Abstract
Structural variants (SVs), including small insertion and deletion variants (indels), are challenging to detect through standard alignment-based variant calling methods. Sequence assembly offers a powerful approach to identifying SVs, but is difficult to apply at scale genome-wide for SV detection due to its computational complexity and the difficulty of extracting SVs from assembly contigs. We describe SvABA, an efficient and accurate method for detecting SVs from short-read sequencing data using genome-wide local assembly with low memory and computing requirements. We evaluated SvABA's performance on the NA12878 human genome and in simulated and real cancer genomes. SvABA demonstrates superior sensitivity and specificity across a large spectrum of SVs and substantially improves detection performance for variants in the 20–300 bp range, compared with existing methods. SvABA also identifies complex somatic rearrangements with chains of short (<1000 bp) templated-sequence insertions copied from distant genomic regions. We applied SvABA to 344 cancer genomes from 11 cancer types and found that short templated-sequence insertions occur in ∼4% of all somatic rearrangements. Finally, we demonstrate that SvABA can identify sites of viral integration and cancer driver alterations containing medium-sized (50–300 bp) SVs.
Structural variants (SVs) are a broad class of genomic alterations that includes deletions, duplications and insertions, inversions, and inter-chromosomal translocations, among other more complex topologies. SVs are an important source of variation in the human population (Sudmant et al. 2015) and substantially alter the structure of the genome in cancer (Beroukhim et al. 2010; Garraway and Lander 2013). In the germline, small to medium-sized events between 10 bp and 10 kbp are the primary source of structural variation (Mullaney et al. 2010). Large events (>10 kbp) and inter-chromosomal translocations are rare in the germline due to stringent selection against gene dosage imbalance, but are prevalent in cancer, where genomes are often unstable and suffer frequent complex events (Stephens et al. 2009). The junctions connecting two SV breakpoints may also involve the insertion of novel sequences created during DNA repair (Mahaney et al. 2009) or insertion of short fragments copied from elsewhere in the genome (Liu et al. 2011; Zhang et al. 2015).
Although inference from short-read alignments forms the core of most variant calling pipelines, properly aligning reads supporting SVs is particularly challenging due to the substantial heterogeneity of SV sizes and topologies. In contrast to single-nucleotide variants (SNVs), which affect only single base pairs, SVs frequently involve long stretches of the genome and are often supported by reads that are completely or partially unaligned (soft-clipped). Alignment is particularly inaccurate at complex junctions that may contain sequences derived from more than two genomic loci and at sites of integration of viral sequences where the viral-supporting reads will be left completely unaligned.
This diversity in SVs and indels has prompted the development of several alignment-based approaches and tools aimed at their detection. Small indels can often be inferred directly from the output of gapped read aligners as in BWA (Li 2013) or from local realignment of candidate reads as in Strelka (Saunders et al. 2012), GATK UnifiedGenotyper (DePristo et al. 2011), and FreeBayes (Garrison and Marth 2012). Longer indel variants can be obtained from direct realignment of clipped and unmapped reads, as in Pindel (Ye et al. 2009), or from targeted sequence assemblies such as in SOAPindel (Li et al. 2013), Scalpel (Narzisi et al. 2014), ScanIndel (Yang et al. 2015), BreaKmer (Abo et al. 2015), and laSV (Zhuang and Weng 2015). For larger SVs, the cornerstone of most alignment-based detection algorithms is the clustering of discordant mates, which are read pairs with insert sizes and relative orientations that differ substantially from expectation based on the physical library preparation (Tuzun et al. 2005; Korbel et al. 2007). Discordant mate clustering is often followed by a more focused step to realign clipped reads at the candidate variant sites, such with DELLY (Rausch et al. 2012), BreakPointer (Drier et al. 2013), and Meerkat (Yang et al. 2013). Some tools like LUMPY perform inference directly from multipart alignments (Layer et al. 2014).
Assembly-based algorithms provide a fundamentally different approach to variant calling, with global whole-genome de novo assembly being the most comprehensive implementation. In principle, longer contigs assembled from short reads can be more accurately aligned to the genome, enabling more sensitive detection of junction-spanning sequences supporting complex indels and SVs. However, whole-genome de novo assembly can be untenable in practice due to the large computational requirements, including significant amounts of memory (>60 GB for a 60× human genome) and substantial CPU time (>1000 h).
In contrast to whole-genome global assembly, local assembly can be used to assemble only reads with an initial alignment to some locus in the reference, significantly reducing computational requirements. Local assembly has been applied in a targeted fashion to detect SVs and indels in exons (Narzisi et al. 2014) and at sites of candidate SVs identified by alignment-based methods (Chen et al. 2014). Local assemblies can also be applied genome-wide by assembling continuous small windows tiled across the entire genome. Genome-wide variant detection from local assemblies has been described for indel and SNP detection with Platypus (Rimmer et al. 2014) and HaplotypeCaller (DePristo et al. 2011) and has recently been described for SVs with novoBreak (Chong et al. 2016) and Manta (Chen et al. 2016).
Here, we describe structural variation analysis by assembly (SvABA), a unified tool to efficiently detect SVs and indels genome-wide using local assembly. The basic idea of this approach is to perform local assembly to create consensus contigs from sequence reads with divergence from the reference and to apply this procedure to every region of the genome. The contigs are then compared to the reference to annotate the variants. By uniting the different classes of variant-supporting reads into a single framework, we further expect that this assembly-first approach would be effective for variants of all sizes and require few parameters. We evaluate the performance of SvABA for detecting both small indels and large SVs, a gap not well covered by current SV analysis methods. We further evaluate SvABA's ability to detect multipart complex rearrangements containing previously unmapped or poorly mapped reads.
Results
Detection of SVs and indels with SvABA
SvABA assembles sequences from multiple classes of read alignments to discover SVs, indels, complex rearrangement junctions, and sites of viral integration (Fig. 1A). Assembly is applied in local 25-kbp assembly windows, which are tiled sequentially with 2-kbp overlaps to cover the entire genome (Fig. 1B). In the initial read retrieval phase within a window, SvABA extracts all sequence reads with significant divergence from the reference, including soft-clipped, gapped, discordant, and highly mismatched alignments, as well as unmapped sequences. These sequences are down-sampled at sites of high-coverage pileups and trimmed to remove low optical-quality bases (Phred score <5) and very low-complexity sequences (repeats ≥30 bp) (Supplemental Fig. S1). Candidate discordant reads are realigned to the reference with an integrated implementation of BWA-MEM (Li 2013) to enumerate all possible alignment sites across the genome for a single read. A discordant read that has a candidate realignment near its mate pair, thereby producing a more parsimonious nonvariant alignment, is discarded as a false positive discordant read. SvABA then uses discordant read clusters to connect two or more local assembly windows together to produce a single collection of reads. This allows variant-supporting reads to be assembled together regardless of which end of the SV breakpoint they align to, thereby increasing the power to detect large and inter-chromosomal variants (Methods).
Figure 1.
Overview of the SvABA structural variation detection tool. (A, left) SvABA uses String Graph Assembler (SGA) to assemble aberrantly aligned sequence reads that may reflect an indel or SV. Such reads include gapped alignments (for indels), clipped alignments (for medium and large SVs), and discordant read pairs (for large SVs). In addition to detecting indels and SVs, SvABA can identify complex rearrangement junctions (middle) and sites of viral integration (right). (B) The workflow for the SvABA pipeline: (1) reads within a small window are extracted from one or multiple BAM files and discordant reads are clustered; (2) discordant reads are realigned to the reference to remove pairs that have a candidate nondiscordant alignment; (3) the discordant read clusters are used to identify additional regions where reads should be extracted; (4) the sequences are error-corrected with BFC and assembled with SGA into contigs, and contigs are immediately aligned to the reference with BWA-MEM; (5) contigs with multipart alignments or gapped alignments are parsed to extract candidate variants; and (6) sequence reads are aligned to the contig and to the reference to establish read support for the reference and alternative haplotypes.
The collection of sequences from one or more joined assembly windows are then error-corrected using BFC (Li 2015) and FM-indexed and assembled with String Graph Assembler (SGA) (Simpson and Durbin 2012). The assemblies are exhaustive so that contigs are produced from each allele, providing for detection of multiple variants in close proximity. The assembled contigs are then aligned to the reference genome with BWA-MEM. Contigs that align to the genome with gapped alignments produce candidate indels, and contigs with multipart alignments produce candidate SVs (Supplemental Fig. S2). The level of support for candidate variants is obtained by aligning the read sequences to their corresponding contigs and determining the number of reads that support the variant haplotype. Variants are then scored based on the number of supporting reads and the quality of the contig alignment to the reference genome (Methods).
SvABA can perform genome-wide local assembly and SV calling on a 30× genome with ∼7 GB of memory and ∼40 CPU hours, orders of magnitude faster than global assemblies. The speed and efficiency of SvABA draws from the fusing and refactoring of several well-established C and C++ tools (SGA, htslib, BWA-MEM) into a single unified process, enabling in-memory manipulation of objects representing sequences, alignments, and assemblies with a minimal RAM and I/O footprint (Wala and Beroukhim 2017). SvABA can process local assembly windows in parallel, enabling a linear increase in speed by using additional CPU cores (set with a simple flag). The minimum input to SvABA consists only of a target reference genome and one or more alignment files (BAM, SAM, or CRAM format) produced by standard alignment algorithms. The primary outputs are variant call files (VCFs) representing the indels and SVs.
Sensitive detection of indels and SVs in NA12878
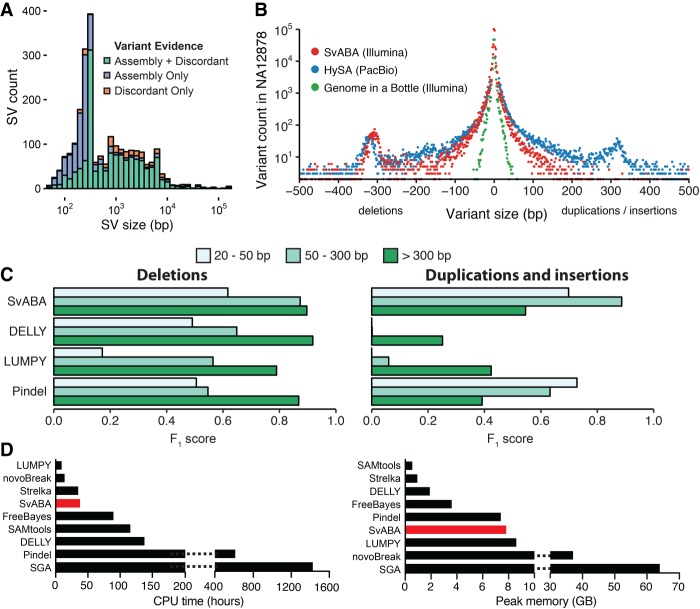
We used the widely studied NA12878 human genome to benchmark variant detection from a single germline sample. We ran SvABA on NA12878 whole-genome data sequenced to a mean coverage of 78.6-fold with 151-bp Illumina paired-end reads. Among variant-supporting contigs, the median contig length was 307 bp for indels (N50 = 330 bp) and 457 bp (N50 = 650) for SVs (Supplemental Fig. S3). SvABA identified 4626 deletions (>50 bp), 2176 duplications/insertions (>50 bp), 634 inversions, 196,068 small insertions (≤50 bp), and 225,801 small deletions (≤50 bp). Among small variants (≤50 bp), 97.1% were represented in the dbSNP database. Agreement with dbSNP was highly size dependent, with 97.6% of variants ≤20 bp represented in the dbSNP database, compared with only 80.3% for variants between 20 and 50 bp (Supplemental Fig. S4). Despite this lower representation, variants of 20–50 bp exhibited nearly the same level of variant-spanning reads (mean 33.7 reads) as small variants (≤20 bp; mean: 38.0 reads), suggesting that true large indels are underreported in dbSNP.
SvABA integrated the different read signals to detect SVs using assembled contig realignment, discordant read clusters, or a combination of both signals. Detection of SVs from multipart alignments of assembled contigs (as opposed to discordant read alignments or gapped alignments) was a particularly important source of evidence for smaller variants (Fig. 2A). Among the SVs below 75 bp, 78.3% were identified through assembly and 21.7% from discordant reads only. We thus find that a substantial amount of small structural variation can be found even in the absence of initial discordant read signals.
Figure 2.
Detection of SVs and indels in the NA12878 human genome. (A) The number of SV events and the types of supporting evidence used by SvABA for detecting SV events of different lengths (indel variants not shown). SVs are detected through realignment of assembly contigs (purple), discordant read clusters (orange), or a combination of both (green). SVs with shorter lengths than the average size of the sequencing fragments are identified almost exclusively through assembly and realignment. (B) The length distributions of indels and small SVs in NA12878 determined from different sequencing and analytical technologies: 151-base paired-end Illumina sequencing by SvABA (red), HySA calls from PacBio sequencing data (blue), and the indel call set of the Genome in a Bottle consortium (green). (C) Comparison of detection accuracy of SvABA, LUMPY, DELLY, and Pindel for deletions (left) and for insertions/duplications (right) across three different length regimes in NA12878. The F1 score is a combined measure of precision and recall and was calculated using the PacBio assemblies and Genome in a Bottle (GIAB) as a truth set. (D) Total CPU and peak memory usage for several indel and SV detection tools applied to a single 33× human genome. SGA CPU and memory usage were estimated using published data (Simpson and Durbin 2012).
We next evaluated the performance of SvABA using two truth sets: indels from the Genome in a Bottle (GIAB) integrated call set (Zook et al. 2014) and SVs and indels from HySA (Fan et al. 2017), which jointly analyzes Pacific Bioscience (PacBio) and Illumina reads. The GIAB integrated call set provides high-confidence short indels by combining 13 different short-read variant callers applied to short reads sequencing data but is not designed to detect larger variants. The substantial length of the PacBio assembly contigs makes them an appropriate platform for detecting larger SVs as well as short indels. We reasoned that the GIAB set and SVs from HySA would provide complementary evidence and cover the full range of variants sizes called by SvABA.
SvABA detected a significantly greater number of small SVs and indels than are present in the GIAB set and exhibited a variant size distribution very similar to that of the HySA variants (Fig. 2B). Among the short indels (≤50 bp) called by SvABA, 77.0% were contained in either the GIAB or HySA call set. The proportion of SvABA indels in the GIAB set was strongly size dependent, with indels <10 bp being nearly four times more likely to be present in the GIAB set than indels >40 bp (Supplemental Fig. S5). Private SvABA calls were significantly enriched for heterozygous variants (P < 0.001, Fisher's exact test), which in many cases appeared to be at sites with only one allele represented by the HySA set.
We next used the HySA and GIAB calls as a truth set to compare the detection performance of SvABA versus three SV callers: LUMPY, Pindel, and DELLY. We selected these callers because of their wide use in the field and because of their different approaches to SV detection, including combinations of inference from discordant reads, split read alignments, and local sequence realignment within each tool. For each of three different size regimes (20–50 bp, 51–300 bp, and 300+ bp), we calculated the F1 score, which is a combined measure of the sensitivity and specificity.
SvABA was sensitive to events across the full range of sizes (Fig. 2C) and exhibited the greatest sensitivity for duplication and insertion variants (Table 1; Supplemental Fig. S6). For medium-sized duplication events between 50 and 300 bp, SvABA detected 1.9-fold as many true events as the next most sensitive method, Pindel. For small deletions (20–50 bp), Pindel and SvABA exhibited the greatest overall sensitivity, with SvABA identifying 1.1-fold more variants than Pindel and with a 1.3-fold lower unvalidated rate. For large deletions, all four methods achieved similar sensitivity and specificity.
Table 1.
SV and large indel detection in NA12878, validated against PacBio assemblies and Genome in a Bottle indels

SvABA uses local read depth and variant read counts to genotype indels and small SVs (<300 bps) using the model described by Li (2011) for SNPs. To evaluate the genotyping accuracy of SvABA, we jointly called variants on a whole-genome sequencing trio from an Ashkenazi Jewish family (Zook et al. 2016). Indels and small SVs comprised 98.9% of all indels and SVs in this trio. SvABA achieved a heterozygous/homozygous ratio of 1.1:1 for variants identified as heterozygous in both parents (theoretical value: 1:1) (Supplemental Fig. S7). Additionally, 97.2% of variants identified as homozygous in each parent were called as homozygous in the son.
SvABA achieved these results without requiring extensive CPU or RAM allocations. A major obstacle to assembly-based variant detection has been the computational requirements needed for de novo assembly. However, SvABA assembled the NA12878 data in 3150 CPU minutes and with 7.7 GB of memory that was primarily used to store the indexed reference genome. Based on a test of a 33× whole genome, SvABA required fewer CPU resources than all other detection tools except LUMPY and novoBreak (Fig. 2D). With the native parallelization in SvABA, we distributed the compute over 12 cores to call variants in just over 5 h of wall-clock time (313 min).
Validation of SvABA using an in silico tumor model
Detection of somatic SVs and indels in cancer poses significant challenges beyond those faced in detecting germline events. Somatic rearrangements involve a higher rate of large and inter-chromosomal events (Yang et al. 2013) and are often clustered tightly with other rearrangements as part of complex events like chromothripsis (Stephens et al. 2011). Somatic variants must also be distinguished from the background of germline variation. These challenges are further amplified by wide variability in relative amounts of normal and tumor cells in tissue samples. We therefore set out to separately validate SvABA as a tool for detecting somatic SVs and indels.
Due to the difficulty of obtaining gold-standard truth sets for somatic SVs from real data, we generated an in silico tumor-normal pair that more closely reflects the unique challenges of SV detection in the cancer genome. The frequency of somatic copy-number events is known to be inversely proportional to the length of the event (Fudenberg et al. 2011; Zack et al. 2013), and we recapitulated this in our simulation by creating a range of indels and rearrangements that mirrored this length distribution (Supplemental Fig. S8). We also spiked in 2000 short (≤10 bp) indels to reflect the high indel rates seen in real tumors. To simulate a sample with low tumor purity, we mixed the simulated tumor reads with real reads from the HCC1143BL lymphoblastic normal cell line at 30× coverage. We then called somatic variants on our in silico tumor using SvABA, FreeBayes, Strelka, DELLY, LUMPY, novoBreak, and Pindel.
SvABA reached the greatest overall sensitivity among the six methods, detecting 87.4% of all variants and achieved the highest F1 score for each size range of variants (Table 2). SvABA was modestly more sensitive overall than FreeBayes and Strelka for indels (1.1-fold increase), but captured a substantially higher number of indels >20 bp (Fig. 3A). Pindel similarly achieved relatively broad coverage across different sizes, but was less sensitive to insertion variants than SvABA. DELLY and LUMPY performed similarly for both medium-sized and large SVs, with LUMPY achieving the lower false positive rate. However, SvABA substantially improved detection (1.6-fold increase) relative to DELLY and LUMPY for medium-sized SVs at similar false positive rates. After SvABA, novoBreak achieved the highest sensitivity for medium-sized SVs with sizes down to 100 bp. For large variants (>300 bp), SvABA, DELLY, novoBreak, and LUMPY exhibited largely similar performance, with SvABA achieving the highest sensitivity by a small margin.
Table 2.
Somatic indel and SV detection performance using an in silico tumor genome

Figure 3.
Benchmarking somatic variants with an in silico tumor. (A) True positive counts for indel calling (y-axis) as a function of variant size (x-axis) for SvABA (red), Pindel (blue), FreeBayes (orange), and Strelka (light blue). All callers achieved similar sensitivities for small somatic indels, while SvABA maintained high sensitivity for larger (>10 bp) indels. (B) Stacked bar chart of the number of SVs detected across all SV types (y-axis) as a function of variant size (x-axis). SvABA maintained sensitivity across variants of all sizes. novoBreak had the second highest sensitivity for medium and large variants after SvABA. Combining calls from a dedicated indel and SV caller (LUMPY and FreeBayes or DELLY and Strelka) improved overall sensitivity, but still left a gap for medium-sized SVs.
We further considered how the detection performance of SvABA would compare with the combined performance of using both an indel caller and an SV caller together. We paired the calls from DELLY and FreeBayes and the calls from LUMPY and Strelka to create two examples of using combined call sets. The two combined call sets reached similar performance as measured by the F1 score (LUMPY + FreeBayes: 0.865; DELLY + Strelka: 0.854), but both were lower than SvABA (0.911). In both cases, the combined callers differed most greatly from SvABA for variants between 50 and 300 bp, in which SvABA detected 1.5-fold more variants than either combined approach (Fig. 3B).
Detection performance in comparison with whole-genome de novo assemblies
We next evaluated the performance of SvABA in real data from a human tumor. We used data from two separate library preparation and sequencing strategies in the HCC1143 breast cancer cell line and its paired lymphoblastic normal line. The first data set was sequenced from libraries prepared with a standard Illumina PCR amplification step and 101-base paired-end reads. The second data set was sequenced from libraries prepared without the PCR amplification and using 250-base paired-end Illumina reads (Kozarewa et al. 2009).
To provide an alternative computational approach using the 250-base PCR-free reads, we performed whole-genome de novo sequence assembly using DISCOVAR de novo, the whole-genome de novo assembly successor to DISCOVAR (Weisenfeld et al. 2014) specifically designed to assemble 250-base reads. We extracted SVs and indels from the DISCOVAR de novo assemblies by aligning the DISCOVAR de novo contigs to the reference with BWA-MEM and then parsed the gapped and multipart alignments to produce variant calls (Supplemental Methods).
The three call sets exhibited substantial overlap, with the main difference being an increased sensitivity from the longer read lengths and a smaller sensitivity gain from the whole-genome de novo assembly as compared with genome-wide local assemblies (Fig. 4A). DISCOVAR detected the highest number of somatic variants (1538), followed by SvABA on the 250-base reads (1409) and then SvABA on the 101-base reads (1016). With the standard 101-base reads, SvABA achieved a high specificity, with 92.8% of variants being rediscovered in the 250-base reads. When comparing SvABA and DISCOVAR calls from the same data set (250-base PCR-free reads), SvABA detected 69.9% of DISCOVAR variants; conversely, 76.3% of SvABA variants were present in the DISCOVAR results. Relaxing the read-support threshold for the DISCOVAR calls increased the support for 250-base SvABA calls to 89.1%.
Figure 4.
Somatic variant detection in the HCC1143 breast cancer cell line using different sequencing and informatics approaches. (A) Comparison of combined somatic SV and indel detection in HCC1143 using: local assembly using SvABA with 101-base paired-end reads (purple), SvABA with 250-base paired-end PCR-free reads (orange), or global assembly using DISCOVAR de novo on 250-base paired-end PCR-free reads and SVlib to extract variants (green). (B) Somatic variant counts (y-axis) for DISCOVAR de novo (250-base PCR-free reads; green) and SvABA using 101-base (purple) or 250-base PCR-free reads (orange), as a function of variant size (x-axis). All methods have similar sensitivities across different sizes, except DISCOVAR de novo was more sensitive to short indels in simple repeats.
Variants detected with DISCOVAR and SvABA show nearly identical size distributions (Fig. 4B). The events that were discovered by DISCOVAR but not SvABA with either data set were highly enriched for events occurring near centromeres (P < 0.01, Fisher's exact test) and in simple repeats (P < 0.01). This is consistent with an improved ability of long reads and global de novo assembly to identify variants in regions of the genome that are difficult to align to, likely resulting from the reduced alignment ambiguity afforded by long sequences.
Somatic rearrangements frequently involve short templated-sequence insertion junctions
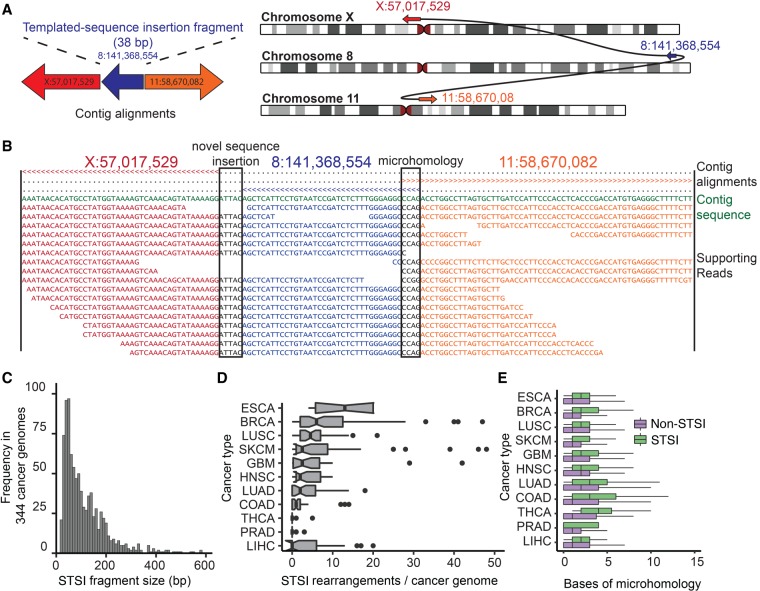
Complex events are increasingly recognized in both germline and cancer genomes (Stephens et al. 2011; Chiang et al. 2012). However, their detection is complicated when neighboring breakpoints are separated by distances on the order of the read length or greater, due to the difficulty in aligning short sequences covering such divergent sequences. We hypothesized that assembly-based methods might have superior sensitivity for such events. While inspecting the DISCOVAR and SvABA contigs from HCC1143, we identified multiple contigs that contained three or more sequences with high-quality alignments to disparate genomic loci, supporting putative complex events with multiple neighboring breakpoints (Fig. 5A).
Figure 5.
SvABA identifies rearrangements with short templated-sequence insertions (STSI) derived from distant genomic loci. (A) Somatic rearrangement between Chr X and Chr 11 in HCC1143 containing a 38-bp fragment of Chr 8. STSI rearrangements are identified by assembly contigs that have multiple non-overlapping alignments to the reference. The direction of the arrows represents the strand that the contig fragment was aligned to (right-facing is forward strand). (B) Partial view of the contig from A showing the multiple alignments of the contig to the reference and the read-to-contig alignments. The top three lines indicate which bp of the contig each of the three BWA-MEM alignments covers (> is forward strand alignment; < is reverse strand alignment). The first two alignments indicate an insertion of 5 bp of novel sequence at the first junction (left), and the second two indicate 4 bp of microhomology at the second junction (right). The middle alignment supports the STSI fragment. These plots are automatically generated by SvABA for each variant (in the *.alignments.txt.gz file). (C) STSI fragment lengths from somatic rearrangements across 344 cancer genomes (mean 86 bp). (D) Prevalence of STSI rearrangements (x-axis) across 11 tumor types (y-axis). (ESAD) esophageal cancer; (BRCA) breast cancer; (LUSC) lung squamous cell carcinoma; (SKCM) melanoma; (GBM) glioblastoma; (HNSC) head and neck squamous cell carcinoma; (LUAD) lung adenocarcinoma; (COAD) colorectal adenocarcinoma; (THCA) thyroid carcinoma; (PRAD) prostate adenocarcinoma; (LIHC) hepatocellular carcinoma. (E) Bases of breakpoint microhomology (x-axis) for different cancer types (y-axis) for somatic STSI rearrangements (green) and somatic non-STSI rearrangements (purple). The STSI rearrangements have a significantly higher degree of breakpoint microhomology than their non-STSI counterparts across all tumor types.
We therefore investigated whether SvABA could systemically discover complex events from the HCC1143 101 base read data. Among the SvABA contigs generated from the 101-base read data, we found eight contigs with high-quality multipart alignments (Supplemental Table S1). These complex contigs were well supported throughout their length by sequence reads, and we found no significant difference in the mean number of breakpoint-supporting reads between simple and complex events (52.4 reads in complex, 50.4 reads in simple, P = 0.69, t-test). There was no significant difference in the mean mapping quality of the alignments between simple and complex rearrangements (55.1 reads in complex, 58.5 reads in simple, P = 0.17, t-test). We concluded that these sequences represent true rearrangements containing short templated-sequence insertions (STSI), which we define as short sequences (<1000 bp) that match a sequence from another genomic locus not immediately contiguous with either rearrangement breakpoint (Fig. 5B). Rearrangements involving templated insertions have been described in the germline at the junctions of larger complex rearrangements (Liu et al. 2011) and in cancer in the context of chromothripsis events (Zhang et al. 2015), but have not been otherwise extensively described in cancer genomes. Therefore, we wished to specifically validate these events in HCC1143 and evaluate their prevalence across other cancers.
The STSIs from single contigs were short (median 56.5 bp), and we hypothesized that additional STSI rearrangements with longer insertions could be found by clustering together chains of rearrangements from multiple contigs with breakpoints separated by <1000 bp. This yielded 30 separate rearrangement clusters, including those from the single-contig rearrangements (Supplemental Table S2; Supplemental Fig. S9). The median fragment size across these clusters was 185 bp. Nine of the clusters involved contiguous chains of multiple STSIs, including cluster 25 that contained five contiguous STSI fragments. This cluster was supported throughout by a DISCOVAR contig.
We also confirmed the predicted sequences for eight STSI junctions by performing PCR spanning the junctions, including ones identified by single and multiple contigs (Supplemental Fig. S10; Supplemental Table S3). The rearrangements were validated as somatic, as none of these junctions were detected by PCR in the HCC1143BL normal cell line. These results confirm both the presence of these rearrangements and provide direct evidence that the multiple breakpoints are present on the same allele. Based on the close proximity of the breakpoints and the results of our validation, it is likely that the remaining clusters represent rearrangements from the same allele. Long-read sequencing would be required to systematically validate genome-wide the phasing of clustered rearrangements and rearrangements with STSI fragments longer than the library fragment size.
These STSIs are not restricted to the HCC1143 cell line but rather appear across a range of cancers. To test whether STSI junctions could be discovered across a range of tumor types, we ran SvABA on 344 TCGA whole-genome tumor-normal pairs (Supplemental Table S4). SvABA called 47,965 rearrangements, including 2124 events harboring STSI junctions (4.4% of all somatic rearrangements) (Fig. 5C). The number of STSIs per tumor was highly correlated with the number of simple events (R2 = 0.38). Esophageal carcinomas exhibited the highest rate of STSI rearrangements, with a median of 13 per sample, followed by breast cancer with six, and lung squamous carcinoma with 4.5 (Fig. 5D). Finally, although somatic rearrangements were largely mediated by nonhomologous repair, somatic STSI rearrangements contained more than twice the amount of breakpoint microhomology as non-STSI junctions (junctions from somatic rearrangements >300 bp without templated-sequence insertions; 2.82 bp in STSI, 1.28 in non-STSI, P < 0.001, t-test) (Fig. 5E). Although somatic rearrangements are thought to be largely mediated by nonhomologous repair, the subtle but significant difference in the length of homology between STSI and non-STSI junctions may reflect different underlying mechanisms generating these events.
The prevalence of STSIs in the cancer genome suggests that they may underlie oncogenic events. As an example of an oncogenic driver alteration formed in part by STSI events, we identified a focal amplification in a glioblastoma of the EGFR receptor tyrosine-kinase containing 52 non-STSI junctions and seven STSI junctions (Supplemental Fig. S11). Many of the reads supporting STSI junctions were initially unmapped and thus rescued by the local assemblies.
Application: identification of viral integration and medium-sized SVs in cancer
Insertion of DNA sequences from viral and bacterial pathogens represents an important mechanism of oncogenesis, but these events will often go undetected because pathogen sequences typically do not align to the reference genome. As a proof of principle of how SvABA might be used to detect foreign sequence insertion sites, we ran SvABA on 16 head and neck carcinomas and used the RefSeq viral sequence database as an alternative genome to look for evidence of integration of the human papillomavirus (HPV). HPV is a known oncovirus in head and neck cancer and is known to integrate into the genome of tumor cells (Parfenov et al. 2014). SvABA identified 16 breakpoints across seven samples where viral sequences were fused with genomic DNA (Supplemental Table S5). All of the viral junctions involved integration of HPV 16 into the genome, and were validated by comparison with Parfenov et al. (2014).
We next examined the contribution of small and medium-sized variants as potential driver events in cancer. Using the Cancer Gene Consensus list of cancer genes, we evaluated the relative burden of somatic indels and SVs in the exons of known cancer genes versus noncancer genes. Across all size regimes, small and medium-sized variants were significantly enriched in cancer genes (P < 0.01, Fisher's exact test) (Supplemental Fig. S12A).
Calling these 21- to 500-bp SVs may be necessary for accurate genotyping of cancer genes. For example, we identified a 62-bp frameshift deletion in exon 34 of NOTCH2 in a breast adenocarcinoma (Supplemental Fig. S12B). C-terminal NOTCH2 alterations have been found to be recurrent in B-cell lymphomas and lead to a gain-of-function product (Lee et al. 2009). We also identified a 44-bp tandem duplication in exon 2 of the TP53 tumor suppressor gene in a lung squamous cell carcinoma (Supplemental Fig. S12C), indicating loss of TP53 function. Based on these findings, we expect that additional driver alterations in this size regime could be discovered in future cancer genome analyses by incorporating genome-wide assembly-based detection methods like SvABA.
Discussion
We found that genome-wide local assembly exhibits broad sensitivity for indels and SVs across a range of variant sizes. Our assembly-based approach was particularly sensitive for variants between 20 and 300 bp and robustly identified complex rearrangement junctions containing short templated-sequence insertions and sites of viral integration. The ability to detect such a broad range of variants within a single framework represents an important advance toward achieving complete characterization of genomes from short-read sequencing data. As a demonstration of how our approach may be used to identify novel biologically relevant variants, we discovered several cases in which complex rearrangement junctions and small SVs contributed toward driver events in cancer.
Despite being primarily an assembly-based detection tool, we found that integrating both assembly and alignment signals improved the overall detection performance relative to either alone. Integrative approaches that combine multiple read signals in one inference framework, notably LUMPY and DELLY, have been previously shown to boost detection performance over any single approach. With SvABA, the addition of genome-wide local assembly provided an important signal for discovering medium-sized and complex variants while providing additional support for large variants with more robust discordant read evidence. In addition to implementing assembly-driven variant detection, SvABA provides several improvements over alignment-based approaches, including realignment of discordant reads and pair-mate region lookups to boost the read support without requiring any BAM preprocessing.
There are limitations to a local assembly-based approach. SvABA relies strongly on having sufficient variant reads to build an assembly contig. For low-coverage genomes or for highly impure tumor samples, the number of reads may not be sufficient to provide for robust assemblies. LUMPY, DELLY, and Meerkat, among other tools, have been specifically tuned to detect variants in genomes with low coverage and may be more sensitive than SvABA for low-coverage data. SvABA detection is based on the discovery of pairs of joined breakpoints and identifies copy-neutral inversions and more complex multipart rearrangements that cannot be inferred from read depth alone. However, read depth–based approaches may provide better genotyping accuracy for large copy-number variants (CNVs) and improve detection of CNVs that arise through homologous recombination where accurate identification of breakpoints is challenging. SvABA also relies on the approximately correct alignment of at least one read in a pair. Read pairs where both ends are unaligned or incorrectly mapped will not be accurately evaluated by SvABA. As such, although SvABA does not rely on any one particular alignment method, the initial alignments should be robust enough to place a sufficient number of read pairs approximately near the breakpoints. Indeed, we observed an increase in sensitivity with whole-genome de novo assemblies using DISCOVAR de novo, although its computational requirements make it currently infeasible to apply to large cohorts. Finally, SvABA uses BWA-MEM for contig alignment and variant calling. Though BWA-MEM enables very rapid alignment of long sequences to the reference, it may be more appropriate to use a more sensitive algorithm for highly divergent queries.
Even with improved informatics approaches like de novo assembly, SV detection in short-read sequencing data is ultimately limited by the read lengths—improved detection requires technologies that produce long-range information that can fully capture variation in repetitive regions or from highly complex rearrangements. Approaches such as PBHoney (English et al. 2014), MultiBreak-SV (Ritz et al. 2014), and HySA have been developed for extracting larger SVs from long sequences. Alternatively, short-reads may be tagged with DNA barcodes to yield libraries of linked-reads (e.g., from 10x Genomics). Linked reads are also particularly useful for long-range haplotyping of detected variants (Zheng et al. 2016).
We found that each of the tools we benchmarked against provided excellent calls within their targeted size regimes. For instance, we found Pindel to be quite sensitive for small deletion variants, whereas LUMPY, DELLY, and novoBreak achieved highly accurate detection of larger SVs. With SvABA, we have provided a single method that achieves high accuracy in both these size regimes and additionally covers the gap in between short indels and larger SVs. An alternative approach to using a single caller is to integrate results from multiples methods covering a variety of detection approaches, as was done by the 1000 Genomes Project to achieve broad SV sensitivity in their recent survey of 2504 human genomes (Sudmant et al. 2015). Integrating call sets is also valuable for increasing specificity, and we expect that SvABA will be a useful addition to large-scale sequencing efforts and consortium SV calling. For instance, SvABA has been recently used to generate somatic variant calls from 2961 cancer whole genomes as part of the International Cancer Genome Consortium (Campbell et al. 2017). We expect that SvABA's low computational burden and ease of running, and hence low cost to operate, coupled with its broad sensitivity and applicability to germline or cancer genomes, will continue to make it a practical and suitable tool for such large-scale analyses.
Methods
Read retrieval and error correction
SvABA extracts the following reads by default: alignments with high-quality clipped bases, discordant reads, unmapped reads, reads with unmapped pair-mates, and reads with deletions or insertions in the CIGAR string. Reads that are marked as PCR duplicates, failed QC reads, and reads with homopolymer repeats >20 bp are removed before assembly. SvABA additionally considers a read a duplicate if it has the same sequence, alignment position, and pair-mate position as another read. Sequencing reads are error-corrected using either BFC (Cibulskis et al. 2013) (default) or SGA (Supplemental Methods).
Discordant read realignment and clustering
The insert-size distribution for each read group is estimated from a sample of five million reads. Only read pairs with a forward-reverse pair orientation are used for estimating the insert size. To exclude read pairs with unusually large or small insert sizes that likely represent misalignment or true variants, the largest and smallest 5% of insert sizes are removed from the insert-size estimation. Reads with nonstandard pair orientations or outlier insert sizes greater than four standard deviations from the expected insert size for that read group are considered candidate discordant reads.
Due to the difficulty of aligning reads in nonunique regions of the genome, most discordant reads can be attributed to mapping artifacts rather than true variation. To reduce the effect of alignment artifacts on generating false positive variant calls, candidate discordant reads are realigned on-the-fly with BWA-MEM to the reference genome. Candidate discordant reads with an available nondiscordant alignment of >70% of the maximum alignment score are removed from discordant read analysis (Fig. 2B, step 2). Reads with more than 20 different high-quality candidate alignments are also removed from discordant read analysis since the true location of the read is ambiguous. The remaining discordant reads are clustered based on their orientation and pair-mate locations. Regardless of the results of the discordant read realignment strategy, the sequences of all candidate discordant reads are used in the local assemblies.
Pair-mate lookup and assembly window merging
To improve the power for detecting large rearrangements and SVs with breakpoints separated by more than the size of the local assembly window, candidate partner loci are identified from the discordant read clusters used to indicate additional genomic loci from which to extract reads prior to assembly. This also provides information about the mapping quality of the pair-mates of discordant reads, which is not typically stored in the alignment records of individual reads. To reduce the number of lookups of candidate partner loci in the BAM, SvABA uses a default threshold of six discordant reads to trigger a candidate lookup or three reads from the case BAM when run in case-control mode.
Contig alignment and candidate variant generation
Following assembly, SvABA aligns the assembly contigs to the full human reference using BWA-MEM and searches for evidence of variant-supporting alignments. The most conservative alignment for a contig is the one that aligns to within the local window from where the reads were extracted and with no candidate variant. To explicitly check for this possibility, the reference sequence from the local assembly window is extracted and indexed with BWA-MEM. Contigs are aligned to this local reference and excluded from further consideration if they have a high-quality nonvariant local alignment with fewer than 30 nonaligned bases and no alignment gaps.
Candidate indels are extracted from contigs that align to the reference with a gapped alignment, and candidate SVs are extracted from contigs with multipart alignments (Fig. 2B). High-quality secondary alignments, where a sequence fragment has multiple possible alignments for the same bases, are retained if they have an alignment score (AS) of >90% of the maximum AS, up to a maximum of 50 alignments. Although these alignments may support true variants, they are inherently ambiguous and likely overlap repetitive elements that are present at more than one copy in the reference genome. SvABA handles these contigs by reporting all the candidate variants, one for each of the possible secondary alignments, in an unfiltered VCF (Supplemental Fig. S2). These candidate rearrangements can then be disambiguated using copy-number data or other genome-wide analyses to select the most likely variant from the set of candidates.
Realignment of sequence reads to assembly contigs
To obtain the read support for a candidate variant, within each assembly window all the reads are aligned to both the contigs and the local reference sequence using BWA-MEM. To be considered a match to a contig, a read must have an AS >90% of the length of the match and have a higher alignment score to the contig than the reference. Clipped read-to-contig alignments are also considered, but only the matched portion is used to indicate read support. Alignment positions and CIGAR strings of the read-to-contig alignments are stored as a tag within with the reads and optionally emitted to a BAM file.
Read-to-contig alignments that span a candidate indel or breakpoint are used to obtain the variant read count. Reads that have an alignment of eight bases to the left and right of a variant site are considered a variant-supporting read. For variants that overlap simple repeats (e.g., homopolymer repeats), this minimum read-to-contig coverage is extended by the length of the repeat. To facilitate rapid review of the evidence for a given contig and variant, the read-to-contigs alignments and contig-to-genome emitted as ASCII plots in the *.alignments.txt.gz file (Fig. 5B).
Rearrangement and breakpoint annotation
SvABA annotates indels and SVs with breakpoint microhomology, the sequences of breakpoint insertions, and whether the contig contains evidence for short templated-sequence insertions (STSI). Microhomology bases are obtained from overlapping BWA alignments on the contig (Fig. 5B, second breakpoint). Conversely, breakpoint insertion bases are called when there is a gap between the two aligned fragments (Fig. 5B, first breakpoint). Rearrangements containing three or more alignments to the reference are annotated with a STSI field in the VCF, and represent STSI rearrangements. To be considered a true STSI rearrangement, both the leftmost and rightmost alignments in the contig coordinates must have a minimum BWA mapping quality of 30 and be supported by at least four breakpoint-spanning reads.
Indel variant scoring, filtering, and genotyping
Candidate indels are initially heuristically filtered to exclude variants from contigs with poor BWA mapping quality (<10), from contig fragments with multiple ambiguous matches, and from contigs with highly uneven coverage of supporting reads (<80% of contig covered by a high-quality read-to-contig alignment). Indels with an allelic fraction of <0.05 are also removed from the final call sets. All candidate indels failing a filter are output in the unfiltered VCF files.
For short-read sequencing by synthesis, the likelihood that a read contains an artificial indel is largely determined by the number of repeats at a site, with large homopolymer stretches being most likely to contain false indels (Ross et al. 2013). To obtain an estimate for the probability that a variant-supporting read is an artifact, SvABA measures the number of repeats in the reference genome immediately to the left and right of indel sites. Repeats are calculated by iteratively moving along the reference genome away from the indel until the repeat pattern is broken, for repeat units up to 5 bp. The repetitive sequences are reported in the VCF files. The total length of the repeat is then converted to an error rate estimate e provided in Ross et al. (2013).
The remaining indels are scored by calculating the log-of-odds (LOD) that a variant has a nonzero allelic fraction f versus homozygous reference (f = 0)
where fMLE is the maximum likelihood estimate for f obtained from the number of variant-supporting reads a divided by the total number of reads k. The default LOD cutoff is 8, or 6 if the variant is present in the dbSNP database.
SvABA will classify an event as germline or somatic if both case (tumor) and control (paired-normal) BAM files are supplied. This functionality can also be used to call de novo variants in trios (mother, father, proband child) or quads. Any number of BAM files can be supplied, and variants will be genotyped for each input sample. To determine if an indel is somatic, we follow a similar approach to the calculations performed by MuTect (Cibulskis et al. 2013). For each candidate somatic indel, the LOD that the indel is homozygous reference in the paired-normal (f = 0), rather than heterozygous (f = 0.5), is calculated from variant and reference read counts as above. The default LOD cutoff for somatic classification is 6.0, or 10.0 if the variant is present at a dbSNP site (and thus more likely to be a germline variant).
Software availability
SvABA is freely available under the GPLv3 license at https://github.com/walaj/svaba (commit a76f160) and as Supplemental Software.
Data access
The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRX3538696 (NA12878), SRX3538871, and SRX3546971 (HCC1143 101 base reads), and SRX3546970 and SRX3546969 (HCC1143 250 base reads). The DISCOVAR de novo assemblies, the simulated tumor genomes, and the somatic and large (>50 bp) variants are available at https://data.broadinstitute.org/svaba/ and as Supplemental Data. The germline variants (<50 bp) have been submitted to dbSNP (https://www.ncbi.nlm.nih.gov/projects/SNP/) with batch number 1062975 (release number B152) and submitter handle BEROUKHIMLAB.
Supplementary Material
Acknowledgments
The National Institutes of Health (T32 HG002295/HG/NHGRI, U54CA143798, and R01CA188228), DFCI-Novartis Drug Discovery Program, Voices Against Brain Cancer, Pediatric Low-Grade Astrocytoma Foundation, the Broad Institute, and the Cure Starts Now Foundation provided financial support. M.I. is supported by a Burroughs Wellcome Fund Career Award for Medical Scientists. We would like to thank Heng Li for helpful comments and as the primary developer of BWA-MEM, and Jared Simpson as the developer of SGA. We would also like to thank Ken Chen and Xian Fan for help with the HySA call set.
Author contributions: J.A.W., M.I., C.Z., M.M., and R.B. wrote the paper. J.A.W. developed SvABA and performed analyses. P.B. and R.O. performed the HCC1143 validation experiments. N.F.G., C.S., Y.L., J.W., P.C., G.G., and S.S. contributed methodological improvements. X.Y. performed the TCGA variant calling. T.S. and C.N. performed the HCC1143 data generation and DISCOVAR de novo assemblies. M.I., C.Z., and R.B. supervised the research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.221028.117.
References
- Abo RP, Ducar M, Garcia EP, Thorner AR, Rojas-Rudilla V, Lin L, Sholl LM, Hahn WC, Meyerson M, Lindeman NI, et al. 2015. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res 43: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Getz G, Stuart JM, Korbel JO, Stein LD, ICGC/TCGA Pan-Cancer of Whole Genomes Net. 2017. Pan-cancer analysis of whole genomes. bioRxiv 10.1101/162784. [DOI]
- Chen K, Chen L, Fan X, Wallis J, Ding L, Weinstock G. 2014. TIGRA: a targeted iterative graph routing assembler for breakpoint assembly. Genome Res 24: 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222. [DOI] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger SR, et al. 2012. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet 44: 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Z, Ruan J, Gao M, Zhou W, Chen T, Fan X, Ding L, Lee AY, Boutros P, Chen J, et al. 2016. novoBreak: local assembly for breakpoint detection in cancer genomes. Nat Methods 14: 65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, Lander ES, Meyerson M, Beroukhim R, Getz G. 2013. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res 23: 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Salerno WJ, Reid JG. 2014. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics 15: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Chaisson M, Nakhleh L, Chen K. 2017. HySA: a Hybrid Structural variant Assembly approach using next-generation and single-molecule sequencing technologies. Genome Res 27: 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Getz G, Meyerson M, Mirny LA. 2011. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat Biotechnol 29: 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. 2013. Lessons from the cancer genome. Cell 153: 17–37. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907 [q-bio.GN].
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. 2009. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat Methods 6: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15: R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Kumano K, Nakazaki K, Sanada M, Matsumoto A, Yamamoto G, Nannya Y, Suzuki R, Ota S, Ota Y, et al. 2009. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci 100: 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997 [q-bio.GN].
- Li H. 2015. BFC: correcting Illumina sequencing errors. Bioinformatics 31: 2885–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li R, Li H, Lu J, Li Y, Bolund L, Schierup MH, Wang J. 2013. SOAPindel: efficient identification of indels from short paired reads. Genome Res 23: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SCS, Dhar SU, Kołodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaney BL, Meek K, Lees-Miller SP. 2009. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 417: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney JM, Mills RE, Pittard WS, Devine SE. 2010. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet 19: R131–R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narzisi G, O'Rawe JA, Iossifov I, Fang H, Lee YH, Wang Z, Wu Y, Lyon GJ, Wigler M, Schatz MC. 2014. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat Methods 11: 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, Lee S, Hadjipanayis AG, Ivanova EV, Wilkerson MD, et al. 2014. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Scis 111: 15544–15549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SR, Wilkie AO, McVean G, Lunter G. 2014. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 46: 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz A, Bashir A, Sindi S, Hsu D, Hajirasouliha I, Raphael BJ. 2014. Characterization of structural variants with single molecule and hybrid sequencing approaches. Bioinformatics 30: 3458–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, Nusbaum C, Jaffe DB. 2013. Characterizing and measuring bias in sequence data. Genome Biol 14: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. 2012. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28: 1811–1817. [DOI] [PubMed] [Google Scholar]
- Simpson JT, Durbin R. 2012. Efficient de novo assembly of large genomes using compressed data structures. Genome Res 22: 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732. [DOI] [PubMed] [Google Scholar]
- Wala J, Beroukhim R. 2017. SeqLib: a C ++ API for rapid BAM manipulation, sequence alignment and sequence assembly. Bioinformatics 33: 751–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Yin S, Sharpe T, Lau B, Hegarty R, Holmes L, Sogoloff B, Tabbaa D, Williams L, Russ C, et al. 2014. Comprehensive variation discovery in single human genomes. Nat Genet 46: 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh CH, Zhang C, Ren X, Protopopov A, Chin L, et al. 2013. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Nelson AC, Henzler C, Thyagarajan B, Silverstein KA. 2015. ScanIndel: a hybrid framework for indel detection via gapped alignment, split reads and de novo assembly. Genome Med 7: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. 2015. Chromothripsis from DNA damage in micronuclei. Nature 522: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, Kyriazopoulou-Panagiotopoulou S, Masquelier DA, Merrill L, Terry JM, et al. 2016. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol 34: 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Weng Z. 2015. Local sequence assembly reveals a high-resolution profile of somatic structural variations in 97 cancer genomes. Nucleic Acids Res 43: 8146–8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, Salit M. 2014. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol 32: 246–251. [DOI] [PubMed] [Google Scholar]
- Zook JM, Catoe D, McDaniel J, Vang L, Spies N, Sidow A, Weng Z, Liu Y, Mason CE, Alexander N, et al. 2016. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data 3: 160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.