorescence was used to confirm induction of transgene prior to harvest.
Chromatin preparation and ChIP-seq
Trypsin-dissociated ESCs (∼107 cells) were washed in ice-cold PBS and fixed for 10 min at 27°C on a benchtop rotator in neutral buffer containing 1% formalin. Fixed, whole cells were washed, lysed, and sonicated in siliconized microfuge tubes in an ice-chilled Diagenode Bioruptor for a total of 35 min, using a 15:15 sec on/off cycle. Chromatin immunoprecipitation was performed with an overnight incubation at 4°C with 5 µg of mouse anti-MASH1 (BD Pharmingen 556604) for ASCL1 ChIP or mouse anti-FLAG (Sigma-Aldrich F1804) antibody for ASCL2 and MYOD1 ChIP. The FLAG fusion moiety in ESCs with inducible ASCL1 was not recognized by FLAG antibodies, so antibodies specific to ASCL1 were used (Nishiyama et al. 2009). Bound DNA fragments were isolated after 4–6 h incubation at 4°C with 25 µg Protein G Dynabeads (Life Technologies 10003D). Purified samples were tested for enrichment of previously identified regions by RT-qPCR. Duplicate ChIP purified samples were pooled prior to library preparation to generate a sufficient template for library generation. Samples were prepared as ChIP libraries using NEB Next library preparation kit and Illumina multiplexing primers. Samples were sequenced using an Illumina HiSeq 2500 line.
RNA preparation
RNA was purified from ESCs (107 cells) in parallel with chromatin preparation using RNA lysis buffer (Zymo Research, R1054). RNA purification was performed using a small volume column elution as per the Zymo Research provided protocol, including 15 min DNase I treatment to remove residual trace DNA prior to column elution (Zymo Research, R1054). Induction of the TF mRNA was verified by RT-qPCR prior to RNA-seq.
Assay for transposase-accessible chromatin (ATAC-seq)
ATAC-seq was performed as per Buenrostro et al. (2013, 2015). All ATAC-seq experiments were performed using 50,000 cells and amplified using a Bio-Rad C1000 thermocycler. Nextera sequencing chemistry was used to allow for demultiplexing of ATAC-seq libraries.
Bioinformatic and computational analysis
Sequencing data were aligned to the mouse mm10 genome (GRCm38) (Kent et al. 2002, 2010) using Bowtie 2 v2.2.6 (Langmead et al. 2009). Peak calling and de novo motif discovery was performed using HOMER (Heinz et al. 2010). Hidden Markov modeling was performed using ChromHMM v1.12 (Ernst and Kellis 2012). Motif spacing analysis was performed using an in-house informatics package. Additional information regarding analysis software and parameters can be found in Supplemental Methods.
Use of previously published data sets
All previously published data sets were processed as described for experiments performed in this study. A compendium of the GEO accession numbers can be found in Supplemental Methods.
Data access
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE97715.
Supplementary Material
Acknowledgments
We acknowledge the many hours of helpful discussions with members of the Johnson laboratory, and critical reading of the manuscript by Drs. Tae-Kyung Kim, Raymond MacDonald, Helen Lai, Zhenzhong Ma, TouYia Vue, and Karine Pozo. We are grateful for the excellent NGS sequencing services provided by the UT Southwestern Microarray/NGS-Sequencing Core (Dr. Wakeland, Director) and the engineered ESCs provided by Dr. Ko, National Institutes of Aging. This work was supported by the National Institute of Neurological Disorders and Stroke (National Institutes of Health) grant R01 NS032817 to J.E.J.
Author contributions: The study was conceptualized by B.H.C. and J.E.J.; methodology was designed by B.H.C., R.K.K., and J.E.J.; software and formal analysis were the responsibility of R.K.K.; the investigation was carried out by B.H.C. and K.P.; J.E.J. was responsible for resources; the original draft was written by B.H.C. and J.E.J.; additional writing, review, and editing were done by R.K.K. and K.P.; supervision and funding acquisition were the responsibility of J.E.J.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.224360.117.
References
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 21: 465–477. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Weintraub H. 1990. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250: 1104–1110. [DOI] [PubMed] [Google Scholar]
- Borromeo MD, Meredith DM, Castro DS, Chang JC, Tung KC, Guillemot F, Johnson JE. 2014. A transcription factor network specifying inhibitory versus excitatory neurons in the dorsal spinal cord. Development 141: 2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. 2012. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10: 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. 2006. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J 25: 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. 2014. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 30: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, et al. 2011. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev 25: 930–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AT, Liu Y, Ayyanathan K, Benner C, Jiang Y, Prokop JW, Paz H, Wang D, Li HR, Fu XD, et al. 2015. An evolutionarily conserved DNA architecture determines target specificity of the TWIST family bHLH transcription factors. Genes Dev 29: 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364: 412–420. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet 27: 187–190. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol 25: 3997–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T, Fass D, Arnaud M, Harrison SC. 1994. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev 8: 970–980. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Church GM, Tonegawa S, Gilbert W. 1985. B lineage–specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science 227: 134–140. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Prendergast GC, Ziff EB, Burley SK. 1993. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363: 38–45. [DOI] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ. 2012. Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev Cell 22: 721–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Johnson NM, Farr GH III, Maves L, Tapscott SJ. 2015. Conversion of MyoD to a neurogenic factor: binding site specificity determines lineage. Cell Rep 10: 1937–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev 11: 436–450. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci 89: 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT. 1988. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem 57: 159–197. [DOI] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 10: 1670–1682. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. 1993. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75: 463–476. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. 1994. Essential role of Mash-2 in extraembryonic development. Nature 371: 333–336. [DOI] [PubMed] [Google Scholar]
- Guturu H, Doxey AC, Wenger AM, Bejerano G. 2013. Structure-aided prediction of mammalian transcription factor complexes in conserved non-coding elements. Philos Trans R Soc Lond B Biol Sci 368: 20130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Birren SJ, Anderson DJ. 1990. Two rat homologues of Drosophila achaete-scute specifically expressed in neuronal precursors. Nature 346: 858–861. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Birren SJ, Saito T, Anderson DJ. 1992. DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc Natl Acad Sci 89: 3596–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26: 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Kyriazopoulou-Panagiotopoulou S, Libbrecht M, Smith CL, Raha D, Winters EE, Johnson SM, Snyder M, Batzoglou S, Sidow A. 2012. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res 22: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlands K, Yin X, Anand G, Prochownik EV. 1997. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J Biol Chem 272: 19785–19793. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Paterson BM, Weintraub H. 1986. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 47: 649–656. [DOI] [PubMed] [Google Scholar]
- Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, et al. 2014. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature 507: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo A, Guanga GP, Rose RB. 2008. Crystal structure of E47–NeuroD1/beta2 bHLH domain–DNA complex: heterodimer selectivity and DNA recognition. Biochemistry 47: 218–229. [DOI] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. 1994. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77: 451–459. [DOI] [PubMed] [Google Scholar]
- Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith DM, Borromeo MD, Deering TG, Casey BH, Savage TK, Mayer PR, Hoang C, Tung KC, Kumar M, Shen C, et al. 2013. Program specificity for Ptf1a in pancreas versus neural tube development correlates with distinct collaborating cofactors and chromatin accessibility. Mol Cell Biol 33: 3166–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, Orkin SH. 1993. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol 13: 3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA. 2016. Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks. Development 143: 2696–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56: 777–783. [DOI] [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. 1994. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta 1218: 129–135. [DOI] [PubMed] [Google Scholar]
- Nakada Y, Hunsaker TL, Henke RM, Johnson JE. 2004. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development 131: 1319–1330. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. 2009. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 5: 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijers J, Junker JP, Mokry M, Hatzis P, Koo BK, Sasselli V, van der Flier LG, Cuppen E, van Oudenaarden A, Clevers H. 2015. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell 16: 158–170. [DOI] [PubMed] [Google Scholar]
- Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CE, van Ijcken WF, Grosveld F, Rudnicki MA. 2012. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell 47: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. 2015. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161: 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13: 613–626. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. 2009. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346: 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22: 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. 2013. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155: 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Lockshon D, Lassar A. 1990. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci 87: 5623–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Binder M, Funk W. 1991. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol 11: 4104–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zhang Y, Li T, Ma Z, Jia H, Chen Q, Zhao Y, Zhai L, Zhong R, Li C, et al. 2017. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat Commun 8: 14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sun H, Wang H. 2016. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci 6: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.