Abstract
Anaplastic lymphoma kinase (ALK) gene rearrangements are present in ∼5% of non-small-cell lung cancers (NSCLCs). These rearrangements occur because of a chromosomal inversion within the short arm of Chromosome 2, which results in the formation of the echinoderm microtubule-associated protein-like 4 (EML4)–ALK fusion oncogene. Whereas NSCLC transformation to SCLC is a rare phenomenon described in epidermal growth factor receptor (EGFR) mutant cancers primarily after treatment with targeted therapy, it is exceedingly rare in ALK-rearranged adenocarcinomas. It is currently unclear what the therapeutic significance of the rearrangement is in this transformed tumor as there is a paucity of medical literature describing follow-up care and outcomes of patients in this rare scenario. We describe a unique case in which a patient with ALK-rearranged adenocarcinoma underwent small-cell transformation at a metastatic site with retained ALK rearrangement and was provided clinical follow-up after treatment with second-generation tyrosine kinase inhibiter (TKI) therapy.
Keywords: neoplasm of the lung
CASE PRESENTATION
A 73-yr-old female, with a remote history of light smoking, was diagnosed with stage 2B non-small-cell lung carcinoma (NSCLC). The lung mass was removed via wedge resection and morphologically examined in its entirety. Histologic examination showed moderately differentiated adenocarcinoma with mucinous and micropapillary histology with focal microscopic visceral pleural invasion and lymphovascular invasion without lymph node involvement. All staging scans were negative for distant metastases. Targeted EGFR analysis and fluorescence in situ hybridization (FISH) of the tumor showed an ALK rearrangement. The patient completed four cycles of chemotherapy with carboplatin/pemetrexed and notably did not receive therapy with an anaplastic lymphoma kinase (ALK) tyrosine kinase inhibitor (TKI).
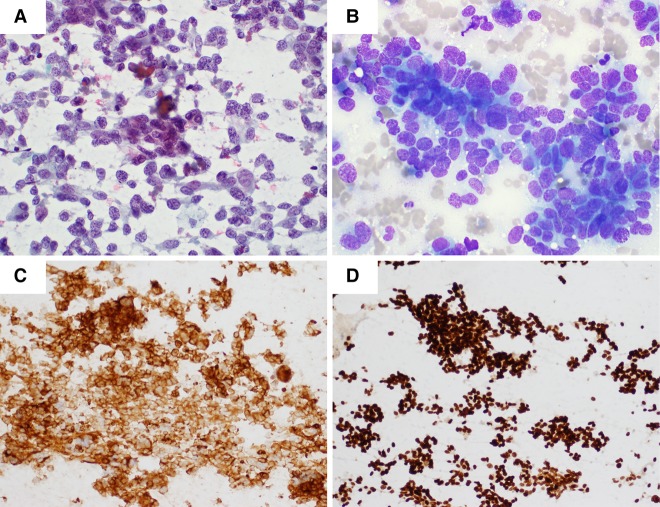
The patient subsequently presented 2 years later with progressive abdominal enlargement and multiple liver metastases without evidence of disease in the thoracic cavity. Biopsy of a liver lesion showed small-cell carcinoma (strongly positive for TTF-1, CK7, and Synaptophysin and negative for CK20) that was morphologically distinct from the original NSCLC (Fig. 1).
Figure 1.
(A) Fine-needle aspiration (FNA) of liver metastasis at time of recurrence (Pap stain). (B) MGG stain. (C) Strong membranous staining with Synaptophysin (27G12, Leica). (D) Strong nuclear staining with TTF-1 (SPT24, Leica). The tumor also demonstrated strong cytoplasmic staining for CK7 (RN7, Leica) and was negative for CK20 (Ks20.8, Leica).
Technical Analysis
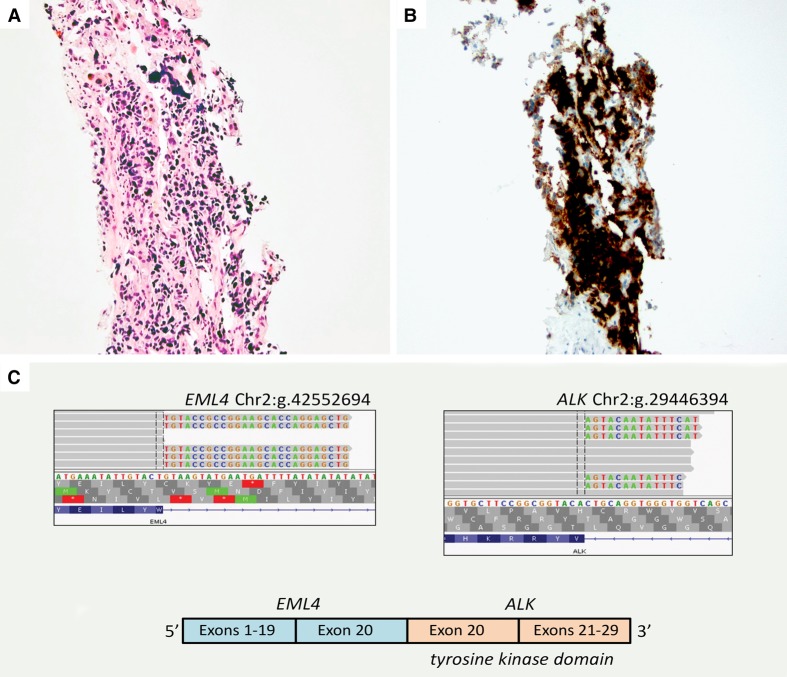
The metastatic tumor was analyzed for the ALK fusion and DNA mutation analysis. ALK rearrangement by FISH was unsuccessful because of technical failure and was thereby determined positive with qualitative immunohistochemistry (IHC) validated to detect ALK expression as a surrogate marker of ALK rearrangement in formalin-fixed paraffin-embedded (FFPE) tissue. The fusion was more specifically confirmed with targeted next-generation sequencing (NGS) of the tumor RNA as the EML4 exon 20 to ALK exon 20 fusion (Fig. 2).
Figure 2.
(A) Hematoxylin and eosin (H&E)-stained section of liver core needle biopsy showing metastatic small-cell carcinoma. (B) ALK IHC (D5F3, Ventana, performed according to manufacturer's instructions) demonstrated the tumor cells to be strongly and diffusely positive in a combined cytoplasmic and membranous pattern. (C) RNA NGS data showing the EML4 exon 20–ALK fusion. Integrated genome viewer (IGV) sequence alignments at the point of mRNA fusion show ALK exon 20 sequence fused to the EML4 exon 20 genomic location (left panel) and EML4 exon 20 sequence fused to ALK exon 20 genomic location (right panel). Schematic of the predicted ELM4–ALK fusion mRNA (bottom).
The DNA from the metastatic tumor was also submitted for targeted genomic profiling for single-nucleotide variants and insertion/deletion mutations using an NGS assay that is clinically validated for use in solid tumors (Illumina MiSeq). The assay uses an Agilent Sure Select XT target probe library custom designed to capture exons and selected intronic regions of 29 genes: AKT1, ALK, BRAF, CDKN2A, CTNNB1, DDR2, EGFR, ERBB2, ESR1, FGFR1, FGFR2, GNA11, GNAQ, IDH1, IDH2, KIT, KRAS, MAP2K1, MAP2K2, MAPK1, MET, MTOR, NRAS, PDGFRA, PIK3CA, PTEN, RET, ROS1 and STK11. Targeted NGS of the tumor DNA did not show any additional clinically actionable mutations. The clinical assays requested did not include analysis of copy number variation, nor did the NGS capture content analyze RB1.
The original tumor was resected at an outside center not affiliated with the University of Vermont Medical Center, and although completely submitted for morphologic evaluation, the tissue was not available for additional testing utilizing NGS (Table 1).
Table 1.
Genomic findings
| Gene/genomic location | Chr | HGVS DNA ref | HGVS protein ref | Variant type | Predicted effect | Allele frequency | Target coverage |
|---|---|---|---|---|---|---|---|
| ALK | 2 | t(2;2)(p23;p21)(hg19 Chr 2:g.42552694::oChr 2:g.29446394) | n/a | EML4–ALK fusion | Oncogenic | n/a | 301× |
Variant Interpretation
The small-cell carcinoma with retained ALK rearrangement was ultimately classified as metastatic recurrent NSCLC that had undergone small-cell transformation. After six cycles of carboplatin and etoposide, computed tomography (CT) scans showed a mixed response with stable bony lesions but innumerable liver lesions and central nervous system involvement. Alectinib was then initiated but with progressive disease at 3 months, it was discontinued with transition to comfort care.
NSCLC transformation to small-cell lung cancer is a rare phenomenon that has been well-described in EGFR-mutant cancers. This transformation typically arises in the setting of treatment with EGFR-targeted TKI therapy and is considered to be a therapeutic resistance mechanism (Zakowski et al. 2006; Morinaga et al. 2007; Alam et al. 2010; Ma et al. 2012; Norkowski et al. 2013; Popat et al. 2013; Watanabe et al. 2013; Jiang et al. 2016). Currently, there are only two published case reports that describe the presence of an ALK rearrangement in small-cell carcinoma (Toyokawa et al. 2012, 2013). One of these reports describes a tumor with mixed histopathology composed of a small-cell component and an adenocarcinoma component; however, unlike our case, ALK fusion was only detected in the small-cell component and the clinical therapeutic experience with TKI therapy was not addressed (Toyokawa et al. 2012).
The case presented herein is unique because the original tumor was completely resected with a complete histologic examination that excluded tumor heterogeneity with a small-cell component, and the patient was not treated with ALK-targeted therapy before undergoing small-cell transformation. This morphologic transformation in a tumor that originally did not exhibit a small-cell component lends support to the hypothesis that both tumor phenotypes, small-cell and non-small-cell, may arise from the same cancer progenitor cell. (D'Angelo and Pietanza 2010; Norkowski et al. 2013; Jiang et al. 2016). Tumor progression of the ALK fusion–positive transformed tumor on a second-generation TKI suggests that progression was more likely driven by genomic alterations more commonly identified in small-cell carcinoma rather than by the ALK fusion classically associated with NSCLC. Biopsy material from only one of many metastatic sites was evaluated and uniformly showed small-cell carcinoma. Although this precludes definitive exclusion of an adenocarcinoma component in the metastatic disease, the patient progressed nonetheless on both conventional chemotherapy and ALK TKI therapy.
Small-cell carcinoma, which typically occurs in heavy smokers and is characterized by aggressive growth and poor prognosis, has a high prevalence of mutations in TP53 and RB1 (Meuwissen et al. 2003; Peifer et al. 2012; Rudin et al. 2012). Additionally, integrative genomic analyses have implicated alterations in genes that encode histone-modifying enzymes including CREBBP, EP300, and MLL along with SOX2 amplification (Peifer et al. 2012). Thus, although numerous recurrent genomic alterations have been documented in de novo small-cell carcinoma, ALK rearrangement currently is not described in the typical genomic profile of this tumor type.
SUMMARY
This case provides evidence that small-cell transformation of ALK-rearranged NSCLC can occur without selective pressure of targeted therapy and suggests these transformed tumors will not be sensitive to ALK-directed TKI therapy. It also demonstrates that transformation can occur after a relatively long, posttherapeutic disease-free interval. This supports a previous observation in transformed, TKI-treated EGFR-mutant NSCLC that small-cell transformation of NSCLC despite “targetable mutations” is associated with a relatively poor prognosis compared to nontransformed conventional small-cell carcinoma of the lung (Jiang et al. 2016). Moreover, progression of metastatic disease despite targeted therapy warrants consideration for therapeutic planning in the scenario of small-cell transformation given that it is hypothesized in this case that the genomic profile driving the tumor progression was not the ALK fusion. Lastly, this case shows there is diagnostic value in performing ALK fusion analysis in cases that are suspicious for small-cell transformation of a tumor originally diagnosed as ALK fusion–positive NSCLC as this ancillary information may provide insight into whether two morphologically distinct tumors share a common lineage. Additional clinical experience and more comprehensive functional genomic analysis of similarly transformed tumors are warranted to further evaluate these hypotheses and observations.
ADDITIONAL INFORMATION
Database Deposition and Access
The variant was submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number SCV000676940.
Ethics Statement
The authors confirm that verbal informed consent was provided by the patient for the publication of case details and any accompanying images. The University of Vermont IRB review found the case experience constituted standard treatment at the University of Vermont Medical Center and, as such, provided a determination of non-research activity requiring no further monitoring by the IRB.
Competing Interest Statement
D.L.A. reports grants from Genentech, personal fees from AstraZeneca, personal fees from Clovis, personal fees from AbbVie, personal fees from Inivata, and personal fees from SeraCare, outside the submitted work. In addition, D.L.A. has a patent Pneumatic cell collection device pending. N.S. reports personal fees from Pierian Dx, outside the submitted work. The remaining authors have declared no conflict of interest.
REFERENCES
- Alam N, Gustafson KS, Ladanyi M, Zakowski MF, Kapoor A, Truskinovsky AM, Dudek AZ. 2010. Small-cell carcinoma with an epidermal growth factor receptor mutation in a never-smoker with gefitinib-responsive adenocarcinoma of the lung. Clin Lung Cancer 11: E1–E4. [DOI] [PubMed] [Google Scholar]
- D'Angelo SP, Pietanza MC. 2010. The molecular pathogenesis of small cell lung cancer. Cancer Biol Ther 10: 1–10. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Zhao J, Wang MZ, Huo Z, Zhang J, Zhong W, Xu Y. 2016. Small-cell lung cancer transformation in patients with pulmonary adenocarcinoma: a case report and review of literature. Medicine (Baltimore) 95: e2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AT, Chan WK, Ma ES, Cheng T, Cheng PN. 2012. Small cell lung cancer with an epidermal growth factor receptor mutation in primary gefitinib-resistant adenocarcinoma of the lung. Acta Oncol 51: 557–559. [DOI] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. 2003. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 4: 181–189. [DOI] [PubMed] [Google Scholar]
- Morinaga R, Okamoto I, Furuta K, Kawano Y, Sekijima M, Dote K, Satou T, Nishio K, Fukuoka M, Nakagawa K. 2007. Sequential occurrence of non–small cell and small cell lung cancer with the same EGFR mutation. Lung Cancer 58: 411–413. [DOI] [PubMed] [Google Scholar]
- Norkowski E, Ghigna MR, Lacroix L, Le Chevalier T, Fadel É, Dartevelle P, Dorfmuller P, Thomas de Montpréville V. 2013. Small-cell carcinoma in the setting of pulmonary adenocarcinoma: new insights in the era of molecular pathology. J Thorac Oncol 8: 1265–1271. [DOI] [PubMed] [Google Scholar]
- Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. 2012. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44: 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popat S, Wotherspoon A, Nutting CM, Gonzalez D, Nicholson AG, O'Brien M. 2013. Transformation to “high grade” neuroendocrine carcinoma as an acquired drug resistance mechanism in EGFR-mutant lung adenocarcinoma. Lung Cancer 80: 1–4. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. 2012. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44: 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokawa G, Taguchi K, Ohba T, Morodomi Y, Takenaka T, Hirai F, Yamaguchi M, Seto T, Takenoyama M, Sugio K, et al. 2012. First case of combined small-cell lung cancer with adenocarcinoma harboring EML4–ALK fusion and an exon 19 EGFR mutation in each histological component. J Thorac Oncol 7: e39–e41. [DOI] [PubMed] [Google Scholar]
- Toyokawa G, Takenoyama M, Taguchi K, Toyozawa R, Inamasu E, Kojo M, Shiraishi Y, Morodomi Y, Takenaka T, Hirai F, et al. 2013. An extremely rare case of small-cell lung cancer harboring variant 2 of the EML4–ALK fusion gene. Lung Cancer 81: 487–490. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Sone T, Matsui T, Yamamura K, Tani M, Okazaki A, Kurokawa K, Tambo Y, Takato H, Ohkura N, et al. 2013. Transformation to small-cell lung cancer following treatment with EGFR tyrosine kinase inhibitors in a patient with lung adenocarcinoma. Lung Cancer 82: 370–372. [DOI] [PubMed] [Google Scholar]
- Zakowski MF, Ladanyi M, Kris MG; Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome Group. 2006. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 355: 213–215. [DOI] [PubMed] [Google Scholar]