Abstract
Multiplex genome editing is the simultaneous introduction of multiple distinct modifications to a given genome. Though in its infancy, maturation of this field will facilitate powerful new biomedical research approaches, and will enable a host of far-reaching biological engineering applications, including new therapeutic modalities and industrial applications, as well as ‘genome writing’ and de-extinction efforts. In this perspective, we focus on multiplex editing of large eukaryotic genomes. We describe the current state of multiplexed genome editing, the current limits of our ability to multiplex edits, and provide perspective on the many applications that fully-realized multiplex editing technologies would enable in higher eukaryotic genomes. We offer a broad look at future directions, covering emergent CRISPR-based technologies, advances in intracellular delivery, and new DNA assembly approaches that may enable future genome editing on a massively multiplexed scale.
Introduction
The facile programmability of CRISPR systems has enabled rapid and widespread adoption, leading to the current revolution in nearly every aspect of genome editing, including multiplex editing. The evolved function of CRISPR systems as multi-pathogen defenses requires a system that is naturally multiplexable and readily adaptable to arbitrary target sequences. CRISPR RNAs functionally combine with CRISPR associated proteins (Cas) to provide anti-viral immunity by targeting foreign nucleic sequences through a complementarity driven mechanism1,2. As a molecular tool, the ability to reprogram a single common effector protein through use of small, trans-acting modular guide RNA (gRNA) sequences that target DNA via nucleobase pairing logic3–6 is an elegant and near ideal solution to the problem of multiplexing, offering affordable and scalable sequence targeting. Older editing technologies required protein-based targeted factors that were large in coding size, and comparatively slow to generate7,8. With the emergence and refinement of gRNA-targeted CRISPR technologies, we now have the prospect of multiplexing on scale previously impossible to consider with pre-CRISPR editing technologies.
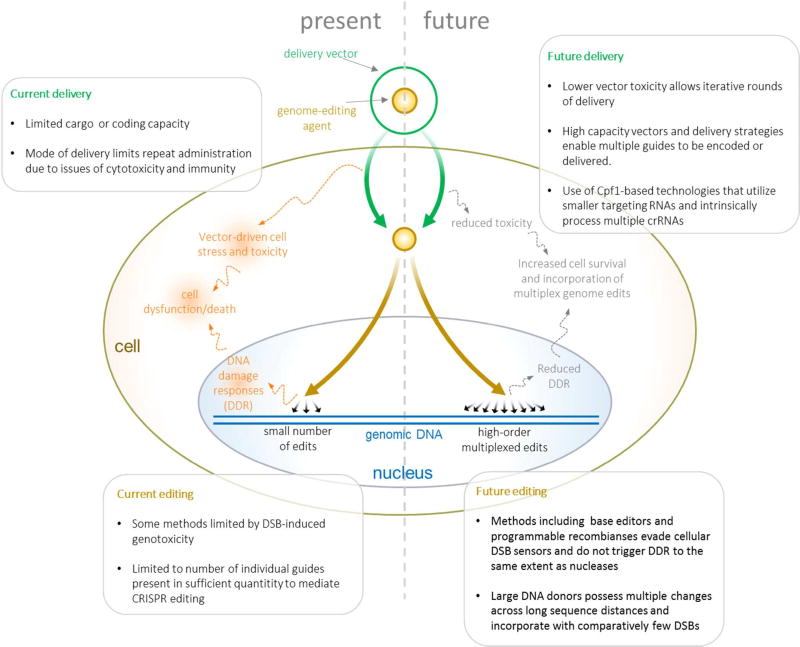
The field of genome editing will need to overcome several limitations that currently prevent highly multiplexed, genome-wide editing of eukaryotic cells (Figure 1). First, new methods that mitigate or avoid the genotoxic stress of multiplex DNA cleavage will need to be developed. Parallel advances will be needed to increase the efficiency of multiplexed delivery while avoiding the cytotoxicity of current multiplex delivery strategies. To edit large portions of a genome, hybrid methods utilizing bacterial DNA assembly methods to produce large CRISPR-targeted donor templates will need to be established, and the delivery of such large DNAs will need to be dramatically improved see application in large-scale genome editing projects.
Figure 1.
The process of multiplex editing, current limitations, and future improvements.
The state of multiplex genome editing technology
Multiplex editing in eukaryotic genomes
The key advantage of CRISPR-Cas based genome editors over previous approaches is the capacity to retarget modifications through use of distinct gRNAs9. The modularity of the CRISPR/cas9 system and the small size of gRNAs, enables a comparatively scalable and rapid production of multiple, distinct genome editing agents. This has not only allowed genome-wide libraries of gRNAs to be screened in parallel as a pooled population10–12, but also enables multiplexing by simply delivering or encoding multiple distinct gRNAs per cell. This overcame many of the design, synthesis, and delivery difficulties when compared to the use zinc finger nucleases (ZFNs) or Transcription activator-like effector nucleases (TALEN).
The intrinsically low barrier to multiplex editing with CRISPR-Cas was recognized from the first reports of its use as an editing tool13,4,14,5,15. Indeed, demonstration of multiplex capability is a common feature of nearly all CRISPR-Cas methodological reports, whether demonstrating novel CRISPR-targeted activities or modes of CRISPR delivery. In addition, the use of CRISPR as a gene expression regulatory tool has also demonstrated the feasibility of a multiplexed CRISPR system through multiplexed activation and/or repression of genes16–18. However, ‘multiplex’ as used with regard to genome editing, thus far describes a very small number of simultaneous edits. Published protocols only demonstrate modification of as many as seven19 distinct genomic targets for SpCas9, and up to four with Cas9 orthologues such as Cpf120. Even considering this small number of multiplex edits, with increasing number of targets, the efficiency at each site decreases when compared to single target editing rates. Moreover, cells can only tolerate a relatively small number of simultaneous double strand breaks (DSBs) due to native DNA damage responses and apoptotic signaling. These factors enforce both procedural and scientific constraints on any effort that invokes multiplex editing.
The largest multiplex CRISPR-based editing effort was recently reported by our group, where targets within 62 porcine endogenous retroviral (PERV) sequences were modified to ablate PERV expression and production, a major barrier to adoption of pig-based organ transplant therapies21. However, this feat was enabled by the high sequence identity between distinct PERV elements, which allowed the use of just two distinct guide sequences with a single guide directing the majority of modifications. While the 62 PERV knockout effort was successful, issues of limited editing efficiency and genotoxicity enforced practical constraints on the development this multiplexed genome editing protocol. First, acceptable modification rates were only achieved by long-term expression of editing agents from transposon-integrated expression units, as standard transfection was ineffective, and lentiviral integrants were silenced. Cas9 and gRNA expression units were integrated into the genome, with expression proceeding over 14 days. Standard approaches such as multiplex transfection or lentiviral integration, and shorter editing windows, failed to not achieve a significant level of editing across multiple PERV loci. Secondly, the presence of multiple DSBs triggered apoptotic responses and limited the number of surviving, completely modified clones. Cells experiencing the most edits were likely depleted from the population via apoptosis. Thus, while a small number of clones with all PERV knockouts were isolated (8% of cells showed 60–100% PERV knockout rates), the overwhelming majority of surviving cells had less than 10% of PERV sequences edited. This genotoxicity-driven selective process raises concerns over the functionality of edited clones. Given the high expected toxicity of multiple DSBs, surviving clones might be expected to carry mutations that enable evasion of genotoxicity-driven apoptotic death, including in p53. Indeed, in cells known to be more sensitive to DSBs, such as pluripotent stem cells where even single DSBs can lead to apoptosis, some CRISPR-editing protocols call for treatment with p53 inhibitory agents22. Even in more robust cell lines, CRISPR nuclease-induced apoptosis may follow introduction of as a few as 4–12 DSBs23,24. Finally, the presence of multiple simultaneous DSBs dramatically increases the chance of non-lethal, but undesirable translocations25. These factors necessitate careful functional, karyotypic or whole genome sequencing-based screening of clones, which further limits the scale and rapidity of multiplex editing efforts.
Though far from ideal, protocols similar to that used in the PERV knockout effort could be adapted for distinct editing efforts of a similar scope. However, current editing approaches simply do not scale beyond a small number of distinct target sequences, and are only practical in scenarios where project goals can tolerate a small number of surviving clones. Hypothetical future applications, whether academic, therapeutic, or industrial, will require substantially higher efficiency and survivability, with genomic modifications multiple orders of magnitude more numerous.
Strategies for multiplex guide RNA expression
When considering edits across multiple distinct loci, multiplex genome editing using CRISPR requires the simultaneous presence of multiple guides inside the cell, which presents a major obstacle to successful multiplex editing in mammalian cells. Though several groups have developed methods which offer CRISPR based multiplex editing, no single method currently exists to effectively deliver or express multiple guides with the efficiency and scale needed for massively multiplexed genome editing goals.
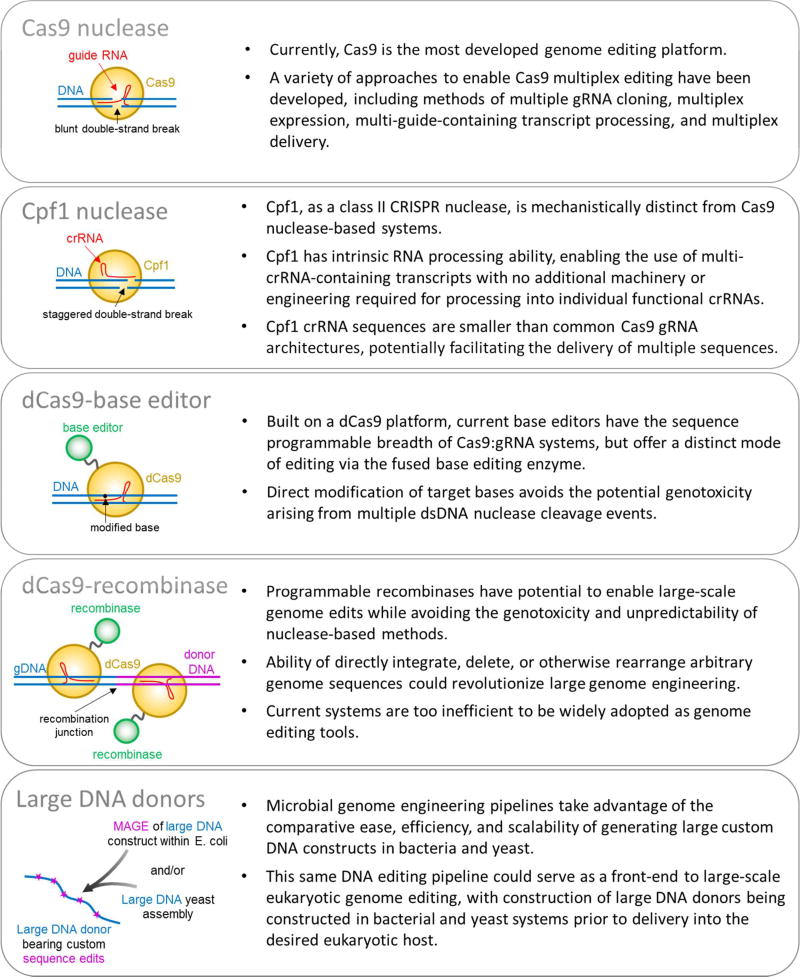
Early reports demonstrated that guide RNAs driven by independent RNA polymerase III transcriptional promoters could be functionally expressed in mammalian cells in a multiplex fashion4,5. Advances in the molecular biology toolbox have greatly simplified assembly of complex expression constructs with techniques such as golden gate cloning which enabled the construction of Cas9 with 7 pol-III regulated gRNAs in a single construct19. Co-transfection of gRNA-encoding material has also shown some success, with an early report demonstrating disruption of as many as five genes following co-transfection of gRNA-encoding PCR products14. Other work has combined the use of multiple distinct promoters with co-transfection techniques, to simultaneously express as many as 12 gRNAs26. However, this method encounters multiplex scalability issues, as with increasing numbers of genomic targets, the repetitive nature of each transcriptional unit results in genetically unstable constructs prone to recombination in E. coli during propagation of plasmid constructs. Moreover, the use of separate promoters imposes an unwanted sequence size burden, further complicating delivery of multiplex expression constructs whether by methods of transfection or viral transduction. An alternative approach drives multiplex gRNA expression from a single transcriptional unit, freeing potentially large amounts of vector capacity. Such methods depend upon enzymatic processing at sites internal to the multi-guide primary transcript to release individual gRNA units. This approach was demonstrated first through co-expression of the Csy4 enzyme, which natively processes CRISPR RNA transcripts27,28,29. More recently, Cpf1 - a class II CRISPR system orthologue of Cas930 - has shown potential as a candidate for multiplex genome editing due to its ability to directly process gRNAs through a DNase-independent RNase domain20. Extensive efforts are being put into expanding the utility of Cpf1 via mutagenesis to alter and expand PAM motif recognition31. As a two-component system, Cpf1 provides an advantage over other single-construct methods, as it does not depend on separate expression of RNA endonucleases, though potentially removing a layer of processing-dependent system control. Other processing methods that can provide higher regulatory ability rely on endogenous processing mechanisms that function in trans such as a gRNA adapted tRNA-processing system32, or in cis through self-processing gRNAs cleaved and catalyzed by ribozymes flanking the gRNA29,33. Lastly, as discussed below, editing strategies that utilize purified CRISPR ribonucleic protein (RNP) complexes offer a simple mode of multiplexing guides though simple mixing of expressed gRNA material34–36. The potential limits of multiplex RNP delivery are currently unknown, though it is likely constrained by the stability of the RNP complex, the intracellular half-life of gRNA, and the number of molecules delivered per cell.
As the demand for CRISPR-based genome editing is increasingly applied to the areas of basic biology, biological engineering, and therapeutics, new methods to enable multiplex editing will be needed. Though all large multiplex efforts will require methods with increased levels of efficiency, scalability, and straightforward experimental implementation, researchers will have to consider the applicability of a given approach to their own system of interest.
Lessons from bacterial genome engineering
To date, the most expansive example multiplex editing of an intact genome have come from bacterial systems and have not utilized CRISPR components. Our lab and others have pushed the development of Multiplexed Automated Genome Engineering, or MAGE37, in multiple prokaryotic organisms. MAGE relies on the introduction of short, single strand DNA (ssDNA) oligos, into cells expressing a single-strand annealing protein (SSAP), such as the lambda phage Beta protein. The SSAP acts at the replication fork to load the ssDNA oligos onto lagging-strand DNA, leading to their sequence incorporation concomitant with replication. The ability to mix a large number of distinct ssDNA species into a single round of MAGE, to select or enrich for edited clones, and the high growth rate of many prokaryotic models enable rapid iteration.
In higher eukaryotic cells however, particularly those that suffer from comparatively slow growth rates and whose DNA replication and repair processes diverge from those of prokaryotes, MAGE may not be a directly transferrable approach. And even if every aspect of MAGE were applicable to a human genetic system, the scale of the editing task would be daunting. Considering coding sequence alone, an effort analogous to the ongoing multi-year ‘RE. coli’ effort38–40, if performed in human cells would require roughly 5-fold greater number of edits, and this in a cellular system which propagates an order of magnitude more slowly. More generally, editing in a higher eukaryotic system must contend with the fact of a roughly 1000-fold larger genome in comparison to E. coli. While more modest multiplex editing would have a profound impact on basic research and therapeutics, the future of genome writing, recoding, and de-extinction research requires molecular tools that can contend with gigabases (gb) of genetic material.
Despite these points of departure, the factors that enable MAGE to scale so effectively to bacterial genome engineering are worth considering. The characteristics that future eukaryotic multiplex editing methods should possess include i) high per-target modification efficiency, ii) high-order, per-round multiplexability, iii) ease of programmability (preferably via base pairing), and iv) short between-round recovery, enabling iterative modification over tractable experimental timescales. Finally, as discussed below, potential future methods of multiplex mammalian genome editing and genome writing may benefit directly from MAGE, with established pipelines of MAGE-based DNA editing serving as a front-end to downstream eukaryotic genome edits.
Applications of multiplex genome editing
New methods of multiplex editing will permit novel applications ranging from basic biological research, to genetic therapeutics, industrial applications in metabolic engineering, and the synthetic biological aims of large scale genome writing and even de-extinction efforts. Practical approaches to safely and efficiently introducing arbitrarily large numbers of edits will be necessary for all of these.
Combinatorial functional genomic methods
The ability to introduce combinations of polymorphic alleles into a genome, whether for specific clonal studies or in library fashion, would enable new methods of studying the genetics of complex traits, with applications in evolutionary biology, population genetics, and in the basic biological study of human diseases. Haplotypes could be manipulated experimentally, ancestral sequences could be reconstructed, and the functional impact of such changes could be evaluated in cell culture or even in animal models bearing homologous sequence changes across multiple variable loci. Sequencing data emerging from the field of cancer genomics has identified an astounding number of mutations arising in tumors, but the functional impact of any given sequence variant, and the interaction between such variants, has been difficult to ascertain. The ability to deconvolute the functional impact of any given set of mutations via editing would greatly enable the field of cancer biology.
Therapeutic application of multiplex editing
Near-term therapeutic applications of multiplex editing could be seen even with relatively few edits. The deletion of unwanted exons in diseases arising from splicing defects41 can be achieved with as few as two multiplex DSBs. The engineering of T-cells for immunotherapeutic applications has been hotly pursued42, with a recent demonstration simultaneously disrupting three target genes whose activities confound the current generation of chimeric antigen receptor (CAR) therapies43. To prevent off-target graft-versus-host responses, reduce host-versus-graft immunity, and block immunosuppressive signaling, the researchers targeted TCR alpha subunit constant (TRAC), human leukocyte antigen class I (HLA-Is), and programmed death-1 (PD-1) genes for Cas9 nuclease disruption, respectively. Future applications may require many more. In the case of CAR T-cell cancer immunotherapies, additional factors that complicate CAR therapeutic potential through non-PD-1 suppression pathways like TIM-3, CTLA-4, or Lag-3, T-cell exhaustion, or suppressive cytokines like IL-10, receptors that mediate graft-versus-host responses and host-versus-graft antigens could be targeted. A fully mature T-cell immunotherapeutic technology could potentially require modification of dozens of sites. A distinct cancer application of multiplex editing technology, as discussed above, may come from the study of mutations arising from cancer genomics. Applied clinically, a pipeline of functional interrogation via multiplex editing of sequence variants found in patient tumor samples, if executed rapidly, could be a powerful diagnostic and predictive tool.
The emerging field of CRISPR-based eukaryotic antiviral therapy44 is another area where advances in multiplex editing provide a clear, near-term benefit. DNA viruses and retroviruses can be inactivated or destroyed via targeted viral genome modification, and this approach has been demonstrated for a number of viral classes, including HIV, HBV, and multiple Herpesviruses45–47. However, as shown with Cas9-targeting of HIV proviral genomes, the ability of viruses to rapidly evolve allows evasion of single-target approaches via mutations conferring resistance to cleavage45,48. However, multiplex antiviral targeting can negate evasion at any single target site. This approach has proven to be effective in a cell culture model of HIV infection45, HCMV, HSV-1, and EBV infection 47, and HBV infection 49. Antiviral activity can be further augmented by combinination of multiplex editing with simultaneous CRISPR-based transcriptional activation of native viral defenses50,51 This stategy may benefit from methods that allow cleavage or transcriptional regulation from a single protein effector52. Advances in multiplex delivery and the safety of multiplex editing will further enable this emerging mode of antiviral therapy.
Finally, modifying host-versus-graft antigens in human-sourced donor tissues is an area with a clear need for more advanced multiplex editing technologies. This need is even more pronounced with efforts to ‘humanize’ non-human donor tissues, potentially requiring many more genome modifications than have been achieved thus far. Depending upon donor material sourcing, the process of editing may be required to turn around edited cells within a short, therapeutically-relevant time scale. And as donor tissues may reside in a recipient for decades, the fidelity of the editing process, and resulting functionality of edited cells, will be of paramount importance.
Genome writing
The announcement of the Genome Project Write (GP-write) consortium in 2016 formally ushered in a new era of genomics that moves beyond sequencing genomes and into ground-up writing of genomes53. The ambition of GP-write is to understand, design, and test living systems through large, truly genome-scale engineering. Pilot projects include engineering cancer- and virus-resistant mammalian cell lines for the production of biologics, immuno-compatible xenotransplantation, and genomes with new functionalities like biocontainment. The complete synthesis of a the small bacterium Mycoplasma genitalium, the engineering and synthesis of large recoded Escherichia coli fragments, and the ongoing global efforts to synthesize the full genome of the yeast Saccharomyces cerevisiae have set groundbreaking precedents that lay the groundwork for the ambitions of GP-write40,54–56. However, projects like GP-write, and so-called ‘de-extinction’ genome-writing efforts, require, in addition to advances in DNA synthesis output and cost-effectiveness, a large degree of multiplexibility in gene editing, assembly, and delivery tools.
The nascent field of de-extinction seeks to revive extinct organisms through genome-writing technologies to engineer viable organisms from the ground-up, beginning from limited genome sequence data. A range of motivations drive these efforts, from understanding the evolutionary history of extinct lineages and the genetic mutations and bottlenecks which accelerate extinction57, to ecological restoration via reintroduction of extinct keystone species, to mitigating the challenges brought about by the ongoing Anthropocene extinction wave. Our lab, in collaboration with Revive and Restore, a non-profit partnership for the genetic rescue of endangered and extinct species, is working on the de-extinction of one of the most iconic Pleistocene animals, Mammuthus primigenius, or the woolly mammoth, an important species in maintaining the Pleistocene steppe ecosystem. Ongoing deextinction efforts around the world now include the Passenger Pigeon and Health Hen, and preservation efforts for endangered species including the Northern White Rhino, and Black-footed Ferret. The development of de-extinction technology would, for the first time in history, mean that bodily extinction of a species is no longer a terminal fate.
Species de-extinction without multiplexing is not feasible. Sequencing data from mammoths reveals a 99.78% identity to the modern Loxodonta africana (African elephant) genome at the level of protein sequence, and 0.6% different at the level of nucleic acid sequence58. Assuming a 3.3 gb size of the elephant genome, between 7.3 and 19.8 million nucleotide changes must be made to achieve full deextinction, intra-species variation notwithstanding. Recent sequencing of several more mammoth specimens placed the mammoth phylogenetically closer to the Asian, rather than the African, elephant, likely decreasing the expected editing burden59. Still the scale of such an editing goal dwarfs any genome engineering effort to date.
From basic biological and evolutionary studies, to ecological engineering and conservation projects, and nascent therapeutic modalities, the future application of genome editing technologies may impact every aspect of our world. To see full application, robust multiplex editing capabilities must be developed. The ability to massively multiplex modifications on a genomic scale will require fundamental improvements to methods of editing, delivery vehicles, and donor DNA construction.
Repetitive genetic elements
One field with an intrinsically low-barrier to multiplex study is that of repetitive genetic elements (RGEs). Though early application of genome editing focused on protein coding genes, there is an increasing interest in developing methods to interrogate the noncoding complement of the genome. Projects such as the encyclopedia of DNA elements60 (ENCODE) presented many intriguing observations linking chromatin structure, gene expression, developmental timing to non-coding loci, including RGEs. Repetitive elements such as Alu, L1 retrotransposons, SVA, and Human endogenous retroviruses (HERVs) may occur with ~2700 to 1 million copies per genome61, representing a non-trivial portion of total genomic sequence, and are a major source of sequence variation. Expression from such elements appears to be highly regulated, indicating they may play important roles biological processes. Such elements are suspected of roles in neurological diseases such as ataxia telangiectasia62, Rett syndrome63, and human cancers64. However, the study of these sequences is hampered in part by the inability to distinguish the effects of individual repeats, and to manipulate them at the level of DNA sequence. Targeting multiple RGEs with a small number of gRNAs may be relatively easy given high sequence conservation. However, such editing, if approached with current nuclease-based CRISPR protocols would result in an exceedingly large number of DSBs, and presumably high genotoxicity. Indeed, one study reported extreme toxicity while attempting to modify a repetitive sequence present at 151 copies in a cancer cell line23. Multiplex editing protocols that are intrinsically less cytotoxic must be developed if RGEs are to be studied with any detail. Improved tools to alter the structure and expression of these elements is required to properly interrogate and assess the function of this major component of eukaryotic genomic structure.
Methods of multiplex genome editing
A host of different genome editing technologies, based primarily around engineered Cas9 systems, but also including other CRISPR components such as Cpf1, and microbial genome engineering methods, have seen rapid development in recent years (Figure 2). Each technology brings specific capabilities and limitations with regard to multiplex editing, which will be explored below.
Figure 2.
Technologies for introducing multiplex genome edits.
Base editing
One potentially powerful method is the recent development of base-editing technologies built upon the CRISPR platform65,66,67 which rely on deaminases to target specific bases for conversion. Deamination of a target cytosine to uracil leads to conversion to a thymidine by cellular DNA repair processes, enabling C→T (or G→A in the complementary strand) transitions of bases within the target window. And with the development of adenine base editors, evolved from a tRNA adenosine deaminase, enabling A→G transitions (or T→C in the complimentary strand), it is now possible to target all single base transitions located an appropriate distance from PAM sequences. As this nascent technology improves, increases in efficiency and target base specificity may be expected.
Base editing offers a number of advantages, including the ability to generate sequence changes in a more defined manner than NHEJ, and with potentially greater efficiency that current methods of HDR. One especially intriguing property of base editor technology is that genomic edits are introduced without the generation of DSBs. This mechanism of editing not only reduces the genotoxicity that arises from cellular sensing of multiple DSBs, but also mitigates the possibilty of genomic translocations caused by improper relegation of mispaired ends during DNA repair. Base editors may thus offer a higher ceiling to multiplex editing, as the process becomes limited more by delivery and editing efficiency than by toxicity.
Programmable recombinases
Research into methods that increase nuclease-targeted homology-directed repair (HDR) in higher eukaryotic cells have been aggressively pursued since the very dawn of the field of genome editing. As an alternative cellular process to NHEJ, which introduces small, random, insertions or deletions following error-prone repair, HDR enables specified sequence changes through incorporation of an exogenous donor DNA template68. The ability to specify arbitrary sequence changes with high efficiency in the DNA of living cells would represent a major break from the already revolutionary capabilities enabled by CRISPR technology. However, in practice HDR rates are generally low in higher eukaryotic cells, precluding many theoretical applications. Despite the potential power of high-efficiency HDR, and the great amount of effort put into solving this problem, there is, as yet, no general solution. As discussed above, targeted base-editing technology holds great potential in generating defined sequence changes, but it is as yet unclear how efficient this approach may be, and at what scale it may be practically multiplexed.
An alternative approach to this problem would use site-specific recombinases or integrases, functioning in a manner analogous to the widely used Cre recombinase69 or piggybac transposase70, but with user-defined sequence specificity. Through the action of paired recombinase sites, potentially any sequence could be enzymatically deleted, inverted, inserted, or exchanged with base pair resolution. This has a number of theoretical advantages.
Whereas nuclease-based editing is mediated by stochastic endogenous DNA repair mechanisms, a recombinase-based approach directly performs targeted strand cleavage, exchange, and re-ligation. Where HDR must compete with NHEJ at the site of repair, modification via a recombinase provides an inherently more predictable sequence outcome. Furthermore, natural rates of HDR vary greatly between genomic loci, are altered during the course of the cell cycle, and differ between cell types. Consistently high rates of homologous recombination are generally only achieved in cells artificially stalled during key points of the cell cycle, or in mutant cell lines that exhibit high basal rates of homologous recombination71,72. A recombinase, as the sole mediator of editing, could potentially normalize modification rates independent of these variables. Additionally, as the majority of cells in the body are post-mitotic and thus exhibit low HDR, donor-based gene conversion approaches may not be applicable in vivo in non-cycling cells72,73. A recombinase approach, or at least one that is fundamentally different from invocation of HDR process –especially when enacted across multiple targets – may be required for in vivo modification of post-mitotic tissue cells.
Finally, whereas multiple DSBs are known to trigger apoptotic cellular responses, the catalytic mechanism of many recombinases may be inherently less prone to induction of DNA damage responses and associated cellular toxicity. During recombination, the recombinase catalytic intermediate is covalently linked the to the DNA backbone, with unlinked free ends held within the tetrameric recombinase complex, preventing detection by DNA damage surveillance proteins74. While prolonged expression of certain recombinases can negatively affect genome stability75, this may be avoided if expressed or delivered transiently as with nuclease-based editors.
Despite the potential of recombinase-based genome editing tools, and a two-decade history of recombinase engineering, even the best demonstrations of programmable recombinases to date have yet to achieve wider adoption as genome editing tools. Ideally, a recombinase technology would be i) fully programmable to arbitrary DNA sequences, ii) offer mechanistic control over recombination directionality, and iii) operate with high efficiency. To date, no single recombinase technology satisfies these criteria.
Simple translation fusions of DNA-binding domains to integrases and transposases have been used to tether integration complexes to a target sequence, thereby increasing integration around that locus. Other efforts have used highly engineered chimeric recombinase fusions that attempt to replicate some aspects of native recombinase functionality, with fusion to zinc fingers76, TALE arrays77, and dCas978 having been demonstrated. However, all of these suffer from a host of defects that prevent their wider adoption, including low efficiency, catalytic domain-constrained sequence preferences, and lack of control over the directionality of the recombined product. As an obligate tetrameric enzymatic process, the orchestration of a recombinase reaction may prove technically challenging74. Not only must catalytic domains be active as chimera, they must also be appropriately spaced in the context of the dimeric recombinase full-site, and need to interact as tetramers in a manner that provides control over directionality of recombinase resolution. And as a further challenge, the catalytic domain itself must impart minimal sequence preferences on the action of the chimeric enzyme. Further insights into catalytic domain sequence preferences, and basic enzymatic mechanisms, should inform future efforts to generate truly programmable recombinases.
In the absence of programmable recombinase technology, existing site-specific enzymes may still have utility in large-scale editing efforts. Recombinases, perhaps best exemplified by the now ubiquitous Cre/loxP and Flp/FRT tyrosine recombinase systems, allow efficient exchange between donor DNA and the genome69. The difficulty in engineering the sequence recognition of recombinases generally limits their utility to cases of large integration or exchange events, and necessitates pre-engineering of the target genome to contain the recombinase target site. For certain genome engineering goals however, this may not be limiting. Through strategic use of expanded panels of orthogonal recombinases, such as the diverse set of characterized serine recombinases79, a pipeline based on orthogonal recombinase exchange events could enable certain bottom-up genome engineering goals.
Large donor DNAs
An approach distinct from triggering multiplex editing events may be to manifest an identical sequence outcome by converting multiple positions simultaneously via large donor DNAs and homology-directed repair processes, triggered by a comparatively small number of DSB events. Alternatively, chromosomal regions could be altered via recombinase-mediated cassette exchange, inserting donor material through the action of engineered recombinases. Regardless of the means of introduction, the engineering of appropriate donor material must proceed at scale relevant to multi-locus editing goals.
DNA synthesis services can today generate sequences kilobases (kb) in length. However, donors of this size may only be of utility in cases where desired sequence edits are closely spaced, as direct synthesis beyond 1 kb can become costly. Thus, methods that can rapidly engineer DNAs on the order of tens to hundreds of kilobases are needed to address sequence variation in higher eukaryotic genomes. When considering the introduction of hundreds to millions of genomic modifications into a eukaryotic genome, experimental pipelines initially developed for prokaryotic genome-engineering efforts take on new relevance40.
Several technologies with a high degree of multiplexing must be combined to streamline whole genome engineering. Thankfully, integrated protocols for introducing thousands of widely-spaced edits into bacterial DNA have already been developed. MAGE (as described above) provides a scalable pipeline for engineering of sequence variants in E. coli. Combined with the availability of BAC libraries for a number of mammalian genomes, a process of high throughput MAGE editing in bacteria, followed by CAGE (conjugative assembly genome engineering) assembly of multiple MAGE-edited fragments could rapidly build up edited donors of hundreds of kilobases prior to delivery into mammalian cells38.
Multiple BAC-sized constructs could even be combined into donor material that approaches megabase (Mb) scale. The exo, beta, and gam genes from the λ-Red phage responsible for bacterial recombineering can process a double stranded DNA fragment to single stranded DNA, which can then be incorporated into the lagging strand during replication. Recently a 100 kb DNA was cut out of the episomal vector via CRISPR/Cas9 and incorporated into the E. coli genome via λ-Red recombineering80. If many of these large fragments can be incorporated in close proximity, then excised via recombinases in a circular form, they can potentially be delivered to mammalian cells. Alternatively, DNA assembly in yeast can generate a complete 580 kb mycobacterium genome from 35 kb sub-genomic fragments54. Yeast assembly may thus be extraordinary useful for large-scale genome engineering.
Generating large ssDNA
Irrespective of how large donors are constructed, the efficiency of integration must be maximized. In the absence of efficient programmable recombinases, or strategic use of their natural recombinase counterparts, HDR rates will need to be augmented. One approach to augmenting DSB-triggered HDR is the use of single-stranded DNA. Single-stranded DNA confers higher modification rates than double-strand DNA (dsDNA) and also minimizes the required length of sequence homology arms81,82.
Short ssDNA oligo deoxynucleotide (ssODN) donors are synthetically accessible at low cost, making their use popular in HDR editing4,5,83 experiments. But given the still low and variable HDR rates, and direct competition of each event with error-prone NHEJ, multiplex HDR with ssODNs is practically unlikely to scale for even modest multiplex editing goals. Larger insertions require DNA donors that may not be as cost-effective to synthesize as ssODNs, or donors that are beyond current DNA synthesis length capabilities. Thus, bacterially-produced plasmid or BAC DNA is typically used, and often in dsDNA form. This is despite the higher efficiency of ssDNA as a donor. The difficulty in isolating sufficient quantities of large, high quality ssDNA has limited the adoption of large ssDNA donor generation methods.
Current methods of producing large ssDNA donors rely on either production of phagemid constructs, primer extension and linear amplification of circular bacterial episome templates, or gel electrophoretic separation of large DNA strands81,84,85. Phagemid constructs are attractive, as standard plasmid cloning, arraying, and library generation methods can be applied directly to phagemid construction. However, excessively long M13 filamentous phage particles result in low production yields, with a practical limit to ssDNA phagemid length of less than 10-kb, especially if phagemid preparations are to be parallelized. Linear ssDNA polymerization methods are similarly flexible and potentially scalable to multiple donor species, but here again donor length is constrained by the processivity of existing polymerases. Finally, though gel electrophoretic separation does allow isolation of large, multi-kilobase ssDNA donors, the procedure is laborious, low-yielding, and practically difficult to parallelize to genome-scale donor library production.
Emerging DNA synthesis technologies continue to push the envelope of synthetic DNA length and cost. If appropriately developed with novel in vitro assembly methods, synthetic DNA inputs could yield large, multikilobase ssDNA donors. Alternatively, novel recombinant DNA techniques that allow input of large dsDNA constructs such as BACs or YACs, and enzymatically manipulate the products to isolate specific strands could be especially powerful.
Programmed genome rearrangement
Looking further afield, there are range of natural processes that appear to generate large-scale genomic rearrangements and reductions in a programmed manner. Large-scale genome rearrangements occur in a number of organisms, and the mechanisms underlying these processes are only beginning to be understood. If the specific factors responsible for orchestrating programmed genome rearrangment can be identified and abstracted for use as molecular tools, then they may enable future genome-scale engineering efforts.
Millions of base pairs are eliminated from somatic tissues of the Lamprey following programmed rearrangement86. And unicellar eukaryotic organisms, including the protozoans tetrahymena and oxytricha, undergo perhaps the most dramatic examples of genome-scale editing. The transcriptionally-active ‘macronucleus’ of these organisms is rearranged on a massive scale in comparison to the germline ‘micronucleus’, with upwards of 225,000 fragments being rearranged during macronuclear formation87. The reproduction and survival of the organism depends upon faithful execution of this program in every generation. Despite the thousands of fragments in play, the correct genomic products are rearranged with basepair resolution, genome wide. There is potential for the processes that mediate natural genome rearrangement to be adapted as genome engineering tools.
Orchestration of this process involves a multitude cellular factors, which may converge non-coding RNAs and RNAi related pathways88,89. This suggests a potential mechanism driven ultimately by the rules of nucleic acid base pairing, and is consistent with the seeming sequence flexibility of the rearrangement process. Regardless of the molecular mechanistic details, the existence of such natural genome rearrangement processes is encouraging evidence that multiplex genetic alterations can occur on a truly genomic scale, and demonstrates one potential route towards that goal. Further study into this mechanism is ongoing at labs around the world, and ultimate elucidation of such protozoan genome-rearrangement pathways may someday lead to a new class of genome engineering tools.
Multiplex delivery
As emerging methods maximize the per-guide efficiency of modifications, and minimize the toxicity of editing itself, our ability to multiplex may be constrained by our ability to deliver a requisitely large number of guides to target cells. Regardless of the specific mechanisms effecting genomic modifications, highly multiplexed editing goals will require methods of delivering more complex cargoes than those established for single or duplex editing experiments. Moreover, as any given method will carry theoretical and practical limits to the number of distinct gRNAs and donors that can be accommodated, large-scale genome editing efforts will likely require approaches to multiplex delivery that are sufficiently fast, cost-effective, and with low cytotoxicity such that they can be iterated over successive rounds of modification within experimentally tractable timescales.
One approach that offers simple multiplexing is delivery of expressed Cas9:gRNA ribonucleoprotein (RNP) complexes, whether by lipid nanoparticles34,36 or electroporation35. The direct delivery of editing material avoids potentially troublesome combinatorial, repeat DNA cloning steps for multiple gRNAs, and allows simple pre-mixing of in vitro transcribed sequences. Another advantage of this approach is the short half-life of the delivered material, cells experience nuclease activity within a short temporal window, which in addition to measurably reducing off-target effects34–36, may also reduce genotoxicity, potentially allowing frequent, repeat-rounds of modification. Though only a small quantity of material is delivered, and is only present for a short time, the irreversible nature of error-prone NHEJ events makes RNP delivery a powerful approach to targeted gene knockout. RNP delivery has also been demonstrated for the Cas9-deaminase BE390. The absence of DSB-triggered genotoxicity from this strategy may provide additional reductions in multiplex delivery toxicity, further enabling the development of iterative, RNP-based multiplex editing protocols.
Current approaches to RNP delivery rely on lipid nanoparticle transfection or electroporation. While effective for a given instance of modification, the cumulative toxicity of such methods may limit repeated rounds of modification over experimentally short times. Recent developments in ex vivo delivery may further enable multiplex editing. These include microfluidic approaches where passing cells at high speed through constrictions smaller than cell diameter results in transient disruption of cell membranes, allowing cargo in solution to pass through91. This approach has been further augmented by application of an electric field that disrupts nuclear membranes, permitting both cytoplasmic and nuclear delivery92. In addition, advances in nanomaterials are providing novel approaches to cell delivery. A substrate-only system includes the use of nanowires coated with molecules that are released when cells are penetrated during culture on the nanowire substrate, allowing co-delivery of proteins and siRNAs93. Another system utilizes a nanofabricated substrate combined with laser pulse-illumination, generating controlled microcavitation bubbles to transiently permeate cell membranes in close proximity to the substrate, allowing delivery of RNP-sized cargo94. In combination with improvements in cell viability compared to traditional methods (e.g. electroporation), these approaches may permit delivery of multiplex gene editing cargos at high efficiency into cells ex vivo, though large DNA cargos remain a delivery challenge.
Viral vectors offer distinct approaches to multiplex ex vivo delivery. Transduction with multiple low-capacity, non-integrating viral vectors at once is one potential route. Choice of viral vector would be key, as production methods must not only scale to either parallel or pooled library production, but high multiplicity of infection must be both achievable (necessitating high production titers) and induce low innate intracellular antiviral responses. This would allow multiple viral genomes to transduce a given cell, and the transduction process might be iterated rapidly. A viral vector such as recombinant adeno-associated virus (rAAV) is one candidate for this, though new methods to augment co-incident transduction events, and to reduce vector immune signaling may be necessary to apply this approach within a pipeline of genome-scale editing. A distinct viral approach would utilize large DNA viral vectors like herpes simplex virus (HSV) amplicon vectors. The ~150-kb packaging capacity of HSV replicons potentially offers the ability to deliver a large number of gRNAs on a single vector. However, if applied to genome-scale editing, upstream assembly of such a vector, and appropriate replication and packaging of such a large repetitive construct, may prove practically difficult.
Finally, where current cell culture and ex vivo approaches to delivery offer higher multiplex capacity, in vivo multiplex delivery capabilities are comparatively limited. General avenues for in vivo delivery of multiple simultaneous gene editors include nanoparticles (lipid, polymer), viral (AAV, lentivirus), and even whole tissue electroporation95. However, nanoparticles are constrained by the bioavailability of individual components across formulation methods, viral vectors are restricted by DNA cargo capacity (AAV) and unpredictable effects of genome integration (lentivirus), and electroporation is limited by physical accessibility of target tissues. Though the inability to introduce a very large number of changes in vivo is unlikely to be a barrier to any near therapeutic application, application of multiplex antiviral defense in vivo, though requiring relatively few edits, will face distinct delivery challenges and necessitate new multiplex delivery methods.
Delivery of large DNAs
As discussed above, the use of large donor DNAs could enable effectively-multiplexed higher eukaryotic genome editing. However, no matter how efficient edited donor generation pipelines become, all donor material must ultimately be delivered to mammalian cells with sufficient efficiency to recover modified clones. Genome-scale engineering may require repeated delivery of DNAs ranging from many hundreds of kilobases to megabases in size. However, established methods of introducing such large DNAs such as microinjection are very low throughput, toxic to recipients, and maybe subject the delivered material to mechanical shearing. Cell-cell fusion-based approaches may stabilize large DNAs during delivery, but are themselves extremely inefficient. Barriers to fusion-based delivery include complications at the level of initial fusion with recipient cell bodies, and subsequent import or incorporation of DNA into the nucleus.
Large human artificial chromosomes (HACs) like chromosome 14 and 21 have been built and are essential to generating humanized animal models or to study phenotypes in the context of a different haplotypes96. Traditionally they have been delivered by microcell-mediated chromosome transfer (MMCT). MMCT is an arduous procedure that first requires transfer and manipulation of DNA into a recombinogenic line, followed by a series of culture treatments that result in condensation of chromatin and envelopment of chromosomal material in membrane-bound cell fragments, which are then fused to recipient cells via fusogens like polyethylene-glycol or virus-mediated agglutinization97. Following this demanding process, the efficiency of incorporation is extremely low, on the order of 1×10−6, precluding routine use as part of a genome-scale engineering effort.
Another delivery alternative recently reported involves the fusion of yeast spheroplasts (or cell-wall free yeast) with cultured mammalian cells98. The advantages of such a delivery system are attractive, as it may interface seamlessly with upstream MAGE and yeast-assembly methods. Though 1000-fold more efficient than MMCT, this yeast-based DNA delivery protocol is currently limited to roughly 0.1% in cultured cells. Higher efficiencies may be needed to cost-effectively apply this approach to genome engineering on the gigabase scale found in higher eukaryotic organisms.
One major bottleneck in this process may be the post-fusion breakdown of the yeast nucleus in recipient cells. Yeast natively have a closed mitosis, and the release of yeast nucleus-borne genetic material, by as-yet undefined processes, is likely extremely inefficient. Moreover, the presence of large DNAs in the cytoplasmic space likely triggers cellular antiviral responses. Finally, the entire yeast nuclear content is delivered to recipient cells, unwanted yeast genomic DNA is incorporated into host cells alongside the desired material.
Future development of this system to enable yeast nuclear breakdown, or nucleus-nucleus fusion, as occurs naturally during yeast sexual reproduction, within the recipient cell, could dramatically increase transfer efficiency. Nucleus-nucleus fusion may also effectively evade certain antiviral responses. Finally, the development of methods to degrade, exclude, or otherwise negate the yeast genomic material in favor of the desired donor material would greatly increase the utility of this approach.
As with bacterial MAGE-based genome engineering, a mammalian genome writing campaign would use a tiered program of parallel engineering efforts, with modified genomic regions being built up in separate lineages, ultimately requiring hybridization of complementarily-editing lines. To avoid the inefficiencies of MMCT at this stage, improvements in cell-cell fusion are especially attractive. Recently, the discovery of fusogenic peptide Myomixer and paired receptor Myomaker has been shown to mediate surprisingly efficient fusion between myoblasts, fibroblasts or myoblast-fibroblast heterotypic cell fusions99. Strategies that augment target cell fusion, when applied to the delivery of large DNAs, could facilitate future genome-scale engineering.
Conclusions
Multiplexed genome editing as enabled by CRISPR-based tools has to potential to transform our ability to study complex biological problems, and enable sophisticated therapeutic modalities. Extending bacterial and yeast genome engineering protocols to the generation of edited mammalian donors may enable actual genome-scale engineering when combined with CRISPR-guided genomic integration. Multiplexing DNA synthesis, editing, assembly, and delivery technologies are at the core of streamlining large genome engineering such as the projects envisioned by GP-write and required for de-extinction efforts. These future applications require fundamental improvements and new developments to the effectors of editing, the production of donor material, and the delivery of both. In coming years, progress on these fronts will foster a new era of genome biology, where researchers gain the ability to systematically alter genomes on a massive scale
Acknowledgments
Authors D.B.T., C.J.S., S.W., and G.M.C. were supported by a DARPA Safe Genes grant, N66001-17-2-4056. S.A. was supported by an NIH T32 Genetics and Genomics PhD Training Grant, GM096911. S.A., O.C., and G.M.C. were also supported by an NIH/NHGRI CEGS CGEO award, 5RM1HG008525-03. E.H. was supported by an award from Charoen Pokphand Group, A27136. O.C. was supported by a Boehringer Ingelheim Fonds fellowship. Finally, D.B.T. was supported by a Wyss Technology Development Fellowship. G.M.C. is a co-founder of Editas Medicine and eGenesis Bio and serves advisory roles in several companies involved in genome editing and engineering. A detailed listing of G.M.C.'s Tech Transfer, Advisory Roles and Funding Sources can be obtained from http://arep.med.harvard.edu/gmc/tech.html.
Citations
- 1.Mojica FJM, Diez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beerli RR, Barbas CF. Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 8.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AN, Gross CA, Dixit A, Parnas O, Regev A, Weissman JS. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882.e21. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J-F, Aach J, Norville JE, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated plant genome editing via guide RNA/Cas9. Nat. Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPRMediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014;4:srep05400. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017;35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Güell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J, Cortazio R, Wilkinson RA, Fishman JA, Church G. Genome-wide inactivation of porcine endogenous retroviruses (PERVs) Science. 2015;350:1101–1104. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- 22.Niu D, Wei H-J, Lin L, George H, Wang T, Lee I-H, Zhao H-Y, Wang Y, Kan Y, Shrock E, Lesha E, Wang G, Luo Y, Qing Y, Jiao D, Zhao H, Zhou X, Wang S, Wei H, Guell M, Church GM, Yang L. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science. 2017:eaan4187. doi: 10.1126/science.aan4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang C-Z, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, Kost-Alimova M, Gill S, Xu H, Ali LD, Jiang G, Pantel S, Lee Y, Goodale A, Cherniack AD, Oh C, Kryukov G, Cowley GS, Garraway LA, Stegmaier K, Roberts CW, Golub TR, Meyerson M, Root DE, Tsherniak A, Hahn WC. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuscu C, Kuscu C, Tufan T, Yang J, Adli M. CRISPR-STOP: Gene silencing through base editing-induced nonsense mutations. Protoc. Exch. 2017 doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, Bao G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. doi: 10.1016/j.stem.2016.07.001. PubMed - NCBI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and Structure-Specific RNA Processing by a CRISPR Endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and Programmable Regulation of Gene Networks with an Integrated RNA and CRISPR/Cas Toolkit in Human Cells. Mol. Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 Is a Single RNAGuided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017;35:789–792. doi: 10.1038/nbt.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 34.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen Z-Y, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient proteinbased genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Kim D, Cho SW, Kim J, Kim J-S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N, Chesnut JD. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 37.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Emig CJ, Bang D, Hwang SJ, Jewett MC, Jacobson JM, Church GM. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lajoie MJ, Rovner AJ, Goodman DB, Aerni H-R, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, et al. Genomically Recoded Organisms Expand Biological Functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, Cervantes N, Zhou M, Singh K, Napolitano MG, Moosburner M, Shrock E, Pruitt BW, Conway N, Goodman DB, Gardner CL, Tyree G, Gonzales A, Wanner BL, Norville JE, Lajoie MJ, Church GM. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 41.Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-Based Genome Editing for Correction of Dystrophin Mutations that Cause Duchenne Muscular Dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim WA, June CH. The Principles of Engineering Immune Cells to Treat Cancer. Cell. 2017;168:724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Zhang Y, Cheng C, Cheng AW, Zhang X, Li N, Xia C, Wei X, Liu X, Wang H. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017;27:154–157. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soppe JA, Lebbink RJ. Antiviral Goes Viral: Harnessing CRISPR/Cas9 to Combat Viruses in Humans. Trends Microbiol. doi: 10.1016/j.tim.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Wang G, Zhao N, Berkhout B, Das AT. CRISPR-Cas9 Can Inhibit HIV-1 Replication but NHEJ Repair Facilitates Virus Escape. Mol. Ther. 2016;24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin S-R, Yang H-C, Kuo Y-T, Liu C-J, Yang T-Y, Sung K-C, Lin Y-Y, Wang H-Y, Wang C-C, Shen Y-C, Wu F-Y, Kao J-H, Chen D-S, Chen P-J. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol. Ther. Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Diemen FR, Kruse EM, Hooykaas MJG, Bruggeling CE, Schurch AC, van Ham PM, Imhof SM, Nijhuis M, Wiertz EJHJ, Lebbink RJ. CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections. PLoS Pathog. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, Wainberg MA, Liang C. CRISPR/Cas9-Derived Mutations Both Inhibit HIV-1 Replication and Accelerate Viral Escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 49.Sakuma T, Masaki K, Abe-Chayama H, Mochida K, Yamamoto T, Chayama K. Highly multiplexed CRISPR-Cas9-nuclease and Cas9-nickase vectors for inactivation of hepatitis B virus. Genes Cells. 2016;21:1253–1262. doi: 10.1111/gtc.12437. [DOI] [PubMed] [Google Scholar]
- 50.Bialek JK, Dunay GA, Voges M, Schäfer C, Spohn M, Stucka R, Hauber J, Lange UC. Targeted HIV-1 Latency Reversal Using CRISPR/Cas9-Derived Transcriptional Activator Systems. PloS One. 2016;11:e0158294. doi: 10.1371/journal.pone.0158294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bogerd HP, Kornepati AVR, Marshall JB, Kennedy EM, Cullen BR. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E7249–7256. doi: 10.1073/pnas.1516305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiani S, Chavez A, Tuttle M, Hall RN, Chari R, Ter-Ovanesyan D, Qian J, Pruitt BW, Beal J, Vora S, Buchthal J, Kowal EJK, Ebrahimkhani MR, Collins JJ, Weiss R, Church G. Cas9 gRNA engineering for genome editing, activation and repression. Nat. Methods. 2015;12:1051–1054. doi: 10.1038/nmeth.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boeke JD, Church G, Hessel A, Kelley NJ, Arkin A, Cai Y, Carlson R, Chakravarti A, Cornish VW, Holt L, Isaacs FJ, Kuiken T, Lajoie M, Lessor T, Lunshof J, Maurano MT, Mitchell LA, Rine J, Rosser S, Sanjana NE, Silver PA, Valle D, Wang H, Way JC, Yang L. GENOME ENGINEERING. The Genome Project-Write. Science. 2016;353:126–127. doi: 10.1126/science.aaf6850. [DOI] [PubMed] [Google Scholar]
- 54.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA, Smith HO. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 55.Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, DiCarlo JE, Lee D, Huang CLV, Chandrasegaran S, Cai Y, Boeke JD, Bader JS. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 56.Mitchell LA, Wang A, Stracquadanio G, Kuang Z, Wang X, Yang K, Richardson S, Martin JA, Zhao Y, Walker R, Luo Y, Dai H, Dong K, Tang Z, Yang Y, Cai Y, Heguy A, Ueberheide B, Fenyo D, Dai J, Bader JS, Boeke JD. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017;355 doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 57.Rogers RL, Slatkin M. Excess of genomic defects in a woolly mammoth on Wrangel island. PLoS Genet. 2017;13:e1006601. doi: 10.1371/journal.pgen.1006601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller W, Drautz DI, Ratan A, Pusey B, Qi J, Lesk AM, Tomsho LP, Packard MD, Zhao F, Sher A, Tikhonov A, Raney B, Patterson N, Lindblad-Toh K, Lander ES, Knight JR, Irzyk GP, Fredrikson KM, Harkins TT, Sheridan S, Pringle T, Schuster SC. Sequencing the nuclear genome of the extinct woolly mammoth. Nature. 2008;456:387–390. doi: 10.1038/nature07446. [DOI] [PubMed] [Google Scholar]
- 59.Rohland N, Reich D, Mallick S, Meyer M, Green RE, Georgiadis NJ, Roca AL, Hofreiter M. Genomic DNA sequences from mastodon and woolly mammoth reveal deep speciation of forest and savanna elephants. PLoS Biol. 2010;8:e1000564. doi: 10.1371/journal.pbio.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA Elements: A Hominid-specific Retroposon Family. J. Mol. Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 62.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muotri AR, Marchetto MCN, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kemp JR, Longworth MS. Crossing the LINE Toward Genomic Instability: LINE-1 Retrotransposition in Cancer. Front. Chem. 2015;3 doi: 10.3389/fchem.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meinke G, Bohm A, Hauber J, Pisabarro MT, Buchholz F. Cre Recombinase and Other Tyrosine Recombinases. Chem. Rev. 2016;116:12785–12820. doi: 10.1021/acs.chemrev.6b00077. [DOI] [PubMed] [Google Scholar]
- 70.Di Matteo M, Mátrai J, Belay E, Firdissa T, Vandendriessche T, Chuah MKL. PiggyBac toolbox. Methods Mol. Biol. Clifton NJ. 2012;859:241–254. doi: 10.1007/978-1-61779-603-6_14. [DOI] [PubMed] [Google Scholar]
- 71.Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem. J. 2009;423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle Georget. Tex. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous endjoining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 75.Janbandhu VC, Moik D, Fässler R. Cre recombinase induces DNA damage and tetraploidy in the absence of loxP sites. Cell Cycle Georget. Tex. 2014;13:462–470. doi: 10.4161/cc.27271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mercer AC, Gaj T, Fuller RP, Barbas CF. Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res. 2012;40:11163–11172. doi: 10.1093/nar/gks875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chaikind B, Bessen JL, Thompson DB, Hu JH, Liu DR. A programmable Cas9- serine recombinase fusion protein that operates on DNA sequences in mammalian cells. Nucleic Acids Res. 2016;44:9758–9770. doi: 10.1093/nar/gkw707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu Z, Thomas L, Davies B, Chalmers R, Smith M, Brown W. Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol. 2013;13:87. doi: 10.1186/1472-6750-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang K, Fredens J, Brunner SF, Kim SH, Chia T, Chin JW. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, Redder R, Richardson GP, Inagaki Y, Sakai D, Buckley SM, Seshacharyulu P, Batra SK, Behlke MA, Zeiner SA, Jacobi AM, Izu Y, Thoreson WB, Urness LD, Mansour SL, Ohtsuka M, Gurumurthy CB. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017;18:92. doi: 10.1186/s13059-017-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paix A, Folkmann A, Goldman DH, Kulaga H, Grzelak M, Rasoloson D, Paidemarry S, Green R, Reed R, Seydoux G. Precision genome editing using synthesis-dependent repair of Cas9- induced DNA breaks. bioRxiv. 2017:161109. doi: 10.1073/pnas.1711979114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Papaioannou I, Simons JP, Owen JS. Oligonucleotide-directed gene-editing technology: mechanisms and future prospects. Expert Opin. Biol. Ther. 2012;12:329–342. doi: 10.1517/14712598.2012.660522. [DOI] [PubMed] [Google Scholar]
- 84.Wrenbeck EE, Klesmith JR, Stapleton JA, Adeniran A, Tyo KEJ, Whitehead TA. Plasmid-based one-pot saturation mutagenesis. Nat. Methods. 2016;13:928–930. doi: 10.1038/nmeth.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hegedüs É, Kókai E, Kotlyar A, Dombrádi V, Szabó G. Separation of 1–23-kb complementary DNA strands by urea–agarose gel electrophoresis. Nucleic Acids Res. 2009;37:e112. doi: 10.1093/nar/gkp539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc. Natl. Acad. Sci. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen X, Bracht JR, Goldman AD, Dolzhenko E, Clay DM, Swart EC, Perlman DH, Doak TG, Stuart A, Amemiya CT, Sebra RP, Landweber LF. The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell. 2014;158:1187–1198. doi: 10.1016/j.cell.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mochizuki K. Developmentally Programmed, RNA-Directed Genome Rearrangement in Tetrahymena. Dev. Growth Differ. 2012;54:108–119. doi: 10.1111/j.1440-169X.2011.01305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yerlici VT, Landweber LF. Programmed Genome Rearrangements in the Ciliate Oxytricha. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0025-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rees HA, Komor AC, Yeh W-H, Caetano-Lopes J, Warman M, Edge ASB, Liu DR. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sharei A, Zoldan J, Adamo A, Sim WY, Cho N, Jackson E, Mao S, Schneider S, Han M-J, Lytton-Jean A, Basto PA, Jhunjhunwala S, Lee J, Heller DA, Kang JW, Hartoularos GC, Kim K-S, Anderson DG, Langer R, Jensen KF. A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2082–2087. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ding X, Stewart MP, Sharei A, Weaver JC, Langer RS, Jensen KF. Highthroughput nuclear delivery and rapid expression of DNA via mechanical and electrical cell-membrane disruption. Nat. Biomed. Eng. 2017;1 doi: 10.1038/s41551-017-0039. s41551-017-0039-017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shalek AK, Robinson JT, Karp ES, Lee JS, Ahn D-R, Yoon M-H, Sutton A, Jorgolli M, Gertner RS, Gujral TS, MacBeath G, Yang EG, Park H. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc. Natl. Acad. Sci. 2010;107:1870–1875. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saklayen N, Huber M, Madrid M, Nuzzo V, Vulis DI, Shen W, Nelson J, McClelland AA, Heisterkamp A, Mazur E. Intracellular Delivery Using Nanosecond-Laser Excitation of Large- Area Plasmonic Substrates. ACS Nano. 2017;11:3671–3680. doi: 10.1021/acsnano.6b08162. [DOI] [PubMed] [Google Scholar]
- 95.Maresch R, Mueller S, Veltkamp C, Ollinger R, Friedrich M, Heid I, Steiger K, Weber J, Engleitner T, Barenboim M, Klein S, Louzada S, Banerjee R, Strong A, Stauber T, Gross N, Geumann U, Lange S, Ringelhan M, Varela I, Unger K, Yang F, Schmid RM, Vassiliou GS, Braren R, Schneider G, Heikenwalder M, Bradley A, Saur D, Rad R. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat. Commun. 2016;7:10770. doi: 10.1038/ncomms10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kazuki Y, Oshimura M. Human artificial chromosomes for gene delivery and the development of animal models. Mol. Ther. J. Am. Soc. Gene Ther. 2011;19:1591–1601. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Doherty AMO, Fisher EMC. Microcell-mediated chromosome transfer (MMCT): small cells with huge potential. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2003;14:583–592. doi: 10.1007/s00335-003-4002-0. [DOI] [PubMed] [Google Scholar]
- 98.Brown DM, Chan YA, Desai PJ, Grzesik P, Oldfield LM, Vashee S, Way JC, Silver PA, Glass JI. Efficient size-independent chromosome delivery from yeast to cultured cell lines. Nucleic Acids Res. 2017;45:e50. doi: 10.1093/nar/gkw1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bi P, Ramirez-Martinez A, Li H, Cannavino J, McAnally JR, Shelton JM, Saánchez-Ortiz E, Bassel-Duby R, Olson EN. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 2017;356:323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]