Abstract
FGFR-TACC fusions, including FGFR3-TACC3, have been identified as potential oncogenic drivers and actionable alterations in a number of different cancer types. The clinical relevance of FGFR3-TACC3 fusions in endometrial cancer has not yet been described. Formalin-fixed, paraffin-embedded metastatic endometrial carcinoma from the spleen and peritoneum were sent for comprehensive genomic profiling (CGP) using the FoundationOne platform as part of a prospective tumor genomic profiling protocol. We report the identification of an FGFR3-TACC3 fusion in a case of metastatic endometrioid endometrial cancer. Other potentially actionable alterations detected in this specimen included PIK3CA T1025S and an uncharacterized rearrangement involving TSC2. The patient initially received an FGFR inhibitor as an investigational agent and experienced stable disease with complete resolution of a pelvic nodule; however, treatment had to be discontinued because of intolerable side effects. A PET/CT scan nearly 3 mo after discontinuation showed disease progression. She subsequently received the mTOR inhibitor, temsirolimus, later accompanied by letrozole, and achieved stable disease. Clinical benefit was attributed to the mTOR inhibitor as tumor stained negative for estrogen receptor. Temsirolimus was discontinued after >17 mo because of disease progression. FGFR inhibitors may have clinical benefit in the treatment of endometrial carcinoma with FGFR3-TACC3 fusions. Additionally, clinical benefit from an mTOR inhibitor may reflect a response to targeting the alteration in PIK3CA or TSC2. More research is needed to understand the activity of FGFR3-TACC3 fusions on tumors and to discover additional therapeutic options for endometrial carcinoma patients with this gene fusion.
Keywords: endometrial carcinoma
INTRODUCTION
Significant progress in the development of comprehensive genomic profiling (CGP) platforms, analyses, and targeted therapies has culminated in the use of these modalities to reclassify and to treat many cancers, including endometrial carcinoma. Endometrial cancer, the most common gynecologic cancer, is predicted to account for 61,380 new cases of gynecologic cancers affecting women in the United States in 2017 (Siegel et al. 2016). Risk factors of endometrial cancer include obesity, diabetes mellitus, high blood pressure, tamoxifen treatment, and unopposed estrogen use (McConechy et al. 2012; Trabert et al. 2015; Morice et al. 2016). Current treatment modalities for women with endometrial cancer are surgery or surgery in combination with radiation therapy and/or chemotherapy, hormone therapy, and biologic therapy (National Cancer Institute 2002; National Comprehensive Cancer Network 2014). However, prognosis is worsened for women with recurrent or metastatic disease, as treatment options are limited and suboptimal (Temkin and Fleming 2009; Rauh-Hain and Del Carmen 2010; Lee and Secord 2014).
Advances in molecular genotyping have not only identified molecular heterogeneity driving tumor behavior (The Cancer Genome Atlas Research Network et al. 2013) but have also shed light on alternative approaches to therapeutically target tumors with specific mutations (Conley 2015; Meric-Bernstam et al. 2015; Tobin et al. 2015). Common alterations seen in endometrial cancers include mutations in the PI3K pathway (occurring in >80% of the endometrioid subtype) (Cheung et al. 2011), copy-number alterations (frequently observed in serous and mixed histology endometrial cancers, but also seen in endometrioid endometrial cancers; seen in such genes as ERBB2, FGFR3, and CCNE1), and frequent TP53 mutations (The Cancer Genome Atlas Research Network et al. 2013). FGFR alterations reported in endometrial cancers most frequently involve FGFR2 (Helsten et al. 2016). According to The Cancer Genome Atlas (TCGA) for data reported in Nature in 2013, the most frequently altered FGFR gene in endometrial cancers was FGFR2, with 12.5% of sequenced samples (n = 248 samples) harboring alterations in this gene. Gene fusions in endometrial cancers, although less common than copy-number alterations and point mutations, involve members of various pathways with recurrent translocations that most frequently involve genes of the BCL-2 family, followed by PI3K, WNT, EGFR, RAS–MAPK, protein kinase A, and retinoblastoma pathways (The Cancer Genome Atlas Research Network et al. 2013).
We report on a woman with a platinum-resistant recurrence of a metastatic endometrioid endometrial carcinoma with poorly differentiated, widespread carcinomatosis and splenic involvement. CGP performed on a tumor specimen revealed several potentially actionable alterations, including both an FGFR3-TACC3 fusion and a PIK3CA-activating mutation. Consensus opinion from our institutional molecular tumor board (MTB) led to enrollment of this patient on a clinical trial including an FGFR inhibitor and subsequent therapy with an mTOR inhibitor, to which she derived a prolonged clinical benefit. This case demonstrates the utility of tumor genomic profiling with the ability to identify gene rearrangements as a method of identifying potentially actionable targets in endometrial cancers.
RESULTS
Clinical Presentation and Family History
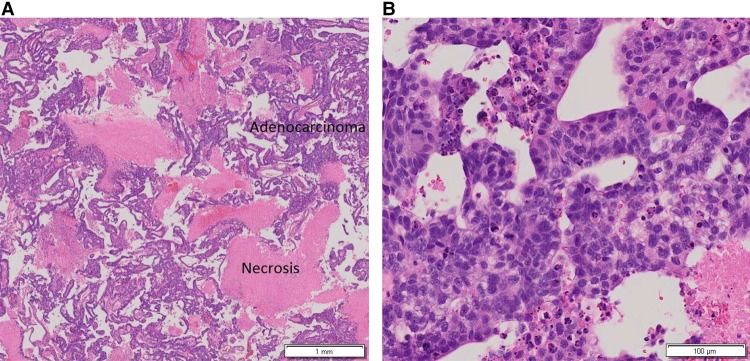
A 57-yr-old nulliparous, postmenopausal female presented with intermittent abnormal vaginal bleeding over 3 yr. Her history was significant for menarche at age 11, menopause at age 50, diabetes managed with metformin, body mass index of 29, and a remote 40 pack/year smoking history where tobacco cessation occurred 20 yr prior to diagnosis. She had no history of exogenous hormone use. She had a dilation and curettage 3 yr prior to the initial cancer diagnosis that showed no pathology. The patient continued to experience abnormal vaginal bleeding, and a second endometrial biopsy revealed moderately differentiated endometrioid adenocarcinoma, FIGO grade II, with extensive necrosis (Fig. 1A,B).
Figure 1.
Hematoxylin and eosin (H&E)-stained endometrial biopsy showing endometrial adenocarcinoma with papillary features and necrosis at low (A) and high (B) magnification, respectively.
She underwent an exploratory laparotomy with total abdominal hysterectomy and bilateral salpingo-oophorectomy. Pathology confirmed a high-grade, 8.5 cm × 8 cm × 5 cm endometrioid adenocarcinoma, FIGO grade II, with extensive necrosis and focal clear cell changes. There was <10% myometrial invasion. No lymphovascular invasion or cervix involvement was noted. Eighteen pelvic and para-aortic lymph nodes showed no metastases. Final staging was 1A (pT1a, N0), based on the 2009 FIGO staging guidelines. Because she had a high risk for recurrence, the patient received whole pelvis radiation therapy and brachytherapy on a clinical trial that was completed 9 mo prior to the presentation of recurrent disease.
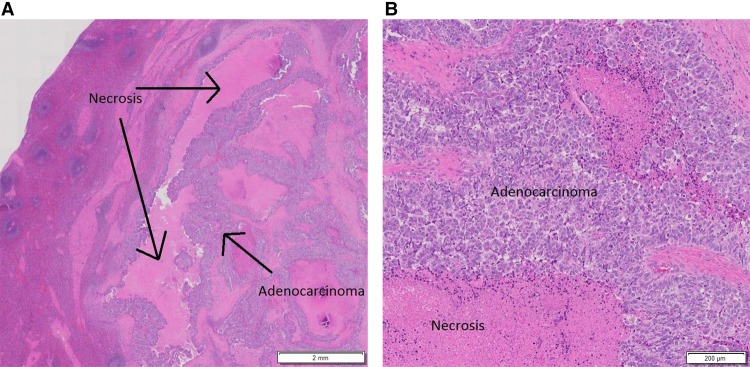
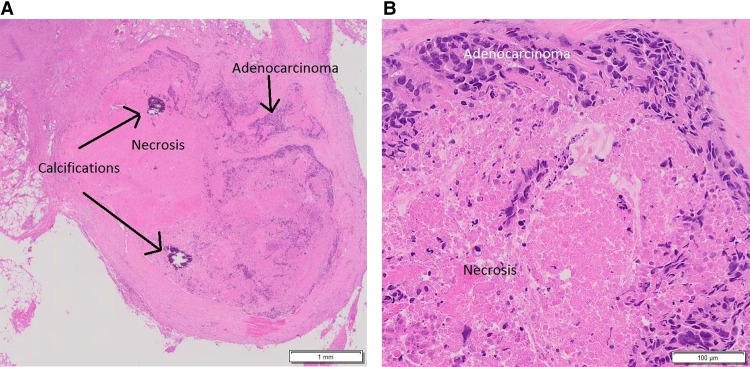
A CT scan of the abdomen and pelvis following completion of radiation showed stable hypodensities in the liver that were too small to characterize. However, 9 mo after completion of the trial, a follow-up CT scan showed a new 8-mm hypodense lesion in the splenic hilum and ring-like enhancement along the posterior margin of spleen not seen on the previous exam. A subsequent PET-CT scan revealed two abnormal hypermetabolic foci in the spleen (SUV 3.2 in the hilum, SUV 4.5 in a 1.2 cm hypodense lesion), stable non-FDG-avid liver lesions, and no pelvic adenopathy. The patient underwent splenectomy, and pathology was consistent with metastasis of the original tumor to the spleen (Fig. 2A,B). She received six cycles of intravenous carboplatin (AUC = 5) in combination with paclitaxel (175 mg/m2). Nearly 14 mo later, a CT scan of the chest, abdomen, and pelvis (CAP) revealed an increase in the size of previously visualized abdominal nodules, which were now suspicious for peritoneal carcinomatosis. A fine needle aspiration of the nodules within the abdomen and pelvis confirmed adenocarcinoma with extensive necrosis. A diagnostic laparoscopy was performed to resect tumor nodules (Fig. 3A,B), which were then sent for comprehensive tumor genomic profiling (CGP) under the auspices of a clinical trial.
Figure 2.
H&E-stained slide of tissue from splenectomy showing metastatic, moderately differentiated adenocarcinoma with necrosis at low (A) and high (B) magnification, respectively.
Figure 3.
H&E-stained slide showing metastatic adenocarcinoma with necrosis and calcifications following diagnostic laparoscopy with removal of the anterior abdominal wall nodule at low (A) and high (B) magnification, respectively; specimen from this procedure was sent for comprehensive genomic profiling.
Genomic Analyses
The patient provided informed consent to participate in the Rutgers CINJ genomic tumor profiling protocol (NCT02688517), which was approved by the Institutional Review Board (IRB) of Rutgers University New Brunswick Health Sciences (Pro2012002075). The specimen from the patient's laparoscopy underwent CGP using the FoundationOne platform, which consisted of whole-exon sequencing of a panel of 236 genes and 47 introns of 19 genes involved in translocations. Tumor genomic profiling revealed an FGFR3-TACC3 fusion (breakpoints at FGFR3 exon 17 and TACC3 exon 8), a missense mutation in PIK3CA (T1025S), and an uncharacterized rearrangement involving TSC2 (Table 1). A subthreshold amplification of the entire coding region of FGFR3 (copy number 6) was also detected, further validating the rearrangement. The results of tumor genomic profiling, clinical course, and pathology were reviewed at the Rutgers Cancer Institute of New Jersey Molecular Tumor Board (RCINJ MTB).
Table 1.
Variants identified from comprehensive genomic profiling
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect |
|---|---|---|---|---|---|
| FGFR3 | 4 | t(4;4) Chr 4 duplication fragment: 5′-FGFR3 (ex1-17 NM_000142)-TACC3 (ex8-16 NM_006342) Breakpoints: FGFR3 exon 17, TACC3 exon 8 |
N/A | Fusion | Functional |
| PIK3CA | 3 | N/A | p.T1025S | Substitution | Predicted functional |
| TSC2 | 16 | t(16;16) ∼206-kbp duplication fragment: 3′-TSC2 (NM_000548), Breakpoints: TSC2 exon 26, ABCA3 intron 25 |
N/A | Fusion | Unknown |
Targeted sequencing of the entire coding sequence was done for 236 genes and 47 introns of 19 genes involved in fusions at a depth of 500–1000× (median exon depth of 792×; 100% of the baited region was sequenced at a depth of at least 100×; sequence alignment error of only 0.27%). The depth of variant call reads was 59×; these 59 chimeric reads equate to an independently validated variant calling method described in Frampton et al. (2013). The depth of wild-type reads was 1155×.
HGVS, Human Genome Variation Society; N/A, not available.
Data suggest that FGFR3 fusion-containing cells are addicted to FGFR kinase activity and are sensitive to FGFR pan-inhibitors, such as pazopanib and PD173074 (Pollock et al. 2007; Lamont et al. 2011; Dieci et al. 2013; Williams et al. 2013). Given the preclinical study results, MTB recommendations to the treating oncologist included consideration of a trial using an FGFR inhibitor, a phase I trial of the combination carboplatin/paclitaxel/pazopanib, a trial with a PIK3CA inhibitor, an mTOR inhibitor, and off-label use of ponatinib or pazopanib.
Intriguingly, several variants of unknown significance were also reported in the tumor of this patient and included a TSC2 rearrangement. The TSC2 rearrangement was a 3′ tandem duplication of TSC2 with breakpoints in exon 26 and ABCA3 intron 25. This is an out of strand fusion event, because TSC2 and ABCA3 are transcribed in the positive and negative directions, respectively. The reciprocal event was not seen for this rearrangement. Additionally, there was a subthreshold amplification in TSC2 exons 26–42 (copy number 7) that further validate the rearrangement.
Treatment Outcomes
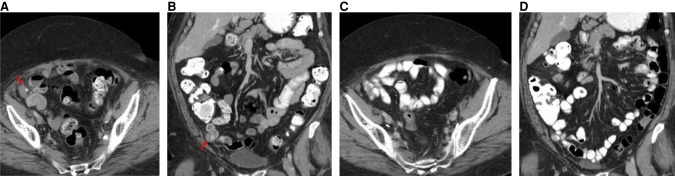
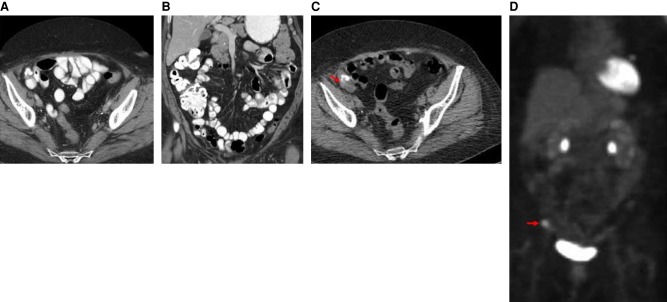
The patient was enrolled into a phase I clinical trial with an FGFR inhibitor. This MTB recommendation was based on the inference that the FGFR3-TACC3 fusion-containing tumor would be addicted to FGFR kinase activity. The patient experienced stable disease over the course of nearly 2 mo with complete resolution of a pelvic nodule, as observed in CT scans (Fig. 4A–D). However, at nearly four full cycles, the patient was removed from the study for grade 3 palmar-plantar erythrodysesthesia. The patient showed disease progression on a PET/CT scan about 3 mo following cessation of the FGFR inhibitor on trial (Fig. 5A–D).
Figure 4.
CT scans of the abdomen and pelvis with contrast prior to initiation of an FGFR inhibitor on trial. (A) Axial CT image with oral and intravenous contrast at the level of the mid pelvis shows a 3-cm soft tissue mass with minimal calcifications (red arrow). (B) Coronal reconstructed CT image with oral and intravenous contrast through the anterior abdomen and pelvis shows the 3-cm soft tissue mass with minimal calcifications (red arrow). CT scans of the abdomen and pelvis with contrast obtained during treatment with FGFR inhibitor on trial. (C) Axial CT image with oral and intravenous contrast at the level of the mid pelvis shows no measurable mass. (D) Coronal reconstructed CT image with oral and intravenous contrast through the anterior abdomen and pelvis shows no measurable mass.
Figure 5.
CT scans of the abdomen and pelvis with contrast shortly following discontinuation of trial with FGFR inhibitor. (A) Axial CT image shows no measurable mass. (B) Coronal reconstructed CT image shows no measurable mass. PET/CT scans 3 mo following cessation of the FGFR inhibitor on trial. (C) Axial CT image without oral or intravenous contrast at the mid pelvis shows a 3-cm mass with calcifications. (D) PET scan with coronal display shows an area of increased abnormal metabolic activity corresponding to the soft tissue mass (red arrow).
The patient was subsequently treated with the mTOR inhibitor temsirolimus based on recommendations from the MTB as an inferred downstream target of the PIK3CA alteration. Recommendations were not based on the TSC2 alteration, as this was a variant of unknown significance. She received temsirolimus for >17 mo with a few instances of the drug being withheld because of fever and neuropathy. Because of stable disease after >8 mo with foci showing mixed response and evidence from the treatment of breast cancer that mTOR inhibitors enhance the response to antiestrogen therapy, letrozole (2.5 mg/d orally) was added to the weekly regimen of temsirolimus (Baselga et al. 2009; Liu et al. 2014). The patient continued on temsirolimus and letrozole for nearly 9 additional months until disease progression was detected.
DISCUSSION
The FGFR family is made up of four active members, FGFR1, FGFR2, FGFR3, and FGFR4 (Turner and Grose 2010). Alterations in these genes, such as amplifications, mutations, or translocations, may lead to the constitutive or increased activation of the tyrosine kinases in their protein products (Dutt et al. 2008; Turner and Grose 2010; Singh et al. 2012). Both in vitro and in vivo studies on cancer cells and tumors containing FGFR fusions suggest oncogenic potential and, therefore, potential growth impairment with FGFR-specific and pan-kinase inhibitors for different tumor subtypes.
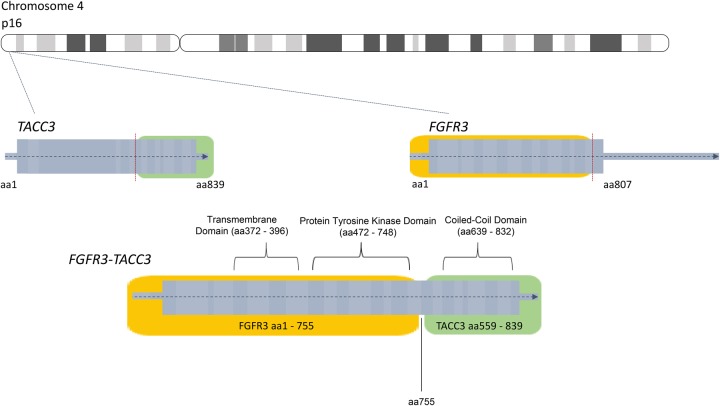
Because of the unusual presentation of recurrent endometrial cancer affecting the spleen, CGP was performed on the tumor in the aforementioned case. This revealed that the tumor contained an FGFR3-TACC3 fusion and a missense mutation in PIK3CA (Table 1). TACC3, or transforming acidic coiled-coil containing protein 3, is involved in mitotic spindle organization and possible stemness properties of cells (Zhou et al. 2015). It is positioned on Chromosome 4 upstream of FGFR3. The FGFR3-TACC3 fusion event is believed to occur through a tandem duplication of a region of the chromosome where both genes exist, 4p16.3 (Parker et al. 2013). The fusion event, through the help of the coiled-coil domain on TACC3, increases autophosphorylation of tyrosine residues on the FGFR3 kinase domain and leads to a constitutively active FGFR3 protein (Fig. 6; Nelson et al. 2016). The frequency of alterations in uterine cancer for the genes altered in our patient case was reported by The Cancer Genome Atlas (TCGA) (Fig. 7; Cerami et al. 2012; Gao et al. 2013).
Figure 6.
Diagram of the formation of the FGFR3-TACC3 fusion protein product.
Figure 7.
Frequency of alterations found in FGFR3, PIK3CA, or TSC2 reported in uterine corpus endometrial carcinoma (TCGA Provisional) for 242 cases. The results shown here are based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Alterations, including amplifications, deletions, and missense and nonsense mutations in FGFR genes, have previously been observed in endometrial cancers (Pollock et al. 2007; Dutt et al. 2008; Byron et al. 2012; Konecny et al. 2013; Helsten et al. 2016). Although the most frequently altered FGFR gene was FGFR2, with ∼15% of cases according to TCGA data, alterations in FGFR1, FGFR3, and FGFR4 have also been observed at a frequency of 6%, 5%, and 5%, respectively, in endometrial cancers (Fig. 8; Cerami et al. 2012; Gao et al. 2013).
Figure 8.
Percentages of uterine cancer cases with alterations in FGFR genes, as reported by TCGA for 242 cases (TCGA Provisional). The results shown here are based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
FGFR fusions, including FGFR-TACC fusions, have been observed among a variety of cancer types. For example, FGFR3-TACC3 fusions have been reported in a subset of bladder cancer (Williams et al. 2013; Wu et al. 2013), low-grade glioma (Granberg et al. 2017), glioblastoma (independently identified and reported by Singh et al. 2012 and Parker et al. 2013), nasopharyngeal carcinoma (Yuan et al. 2014), cervical cancer (Carneiro et al. 2015), head and neck squamous cell carcinoma (Wu et al. 2013), lung squamous cell carcinoma (Wu et al. 2013), and oral cancer (Wu et al. 2013). Helsten et al. (2016) reviewed the genomic profiling of nearly 5000 tumor samples from a variety of cancer types. They found one case out of 80 cases of endometrial cancer with an FGFR3-TACC3 fusion (Helsten et al. 2016). Additionally, data from Foundation Medicine, Inc. found four cases of endometrial cancer that contained an FGFR3-TACC3 fusion. Among 813 patients with endometrial adenocarcinoma (NOS), two patients (0.25%) had an FGFR3-TACC3 fusion. One of these patients included the patient featured in our case study. Additionally, one out of 64 patients (1.60%) with clear cell endometrial adenocarcinoma and one out of 419 patients (0.24%) with papillary serous endometrial adenocarcinoma had an FGFR3-TACC3 fusion. These data highlight the unique nature of these fusions in endometrial cancers. Details on the genomic breakpoints involved in these fusions are listed in Table 2. Our case sheds additional light on this rare but significant fusion event in endometrial cancer and follows the patient through treatment with targeted therapy.
Table 2.
Descriptions of the genomic breakpoints for the patients provided by Foundation Medicine, Inc.
| Disease ontologya | Genomic breakpoints |
|---|---|
| Uterus endometrial adenocarcinoma (NOS) | Chr 4 duplication fragment: 5′-FGFR3(ex1-18 UTR NM_000142)-TACC3(ex10-16 NM_006342) Breakpoints FGFR3 exon 18, TACC3 intron 9 |
| aUterus endometrial adenocarcinoma (NOS) | Chr 4 duplication fragment: 5′-FGFR3(ex1-17 NM_000142)-TACC3(ex8-16 NM_006342) Breakpoints FGFR3 exon 17, TACC3 exon 8 |
| Uterus endometrial adenocarcinoma clear cell | Chr 4 duplication fragment: 5′-FGFR3(ex1-2 NM_000142)-TACC3(ex7-16 NM_006342) Breakpoints FGFR3 exon 2, TACC3 intron 6 |
| Uterus endometrial adenocarcinoma papillary serous | Chr 4 duplication fragment: 5′-FGFR3(ex1-18 UTR NM_000142)-TACC3(ex10-16 NM_006342) Breakpoints FGFR3 exon 18, TACC3 intron 9 |
aHighlighted case results.
Di Stefano et al. (2015) illustrated differential effectiveness of the FGFR inhibitor JNJ-42756493 on mouse astrocytes and human glioma stem cells expressing the FGFR3-TACC3 gene fusion versus similar cells lacking this fusion. The authors reported on two patients with recurrent glioblastoma who were found to have FGFR3-TACC3 fusions (breakpoints: FGFR3 exon 17 and TACC3 exon 6; FGFR3 exon 17 and TACC3 exon 8). Following treatment with the FGFR inhibitor JNJ-42756493 in the context of a clinical trial, stabilization of disease was observed for ∼16 and ∼19 wk, respectively, where 2 wk was equivalent to one cycle. Progression of disease was subsequently observed (Di Stefano et al. 2015). This drug has also been shown to lead to partial responses in patients with an FGFR3-TACC3-containing urothelial cancer and a decrease in tumor size in a patient with an adrenal cancer harboring this fusion (Tabernero et al. 2015).
Before developing toxicity, the patient in the present report was benefiting from the FGFR inhibitor within the context of a clinical trial. Other FGFR-specific inhibitors have been developed, though none has yet been FDA-approved for the treatment of solid tumors. The FGFR inhibitor PD173074 reduced the viability of nasopharyngeal carcinoma cells harboring a FGFR3-TACC3 fusion in a dose-dependent manner (Yuan et al. 2014). FGFR3-TACC3 showed constitutive FGFR kinase activity, followed by the inhibition of this activity with PD173074 in transduced human astrocytes and mouse glioma stemlike cells (Singh et al. 2012). NVP-BGJ398 demonstrated FGFR1-4 inhibition in human embryonic kidney cells (Guagnano et al. 2011), whereas LY2874455 exhibited inhibition of FGFR2 in human gastric carcinoma cell lines (Zhao et al. 2011). These studies and our present case provide the rationale for considering FGFR inhibitors for patients whose tumor genomic profiles indicate this fusion. More research is needed to understand the activity of FGFR3-TACC3 fusions on tumors and, consequently, to discover additional clinically relevant therapeutic options for endometrial cancer patients with this gene fusion.
This patient's tumor also harbored a PIK3CA mutation, which is another putative targetable alteration. PIK3CA is a gene encoding for a catalytic subunit within the PI3K molecule (Volinia et al. 1994). Alterations in PIK3CA are implicated in 41%–52% of endometrioid endometrial carcinomas and between 33% and 38% of nonendometrioid endometrial carcinomas (e.g., papillary serous histology, clear cell histology) (Slomovitz and Coleman 2012). PI3K inhibitors, although still experimental or in clinical trials, have been developed (Khan et al. 2013).
Tumors harboring PIK3CA mutations may benefit from mTOR inhibitors, as mTOR is a target downstream from PI3K (Khan et al. 2013). mTOR inhibitors are a class of drugs that target mutations in the PI3K/AKT/mTOR pathway and limit both tumor cell proliferation and progression through the cell cycle (Husseinzadeh and Husseinzadeh 2014). The predictive value of a PIK3CA mutation for mTOR inhibitor response has been inconsistent across tumor types. Other alterations in this tumor may account for drug response. For example, this tumor also carried a TSC2 rearrangement with a breakpoint in exon 26 but was reported as a variant of unknown significance. Germline mutations in TSC2 result in developmental defects seen in tuberous sclerosis (Jones et al. 1997; Dabora et al. 2001; Langkau et al. 2002). Although tuberous sclerosis is an autosomal dominant disease, the hamartomatous growths associated with this disease often include the loss of heterozygosity of TSC2 (or, less commonly, TSC1) (Carbonara et al. 1996; Kwiatkowski 2003). It is not known if this tumor displays loss of heterozygosity. This alteration disrupts TSC2 upstream of the GTPase-activating protein (GAP)-related domain (exons 34–38) and may confer relevant loss of function (Maheshwar et al. 1997). With loss-of-function mutations in TSC2, the protein is phosphorylated and inhibited by AKT, stimulating Rheb GTPase activity, and leading to the activation of mTOR (Maheshwar et al. 1997; Kwiatkowski 2003; Li et al. 2004; Dibble and Cantley 2015; Huynh et al. 2015). Exceptional responses have been observed in patients with otherwise aggressive, TSC2-mutated tumors with mTOR inhibitor therapy (Wagle et al. 2014). Because retrospective analysis of this tumor did not reveal expression of estrogen receptor (not shown), the 17-mo clinical benefit experienced by this patient was likely due to mTOR inhibition. The observed clinical benefit strongly implicates the TSC2 rearrangement as being functional, although a benefit due to other upstream alterations cannot be ruled out.
METHODS
This patient was evaluated at the Rutgers CINJ and provided informed consent to participate in a prospective study trial for tumor genomic profiling (NCT02688517). This trial was approved by the Rutgers University New Brunswick Health Sciences IRB (Pro2012002075). Clinical records were abstracted for relevant patient history and tumor characteristics; data were abstracted until the end of October 2015, therefore, subsequent data were not reflected in this manuscript. Medical history, pathology, radiology, clinical course, and genomic profiling results were reviewed in an anonymized fashion at a formal MTB. Profiling results were discussed in the context of clinical course, tumor type, mutational frequency, role in cell pathways contributing to cancer biology, and considerations for future therapy including consensus recommendations for clinical trials, FDA-approved therapies (on or off label), and genetic counseling, if appropriate. The patient was followed prospectively for clinical course. The protocol for tumor genomic profiling was approved by the Institutional Review Board of Rutgers Robert Wood Johnson Medical School.
For the additional patients provided by Foundation Medicine, Inc., approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817).
Comprehensive Genomic Profiling
Details on the sequencing methodology used in this study can be found in Frampton et al. (2013). Routine formalin-fixed, paraffin-embedded (FFPE) tissue was sent to Foundation Medicine, Inc. for CGP using the CLIA-certified FoundationOne platform (Frampton et al. 2013). Targeted sequencing of the entire coding sequence was done for 236 genes and 47 introns of 19 genes involved in fusions at a depth of 500×–1000× (median exon depth of 792×; 100% of the baited region was sequenced at a depth of at least 100×; sequence alignment error of only 0.27%). The depth of variant call reads was 59×; these 59 chimeric reads equate to an independently validated variant calling method described in Frampton et al. (2013). The depth of wild-type reads was 1155× (please see Supplemental Table S1).
Pathology
H&E-stained slides of tissue samples at various time points in the clinical care of the case patient were reviewed by Pathology.
Radiology
CT scans of the abdomen and pelvis (axial and coronal reconstructed), with or without oral/intravenous contrast, were performed at various time points in the clinical care of the case patient. A PET scan with coronal display was also performed. The scans were reviewed by J.K.A. and L.R.-R. RECIST criteria were used to determine response based on CT scan imaging (Eisenhauer et al. 2009).
ADDITIONAL INFORMATION
Data Deposition and Access
The genomic variants for our patient case were deposited into ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000693748–SCV000693750.
Ethics Statement
The patient provided informed consent to participate in the Rutgers CINJ genomic tumor profiling protocol (NCT02688517), which was approved by the Institutional Review Board (IRB) of Rutgers University New Brunswick Health Sciences (Pro2012002075).
Acknowledgments
We thank the patient, Natasha Nicholson, Frances DiClemente, Joseph Miktus, and service provided by the Biospecimen Repository Service Core Facility. We thank Jennifer Hostettler for assistance with drafting and editing.
Author Contributions
J.D. contributed to manuscript preparation; K.M.H. contributed to genomic data interpretation and manuscript preparation; S.G. contributed to genomic data interpretation and manuscript preparation; M.H. contributed to patient care and manuscript preparation; V.R. contributed to manuscript preparation; J.K.A. contributed to manuscript preparation and data interpretation; G.M.R. contributed to manuscript preparation and data interpretation; H.Z. contributed to manuscript preparation and data interpretation; S.M.A. contributed to comprehensive genomic profiling, data analysis, and manuscript preparation; D.P. contributed to comprehensive genomic profiling, data analysis, and manuscript preparation; J.A.E. contributed to comprehensive genomic profiling, data analysis, and manuscript preparation; L.R.-R. contributed to patient care, genomic data interpretation, and manuscript preparation.
Funding
This research was supported by a generous gift to the Genetics Diagnostics to Cancer Treatment Program of the Rutgers Cancer Institute of New Jersey and RUCDR Infinite Biologics and by the National Institutes of Health (NIH) P30CA072720.
Competing Interest Statement
S.G. serves on a scientific advisory board and as consultant to Inspirata, Inc., and has equity in Inspirata, Inc.; serves on an advisory board for Novartis Pharmaceuticals; and has patents on digital imaging technology licensed to Inspirata, Inc. S.M.A. is an employee of Foundation Medicine Inc. D.P. is an employee of Foundation Medicine Inc. J.A.E. is an employee of Foundation Medicine Inc.
Referees
Wei Zhang
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Baselga J, Semiglazov V, van Dam P, Manikhas A, Bellet M, Mayordomo J, Campone M, Kubista E, Greil R, Bianchi G, et al. 2009. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. J Clin Oncol 27: 2630–2637. [DOI] [PubMed] [Google Scholar]
- Byron SA, Gartside M, Powell MA, Wellens CL, Gao F, Mutch DG, Goodfellow PJ, Pollock PM. 2012. FGFR2 point mutations in 466 endometrioid endometrial tumors: relationship with MSI, KRAS, PIK3CA, CTNNB1 mutations and clinicopathological features. PLoS One 7: e30801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonara C, Longa L, Grosso E, Mazzucco G, Borrone C, Garrè ML, Brisigotti M, Filippi G, Scabar A, Giannotti A, et al. 1996. Apparent preferential loss of heterozygosity at TSC2 over TSC1 chromosomal region in tuberous sclerosis hamartomas. Genes Chromosomes Cancer 15: 18–25. [DOI] [PubMed] [Google Scholar]
- Carneiro BA, Elvin JA, Kamath SD, Ali SM, Paintal AS, Restrepo A, Berry E, Giles FJ, Johnson ML. 2015. FGFR3–TACC3: a novel gene fusion in cervical cancer. Gynecol Oncol Rep 13: 53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, Lu Y, Stemke-Hale K, Dyer MD, Zhang F, et al. 2011. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov 1: 170–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley BA. 2015. Genomically guided cancer treatments: from “promising” to “clinically useful”. J Natl Cancer Inst 107: djv168. [DOI] [PubMed] [Google Scholar]
- Dabora SL, Jozwiak S, Franz DN, Roberts PS, Nieto A, Chung J, Choy YS, Reeve MP, Thiele E, Egelhoff JC, et al. 2001. Mutational analysis in a cohort of 224 tuberous sclerosis patients indicates increased severity of TSC2, compared with TSC1, disease in multiple organs. Am J Hum Genet 68: 64–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano AL, Fucci A, Frattini V, Labussiere M, Mokhtari K, Zoppoli P, Marie Y, Bruno A, Boisselier B, Giry M, et al. 2015. Detection, characterization, and inhibition of FGFR–TACC fusions in IDH wild-type glioma. Clin Cancer Res 21: 3307–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Cantley LC. 2015. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol 25: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci MV, Arnedos M, Andre F, Soria JC. 2013. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov 3: 264–279. [DOI] [PubMed] [Google Scholar]
- Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, Nicoletti R, Winckler W, Grewal R, Hanna M, et al. 2008. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci 105: 8713–8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. 2009. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45: 228–247. [DOI] [PubMed] [Google Scholar]
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, et al. 2013. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31: 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granberg KJ, Annala M, Lehtinen B, Kesseli J, Haapasalo J, Ruusuvuori P, Yli-Harja O, Visakorpi T, Haapasalo H, Nykter M, et al. 2017. Strong FGFR3 staining is a marker for FGFR3 fusions in diffuse gliomas. Neuro Oncol 19: 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, Brueggen J, Jensen MR, Schnell C, Schmid H, et al. 2011. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J Med Chem 54: 7066–7083. [DOI] [PubMed] [Google Scholar]
- Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. 2016. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res 22: 259–267. [DOI] [PubMed] [Google Scholar]
- Husseinzadeh N, Husseinzadeh HD. 2014. mTOR inhibitors and their clinical application in cervical, endometrial and ovarian cancers: a critical review. Gynecol Oncol 133: 375–381. [DOI] [PubMed] [Google Scholar]
- Huynh H, Hao HX, Chan SL, Chen D, Ong R, Soo KC, Pochanard P, Yang D, Ruddy D, Liu M, et al. 2015. Loss of tuberous sclerosis complex 2 (TSC2) is frequent in hepatocellular carcinoma and predicts response to mTORC1 inhibitor everolimus. Mol Cancer Ther 14: 1224–1235. [DOI] [PubMed] [Google Scholar]
- Jones AC, Daniells CE, Snell RG, Tachataki M, Idziaszczyk SA, Krawczak M, Sampson JR, Cheadle JP. 1997. Molecular genetic and phenotypic analysis reveals differences between TSC1 and TSC2 associated familial and sporadic tuberous sclerosis. Hum Mol Genet 6: 2155–2161. [DOI] [PubMed] [Google Scholar]
- Khan KH, Yap TA, Yan L, Cunningham D. 2013. Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin J Cancer 32: 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecny GE, Kolarova T, O'Brien NA, Winterhoff B, Yang G, Qi J, Qi Z, Venkatesan N, Ayala R, Luo T, et al. 2013. Activity of the fibroblast growth factor receptor inhibitors dovitinib (TKI258) and NVP-BGJ398 in human endometrial cancer cells. Mol Cancer Ther 12: 632–642. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ. 2003. Rhebbing up mTOR: new insights on TSC1 and TSC2, and the pathogenesis of tuberous sclerosis. Cancer Biol Ther 2: 471–476. [DOI] [PubMed] [Google Scholar]
- Lamont FR, Tomlinson DC, Cooper PA, Shnyder SD, Chester JD, Knowles MA. 2011. Small molecule FGF receptor inhibitors block FGFR-dependent urothelial carcinoma growth in vitro and in vivo. Br J Cancer 104: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langkau N, Martin N, Brandt R, Zugge K, Quast S, Wiegele G, Jauch A, Rehm M, Kuhl A, Mack-Vetter M, et al. 2002. TSC1 and TSC2 mutations in tuberous sclerosis, the associated phenotypes and a model to explain observed TSC1/TSC2 frequency ratios. Eur J Pediatr 161: 393–402. [DOI] [PubMed] [Google Scholar]
- Lee PS, Secord AA. 2014. Targeting molecular pathways in endometrial cancer: a focus on the FGFR pathway. Cancer Treat Rev 40: 507–512. [DOI] [PubMed] [Google Scholar]
- Li Y, Corradetti MN, Inoki K, Guan KL. 2004. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem Sci 29: 32–38. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang X, Liu J, Hou G, Zhang S, Zhang J. 2014. Everolimus in combination with letrozole inhibit human breast cancer MCF-7/Aro stem cells via PI3K/mTOR pathway: an experimental study. Tumour Biol 35: 1275–1286. [DOI] [PubMed] [Google Scholar]
- Maheshwar MM, Cheadle JP, Jones AC, Myring J, Fryer AE, Harris PC, Sampson JR. 1997. The GAP-related domain of tuberin, the product of the TSC2 gene, is a target for missense mutations in tuberous sclerosis. Hum Mol Genet 6: 1991–1996. [DOI] [PubMed] [Google Scholar]
- McConechy MK, Ding J, Cheang MC, Wiegand KC, Senz J, Tone AA, Yang W, Prentice LM, Tse K, Zeng T, et al. 2012. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol 228: 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F, Johnson A, Holla V, Bailey AM, Brusco L, Chen K, Routbort M, Patel KP, Zeng J, Kopetz S, et al. 2015. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst 107: djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. 2016. Endometrial cancer. Lancet 387: 1094–1108. [DOI] [PubMed] [Google Scholar]
- National Cancer Institute. 2002. Endometrial cancer treatment (PDQ®): patient version. In PDQ cancer information summaries, http://www.ncbi.nlm.nih.gov/books/NBK65896/?report=printable. [Google Scholar]
- National Comprehensive Cancer Network. 2014. NCCN clinical practice guidelines in oncology (NCCN guidelines®): uterine neoplasms. Version 2.2015 National Comprehensive Cancer Network, Fort Washington, PA. [Google Scholar]
- Nelson KN, Meyer AN, Siari A, Campos AR, Motamedchaboki K, Donoghue DJ. 2016. Oncogenic gene fusion FGFR3-TACC3 is regulated by tyrosine phosphorylation. Mol Cancer Res 14: 458–469. [DOI] [PubMed] [Google Scholar]
- Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, Li X, Gumin J, Zheng H, Hu L, et al. 2013. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest 123: 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock PM, Gartside MG, Dejeza LC, Powell MA, Mallon MA, Cancer Genome Project, Davies H, Mohammadi M, Futreal PA, Stratton MR, Trent JM, et al. 2007. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene 26: 7158–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh-Hain JA, Del Carmen MG. 2010. Treatment for advanced and recurrent endometrial carcinoma: combined modalities. Oncologist 15: 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2016. Cancer statistics, 2016. CA Cancer J Clin 66: 7–30. [DOI] [PubMed] [Google Scholar]
- Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, et al. 2012. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337: 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovitz BM, Coleman RL. 2012. The PI3K/AKT/mTOR pathway as a therapeutic target in endometrial cancer. Clin Cancer Res 18: 5856–5864. [DOI] [PubMed] [Google Scholar]
- Tabernero J, Bahleda R, Dienstmann R, Infante JR, Mita A, Italiano A, Calvo E, Moreno V, Adamo B, Gazzah A, et al. 2015. Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol 33: 3401–3408. [DOI] [PubMed] [Google Scholar]
- Temkin SM, Fleming G. 2009. Current treatment of metastatic endometrial cancer. Cancer Control 16: 38–45. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. 2013. Integrated genomic characterization of endometrial carcinoma. Nature 497: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin NP, Foukakis T, De Petris L, Bergh J. 2015. The importance of molecular markers for diagnosis and selection of targeted treatments in patients with cancer. J Intern Med 278: 545–570. [DOI] [PubMed] [Google Scholar]
- Trabert B, Wentzensen N, Felix AS, Yang HP, Sherman ME, Brinton LA. 2015. Metabolic syndrome and risk of endometrial cancer in the United States: a study in the SEER-medicare linked database. Cancer Epidemiol Biomarkers Prev 24: 261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Grose R. 2010. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10: 116–129. [DOI] [PubMed] [Google Scholar]
- Volinia S, Hiles I, Ormondroyd E, Nizetic D, Antonacci R, Rocchi M, Waterfield MD. 1994. Molecular cloning, cDNA sequence, and chromosomal localization of the human phosphatidylinositol 3-kinase p110 α (PIK3CA) gene. Genomics 24: 472–477. [DOI] [PubMed] [Google Scholar]
- Wagle N, Grabiner BC, Van Allen EM, Amin-Mansour A, Taylor-Weiner A, Rosenberg M, Gray N, Barletta JA, Guo Y, Swanson SJ, et al. 2014. Response and acquired resistance to everolimus in anaplastic thyroid cancer. N Engl J Med 371: 1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SV, Hurst CD, Knowles MA. 2013. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 22: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, et al. 2013. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 3: 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu ZH, Lin ZR, Xu LH, Zhong Q, Zeng MS. 2014. Recurrent FGFR3-TACC3 fusion gene in nasopharyngeal carcinoma. Cancer Biol Ther 15: 1613–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Li WY, Chen D, Henry JR, Li HY, Chen Z, Zia-Ebrahimi M, Bloem L, Zhai Y, Huss K, et al. 2011. A novel, selective inhibitor of fibroblast growth factor receptors that shows a potent broad spectrum of antitumor activity in several tumor xenograft models. Mol Cancer Ther 10: 2200–2210. [DOI] [PubMed] [Google Scholar]
- Zhou DS, Wang HB, Zhou ZG, Zhang YJ, Zhong Q, Xu L, Huang YH, Yeung SC, Chen MS, Zeng MS. 2015. TACC3 promotes stemness and is a potential therapeutic target in hepatocellular carcinoma. Oncotarget 6: 24163–24177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.