Abstract
Activation of the lysosomal ceramide-producing enzyme, acid sphingomyelinase (ASM), by various stresses is centrally involved in cell death and has been implicated in autophagy. We set out to investigate the role of the baseline ASM activity in maintaining physiological functions of lysosomes, focusing on the lysosomal nutrient-sensing complex (LYNUS), a lysosomal membrane-anchored multiprotein complex that includes mammalian target of rapamycin (mTOR) and transcription factor EB (TFEB). ASM inhibition with imipramine or sphingomyelin phosphodiesterase 1 (SMPD1) siRNA in human lung cells, or by transgenic Smpd1+/− haploinsufficiency of mouse lungs, markedly reduced mTOR- and P70-S6 kinase (Thr 389)-phosphorylation and modified TFEB in a pattern consistent with its activation. Inhibition of baseline ASM activity significantly increased autophagy with preserved degradative potential. Pulse labeling of sphingolipid metabolites revealed that ASM inhibition markedly decreased sphingosine (Sph) and Sph-1-phosphate (S1P) levels at the level of ceramide hydrolysis. These findings suggest that ASM functions to maintain physiological mTOR signaling and inhibit autophagy and implicate Sph and/or S1P in the control of lysosomal function.
Keywords: lysosome, sphingolipids, membrane, endothelial cells, lung, sphingosine, mammalian target of rapamycin, lysosomal nutrient-sensing complex
The activation above homeostatic levels of acid sphingomyelinase (ASM), a lysosomal phosphodiesterase typically involved in apoptosis via production of ceramide, has recently been implicated in stress-induced macro-autophagy (referred to as autophagy, henceforth) (1, 2). Despite increased understanding of the involvement of ASM’s brisk activation in the stress response, little is known about the role of baseline ASM activity in maintaining lysosomal function during physiological conditions. The causal link between loss of ASM function and the lipid storage disorder, Niemann-Pick disease, revealed a requirement of ASM for proper cellular function and highlighted the essential role of ASM in sphingomyelin breakdown and lysosomal membrane homeostasis (3). Docked at the lysosomal membrane, the lysosomal nutrient-sensing complex (LYNUS) controls protein synthesis and autophagy, an essential mechanism of cellular survival during stress and starvation (4). In turn, aberrant autophagy has been implicated in the pathogenesis of several lung diseases, including chronic obstructive pulmonary diseases (5). Therefore, understanding the molecular determinants of autophagy is of clinical importance.
The lysosomal and secreted forms of ASM, both being encoded by the sphingomyelin phosphodiesterase 1 (SMPD1) gene, catalyze, optimally at acidic pH, the hydrolysis of sphingomyelin into ceramide and phosphorylcholine. ASM activity is rapidly increased during stress, generating excess ceramide that, in turn, interacts with specific signaling molecules to trigger second messenger effects. In addition, ceramide incorporates and alters the biophysical properties of membrane bilayers, such as fluidity and stiffness, by dislodging cholesterol from membrane lipid rafts, impacting acyl chain order parameters of neighboring lipids, and modifying membrane bilayer composition (6). Because ceramides produced in the lysosome by ASM are not shuttled to other subcellular compartments, they may directly influence the coalescence of microdomains required for lysosomal signaling (7). However, ASM forms a molecular complex with acid ceramidase, which metabolizes ceramide to sphingosine (Sph), a molecule that is, itself, implicated in autophagy via alterations of lysosomal calcium efflux (8). We investigated to determine whether ASM is required for LYNUS function of inhibition of autophagy during homeostatic conditions, hypothesizing that inhibition of ASM is sufficient to trigger autophagy associated with disrupted LYNUS signaling.
During plentitude of ATP, amino acids, and growth signals, the mammalian target of rapamycin (mTOR), a serine/threonine kinase component of LYNUS, stimulates protein synthesis (4). This fundamental function of LYNUS requires docking to the lysosomal membrane, ensured by its farnesylated component, the Ras homolog enriched in brain (RHEB) (4). Upon loss of growth signals, mTOR becomes inactive and one of its phosphorylation targets in LYNUS, transcription factor EB (TFEB), translocates to the nucleus, where it upregulates transcription of genes involved in lysosomal biogenesis and autophagy, including its own (9). When activated above homeostatic levels, such as during stress, ASM triggers autophagy with impaired lysosomal degradation (decreased autophagic flux) coupled with decreased mRNA transcripts for TFEB targets (2). Whether this effect is due to production of excess ceramide or to other sphingolipid perturbations remains unclear. Such alterations in sphingolipids may either have second messenger signaling effects, e.g., excess ceramide engages signaling similar to that induced by amino acid depletion (10), or could alter the lysosomal membrane lipid composition and fluidity, potentially affecting LYNUS anchoring.
Utilizing human primary lung endothelial cells, human lung epithelial cells, and murine lungs, we show that inhibition of homeostatic ASM activity is sufficient to inactivate signaling of LYNUS components and to initiate autophagy with degradative potential associated with marked reductions in Sph, rather than ceramide, levels.
MATERIALS AND METHODS
Reagents
Unless otherwise stated, all reagents were purchased from Sigma-Aldrich.
Cell culture
All experiments were performed in full (containing all growth supplements) medium unless otherwise stated. Primary human pulmonary artery endothelial cells (HPAECs) (Invitrogen, C0085C) were cultured in Medium 200 (Invitrogen, M200500) with low serum growth supplement (Invitrogen, S00310). Primary human lung microvascular endothelial cells (HLMVECs) (Invitrogen, CC2527) were cultured in an EGM-2MV BulletKit (Invitrogen, CC3202). Primary human umbilical vein endothelial cells (HUVECs), a kind gift from Dr. Mathias Clauss (Indiana University), were cultured in VascuLife VEGF medium complete kit (LifeLine, LL0003). THP-1 monocytes (ATCC, TIB202) were cultured in RPMI (Invitrogen, 11875119) supplemented with 10% FBS (Thermo Fisher Scientific, sh30910-03HI). Immortalized bronchial epithelial cells (BEAS2b cells) were cultured in DMEM with high glucose and pyruvate (Invitrogen, 11995-073), supplemented with 10% FBS and 1% penicillin/streptomycin (Sigma, P4333-100ML). All cells were incubated at 37°C with 5% CO2 and 100% humidity.
ASM inhibition with imipramine
When cells were 80–90% confluent, the medium was replaced and imipramine or vehicle was added for the indicated time, when cells were washed with 5 ml PBS at 4°C and scraped in 1.5 ml PBS at 4°C. Cells were then spun at 16,000 g for 10 min and the cell pellets were snap-frozen in liquid nitrogen until assayed.
ASM activity
Lysosomal ASM activity was measured with an Amplex Red sphingomyelinase activity kit (Invitrogen, A12220) or radioactivity. Briefly, the kit utilizes an indirect two-step reaction that produces a fluorescent readout of ASM activity. Cells were washed in ice-cold PBS at the end of treatment, scraped in ice-cold PBS, and centrifuged at 16,000 g for 10 min. Supernatant was removed, and the remaining cell pellet was snap-frozen in liquid nitrogen. At the time of assay, pellets were hydrated with a reaction buffer for lysosomal ASM activity, incubated with 5 mM sphingomyelin, 2 U/ml HRP, and 8 U/ml alkaline phosphatase. Kinetic fluorescence measurements were read using a wavelength of 590 nm over a period of 2 h at 37°C in a SpectraMax m2e microplate reader using SoftMax Pro software.
For the radioactive assay, cells pellets were lysed in 75 μl of a buffer containing 25 mM Tris (pH 7.6), 5 mM EDTA, 0.2% Triton-X, phosphatase inhibitors, and protease inhibitors. To 4 ml of the same buffer used to lyse cells, 3.08 μl of 14C-choline methyl sphingomyelin at 55 mCi/mmol and 0.1 mCi/ml were added to create a substrate buffer (1.4 pmol/μl). To 103 μl of the reaction buffer, 35 μl of lysate, 12 μl of 0.2 M acetic acid, and 50 μl of substrate buffer were added, mixed by vortex, and allowed to react at 37°C for 2.3 h. To stop the reaction, 250 μl of a 2:1 (chloroform:methanol) solution were added and vortexed. To this, 800 μl of a 2:1 (methanol:chloroform) solution plus 250 μl of water were added and vortexed. From the aqueous phase, 200 μl were extracted and added to 800 μl of MicroScint PS (Perkin-Elmer, 6013631) and radioactivity quantified on a Topcount scintillation counter.
Western blotting
Cells were washed once with ice-cold PBS, gently scraped into ice-cold PBS, and centrifuged at 16,000 g for 10 min. Then, the supernatant was removed and the pellets were snap-frozen in liquid nitrogen. Cell pellets were thawed at 4°C in cell lysis buffer containing 1% Triton-X (EMD, 9002931), 150 mM NaCl (Thermo Fisher, BP3581), and 50 mM Tris (pH 7.6; Invitrogen, 15504020) and then were vigorously vortexed five times during a 1 h period, followed by centrifugation at 4°C for 10 min and the use of the supernatant for assays as whole cell lysate.
Protein concentration was determined using BCA (Pierce, 23227). Equal amounts of protein (2–20 μg) were diluted in Laemmli 4× buffer (reducing) (Boston Bio Products, NC9099736) and resolved in Criterion 12+2-well 4–20% TGX gels (Bio-Rad, 5671093). A semi-dry transfer apparatus (Bio-Rad, 1703848) was employed to transfer proteins to a polyvinylidene fluoride membrane (EMD, IPVH00010). Membranes were probed with the following antibodies: anti-β-actin (A5441), anti-microtubule-associated protein 1 light chain 3β (LC3B) (Sigma, L7543), anti-vinculin (Abcam, ab10858), anti-GAPDH (Abcam, ab9485), anti-phospho P70-S6 kinase (P70-S6k) (Thr 389) (Cell Signaling Technologies, 9205), and anti-phospho mTOR (Ser 2448) (p-mTOR) (Cell Signaling Technologies, 5536). Appropriate secondary antibodies (goat anti-rabbit/mouse, HRP conjugate) (GE Healthcare, 45001175/45001187) were used in conjunction with ECL Prime (Thermo Fisher, RPN2232) or Luminata Forte (EMDMillipore, WBLUF0500) for chemiluminescent reaction. Images were taken with a ChemiDoc (Bio-Rad) XRS system with ImageLab software.
Densitometry
Density quantification of protein bands in Western blots was performed with ImageJ software (The National Institutes of Health, Bethesda, MD; http://imagej.nih.gov/ij/). Quantification of proteins of interest was performed relative to the intensity of the respective loading controls and this ratio was set equal to one for the experimental control (e.g., vehicle-treated or untreated) group.
Transfections
Nucleofector kits for primary endothelial cells (Lonza, VVPI-1001) in conjunction with Amaxa Nucleofector to transfect HPAECs with siGenome siRNA (Dharmacon, siGlo/non-targeting pool 1/non-targeting pool 2/SMPD1, D0016300205/D-001206-13/D-001206-14/M-006676-01-0020) using the U-017 program per the manufacturer’s protocol. The concentration of siRNA listed was that achieved in 150 μl of the inside cuvette prior to electroporation. After electroporation, cells were added to tissue culture dishes in 10 ml of full (containing all growth supplements) medium. The medium was changed 18 to 24 h after electroporation. Cells were harvested at 48 or 72 h after transfection, as indicated.
Quantitative PCR
Cells were washed with ice-cold PBS, gently scraped in ice-cold PBS, centrifuged at high speed for 10 min, supernatant removed, and pellet snap-frozen in liquid nitrogen. RNA was extracted with a GenElute mammalian total RNA mini kit (Sigma, RTN70-1KT) per the manufacturer’s protocol, and cDNA was reverse transcribed with a high capacity cDNA reverse transcription kit with RNase inhibitor (Applied Biosystems, 4374966). Using SYBR Select Master Mix (Applied Biosystems, 4472908) and SMPD1/PGK1 primers (Qiagen, PPH02494A/PPH02049A), 25 ng of cDNA was amplified on an ABI 7500 real-time PCR system (Applied Biosystems).
Immunofluorescence
HPAECs were cultured (125,000 cells per well) on 18 mm glass coverslips (VWR, 48380046) residing in 12-well dishes (Costar, 3513) coated with a gelatin-based coating solution (Cell Biologics, 6950) and were, after treatment and fixation with formaldehyde, exposed to rabbit anti-TFEB polyclonal antibody, Alexa Fluor 555 conjugated (Bioss, bs-5137R-A555), and mounted with SlowFade Gold Antifade reagent with DAPI (Invitrogen, S36939) to microscope slides (Thermo Fisher, 12-544-3). Images were taken on a Nikon Eclipse 80i microscope.
BEAS2b cells that stably express eGFP-LC3B were cultured in 4-well Lab-Tek (Thermo Fisher Scientific, 154526) slides (50,000 cells per well). After treatment and fixation with formaldehyde, they were probed for lysosome-associated membrane protein 1 (LAMP1) by being exposed to rabbit anti-LAMP1 (Cell Signaling Technology, 9091), Alexa Fluor 594 (Thermo Fisher, A20185), and mounted with SlowFade Gold Antifade reagent with DAPI (Invitrogen, S36939) to coverslips (Thermo Fisher, 12-544-14) and imaged on a Nikon Eclipse 80i microscope.
Electron microscopy
After washing with ice-cold PBS (Fisher), cells were fixed with glutaraldehyde (Sigma, G7776) and formaldehyde (Thermo Fisher, 28908), both at 2% in 0.1 M phosphate buffer for 30 min at room temperature. After two rounds of washing with PBS, cells were gently scraped in PBS, centrifuged for 10 min and further processed by the Electron Microscopy Core Facility, Indiana University School of Medicine, which included post fixing, embedding, cutting, and mounting on slides. Images were taken with a Tecnai G2 12 Bio Twin (FEI, Hillsboro) equipped with an AMR CCD (Advanced Microscopy Techniques).
Stable clone generation
BEAS2b cells were cotransfected (Amaxa program U-017) with mammalian expression plasmid (pEF6/V5-His) expressing C-terminally tagged ASM (11), a kind gift from Dr. Yusuf Hannun (Stony Brook University) and mammalian expression plasmid expressing N-terminally tagged eGFP-LC3B under the control of a cytomegalovirus promoter (12) (a kind gift from William Jackson, Stanford University). To verify punctate formation with autophagic stimuli, cells were transiently transfected with eGFP-LC3B plasmid and treated with chloroquine (supplemental Fig. S1A). Selection pressure was applied with Geneticin (Santa Cruz, sc29065) for several passages. The cotransfected cells were passaged several times and the overexpression of eGFP-LC3B confirmed by immunoblot (supplemental Fig. S1).
Differential centrifugation
Cells were cultured until 80–90% confluent and then treated as indicated, followed by medium removal and ice-cold Accutase (10 ml) addition to dishes. All reagents and centrifuges were kept at 4°C during all procedures. Cells were collected by pipetting and were spun at 500 g for 5 min. Pellets were suspended in 1 ml of PBS and spun again at 500 g for 5 min. After removing supernatants, the pellets were suspended in solution B from Axis-Shield Application S53 plus phosphatase and protease inhibitors. An aliquot was immediately taken and spun at 500 g for 5 min to prepare whole cell samples. After 25 min, each of the 10 samples generated was homogenized separately (30 strokes) in a 1 ml Dounce homogenizer with pestle B. Samples were then spun at 16,000 g for 5 min. The supernatant was labeled cytosolic fraction and the remaining pellet was labeled organelle fraction. The organelle fraction was then resuspended in solution C, as per Axis-Shield Application S53. A 14 ml gradient with 2 ml discontinuous steps was made with v/v dilutions of OptiPrep as follows: 10% (sample), 12%, 14%, 16%, 18%, 20%, and 25%, and spun at 150,000 g for 20 h. Fractions were collected from the bottom using a 3 ml syringe with a 4 inch 16 gauge blunt needle. Six fractions were collected, each 150 μl, starting with fraction six and either added to 4× Laemmli (Boston Bioproducts, NC9099736) for Western blotting or diluted with 10 ml of PBS and spun at 150,000 g for 10 min. The supernatant was decanted and 1 ml of methanol was used to prepare the pellet for lipid extraction.
Lipid extraction
Methanol, water, and acetonitrile (HPLC grade) were purchased from Burdick and Jackson (Muskegon, MI). Sph, dihydro (DH)Sph, a 17-carbon analog of Sph (C17-Sph), S1P, DHS1P, a 17-carbon analog of S1P (C17-S1P), N-acylated (C14:0, C16:0, C18:1, C18:0, C20:0, C24:1, C24:0) Sphs (Ceramides, Cer), N-heptadecanoyl-Sph (C17:0-Cer), N-acylated sphingosylphosphorylcholines (sphingomyelins, N-12:0, N-16:0, N-18:0, N-24:1, and N-24:0), and glucosylceramides (N-16:0, N-24:1, N-24:0) were obtained from Avanti Polar Lipids (Alabaster, AL). N-D3-16:0-glucosylceramide and N-D3-16:0-lactosylceramide were from Matreya LLC (State College, PA). The standards were dissolved in methanol (sphingoid base phosphates were dissolved with the addition of a trace amount of concentrated hydrochloric acid) and stored at −20°C. Pyridine (acetylation grade) and acetic anhydride were products of Alltech Associates (Deerfield, IL).
Cellular lipids were extracted with a modified Bligh and Dyer procedure with the use of 0.1 N HCl for phase separation (13). C17-S1P (30 pmols), C17-Sph (30 pmols), N-C17:0-Cer (30 pmols), N-12:0-sphingosylphosphorylcholine (12:0-SM, 100 pmols), N-D3-16:0-glucosylceramide (25 pmol), and N-D3-16:0-lactosylceramide (25 pmol), employed as internal standards, were added during the initial step of lipid extraction. The extracted lipids were dissolved in ethanol and aliquots were taken out to determine total phospholipid content, as described by (13). Samples were concentrated under a stream of nitrogen, transferred to autosampler vials, and subjected to LC-MS/MS.
Pulse labeling of sphingolipid metabolites
Cells (HPAECs) were incubated (for 5 or 8 h, as indicated) in complete EBM-2 medium supplemented with L-[U-13C,15N]serine (50 mg/l; Cambridge Isotope Laboratories, Andover, MA). Imipramine was added to cells at 3 or 5 h after the beginning of cell labeling, respectively. At the end of incubation, the medium was removed and lipids were extracted as described above. Quantitation of label-incorporated sphingolipids was achieved by LC-MS/MS by monitoring [M+3] isotopes in precursor and product ions, as described below.
LC-MS/MS
Analyses of sphingoid base-1-phosphates, ceramides, sphingoid bases, sphingomyelins, glycosylceramides, and lactosylceramides were performed by ESI-LC/MS/MS. The instrumentation employed was a Sciex 6500 QTRAP hybrid triple quadrupole linear ion-trap mass spectrometer (AB Sciex, Redwood City, CA) equipped with an Ion Drive Turbo V ionspray ionization source interfaced with a Shimadzu Nexera X2 UHPLC system. All lipid molecules and their derivatives were separated using an Ascentis Express RP-Amide 2.7 μm 2.1 × 50 mm column and gradient elution from methanol:water:formic acid (65:35:0.5, 5 mM ammonium formate) to methanol:chloroform:water:formic acid (90:10:0.5:0.5, 5 mM ammonium formate). S1P and DHS1P were analyzed as bis-acetylated derivatives with C17-S1P as the internal standard employing negative ion ESI and multiple reaction monitoring (MRM) analysis, as described (14). Ceramides and sphingoid bases were analyzed with C17:0-Cer and C17-Sph as internal standards using positive ion ESI and MRM analysis (13). To facilitate sphingomyelin analysis and to avoid stable isotope overlap between phosphatidylcholines and sphingomyelins, lipids were hydrolyzed using a methylamine reagent for 2 h at 55°C (15). Reagents were evaporated with a nitrogen stream. The residual nonsaponified lipids were then dissolved in 0.2 ml of methanol and subjected to LC-MS/MS analysis of sphingomyelins. Sphingomyelins were detected as positive ions in MRM mode by a transition from the corresponding molecular ion to the m/z of 184 (phosphocholine). Sphingomyelin quantification was achieved by creating standard curves of variable amounts of sphingomyelin standards (N-16:0-, 18:0-, 24:1-, and 24:0-sphingomyelins) versus a fixed amount of N-12:0-sphingomyelin (internal standard). The linearity and the correlation coefficients of the standard curves were obtained via linear regression analysis. Quantification of sphingolipid molecular species for which there were no standards available was performed using the best approximation from the available analogs with the most similar structure.
Mouse experiments
All experiments were approved by the IACUC (Indiana University School of Medicine, Indianapolis, IN) and conformed to the Public Health Service policy on humane care and use of laboratory animals as outlined by the Institute for Laboratory Animal Research guide of care and use of laboratory animals. Three-month-old female Smpd1+/+, Smpd1+/−, or Smpd1−/− mice (C57Bl/6 background strain), obtained from Dr. Edward Schuchman and Dr. Erich Gulbins, were used to harvest perfused lungs, which were snap-frozen in liquid nitrogen. Lung tissue was homogenized in the cell lysis buffer used for Western blotting. Homogenized tissue was sonicated for 2 s at 30% power, centrifuged at high speed, and the supernatant was removed as whole lung lysate.
Statistical analysis
All statistical analyses were performed using GraphPad Prism (GraphPad Software, La Jolla, CA).
RESULTS
Effect of ASM inhibition on LYNUS signaling
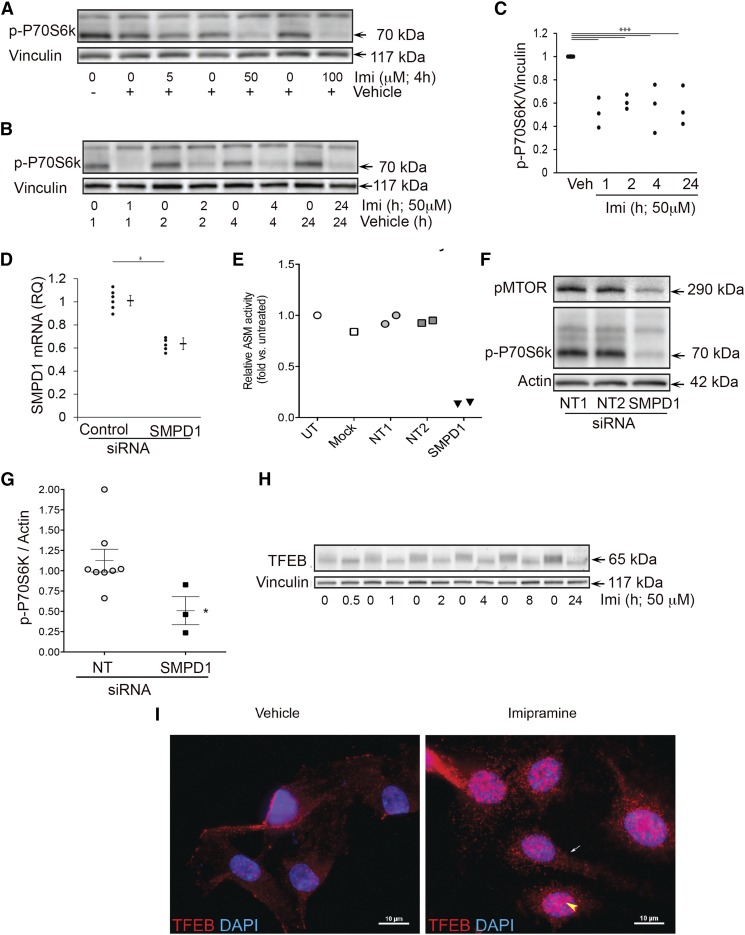
First, using a fluorometric ASM activity assay, we measured the baseline activity of ASM in HPAECs and confirmed the marked inhibitory effect of imipramine (50 μM, 4 h) (supplemental Fig. S2), a tertiary amine tricyclic antidepressant known to increase the proteolytic degradation of lysosomal ASM (16). Pharmacological inhibition of ASM with imipramine significantly decreased the phosphorylation status of P70-S6k rapidly (within 1 h) and sustainably (for up to 24 h), as visualized by Western blot and quantified by densitometry (Fig. 1A–C). ASM inhibition also reduced mTOR phosphorylation (supplemental Fig. S3A), but did not affect the phosphorylation of Akt (supplemental Fig. S3B). Because ASM inhibition with imipramine may have nonspecific effects, we corroborated these results by targeting the transcript of the sphingomyelin phosphodiesterase gene (SMPD1) with siRNA (1–3.33 μM, 48–72 h). Compared with three nontargeting pools of siRNA that included fluorescently labeled siRNA (siGLO), SMPD1 siRNA reduced SMPD1 transcript levels by ∼50%, as measured by quantitative (q)PCR (Fig. 1D) and markedly inhibited ASM activity measured fluorimetrically (by ∼85%, Fig. 1E) or by radioactive assay (by ∼65%, data not shown). ASM knockdown with SMPD1 siRNA reduced the phosphorylation of both mTOR and P70-S6k, as detected by Western blot and quantified (P70-S6k) by densitometry (Fig. 1F, G). We next interrogated whether ASM inhibition activated TFEB, a transcription factor docked at LYNUS that requires dephosphorylation for translocation to the nucleus. ASM inhibition was associated with loss of the “smear” appearance of immunoblotted TFEB seen in control conditions, with the appearance of a lower molecular weight more resolved band, indicating loss of phosphorylation (Fig. 1H). ASM inhibition was associated with an apparent increase in both cytosolic and nuclear TFEB immunostaining, as detected by epifluorescence (Fig. 1I). Together, the inhibition of mTOR and its downstream target P70-S6k, TFEB dephosphorylation, and increased TFEB in the nucleus indicate that ASM inhibition decreases LYNUS function.
Fig. 1.
Effects of ASM inhibition on LYNUS signaling in HPAECs. A–C: Representative Western blots (A, B) and quantification by densitometry (C) of phosphorylated (p)-P70-S6k and vinculin (loading control) following treatment with the ASM inhibitor, imipramine, or vehicle (water) for the indicated time (hours) and concentrations (micromoles). D, E: SMPD1 mRNA expression measured by qPCR and ASM activity detected by fluorimetry using a fluorescently labeled substrate (Amplex Red) following treatment with transfection reagent (mock), control siRNA [siGlo (n = 1)], nontargeting (NT) pools 1 or 2 siRNAs, or SMPD1 siRNA (each 3.33 μM; 72 h) or left untreated (UT). F, G: Phosphorylated mTOR (pMTOR; at Ser 2448) and p-P70-S6k abundance detected by Western blotting (F) and quantified by densitometry relative to actin loading control (G) following the indicated siRNA. Graphs show individual data points of independent experiments, means, and SEM; *** P ≤ 0.001 (ANOVA; Tukey’s post hoc test); * P ≤ 0.05 (Student’s t-test), respectively. H: Western blot of TFEB following imipramine treatment (50 μM; for the indicated time). Note the absence of smear-like TFEB protein detection in ASM-inhibited cells. I: Representative fluorescence micrographs following immunostaining for TFEB (red) and nuclear staining (DAPI, blue; n = 3). Note the increased TFEB immunostaining in both the nucleus (yellow arrowhead) and cytoplasm (white arrow) in ASM-inhibited cells at 4 h following the addition of imipramine.
ASM inhibition triggers autophagy
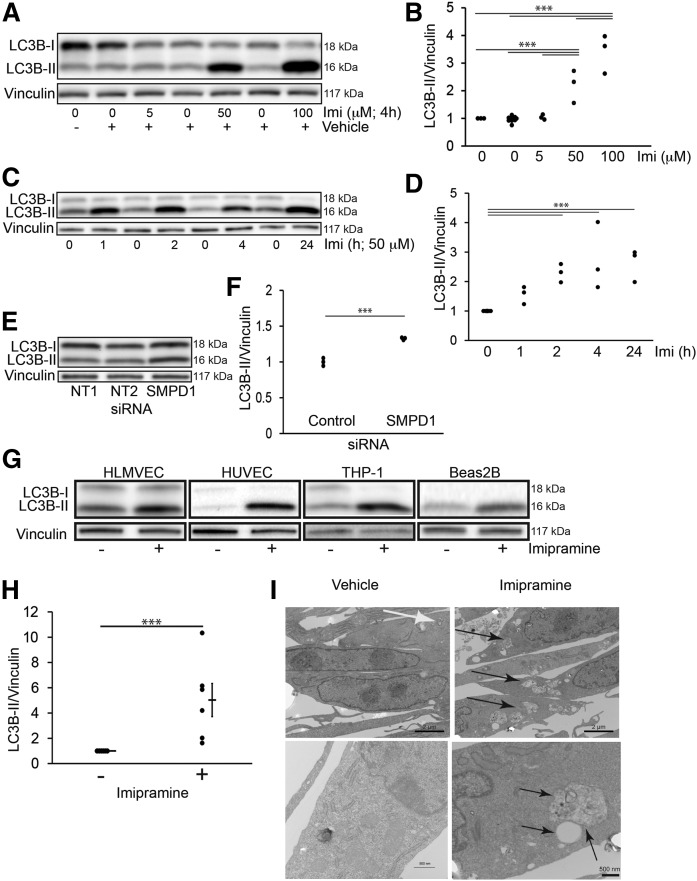
Because mTOR inhibits autophagy, we next determined whether baseline ASM activity in normal growth conditions is required not only to maintain LYNUS activity, but also to inhibit autophagy. Autophagosome abundance was first measured by the amount of intracellular LC3B-II. Pharmacological inhibition of ASM with imipramine (50–100 μM for 2–24 h) increased LC3B-II levels, as detected by Western blot and quantified by densitometry (Fig. 2A–D). Similarly, ASM knockdown with siRNA led to significant increases in LC3B-II, as detected by Western blot and quantified by densitometry (Fig. 2E, F). To determine whether this effect extended to other types of cells than HPAECs, we used similar conditions of ASM inhibition (imipramine, 50 μM, 4 h) to treat other human endothelial cells, such as HLMVECs, HUVECs, human THP-1 monocyte cells, and the lung epithelial cell line, BEAS2b, and noted that all exhibited similar increases in LC3B-II (Fig. 2G, H). In human BEAS2b cells, we utilized electron microscopy to visualize morphological changes triggered by ASM inhibition. Cells exhibited relatively few intra-cytoplasmic autophagic bodies during baseline conditions (Fig. 2I). In contrast, cells treated with the ASM inhibitor accumulated multiple large autolysosomal bodies with more heterogeneous content (Fig. 2I).
Fig. 2.
Effect of ASM inhibition on autophagy. A–E: Representative Western blots (A, C, E) and quantification by densitometry (B, D, F) of LC3B-I and -II protein abundance relative to vinculin (loading control) in HPAECs following ASM inhibition with imipramine at the indicated concentrations (A, B: 4 h) and time (C, D; 50 μM), or with SMPD1 siRNA (E, F: 3.33 μM, 72 h). G: LC3B-I and -II and vinculin (loading control) detected by Western blot in HLMVECs, HUVECs, human monocytes (THP-1), or BEAS2b cells following imipramine treatment (50 μM; 4 h). H: Relative abundance of LC3B-II (vs. vinculin) in BEAS2b cells treated with imipramine (+) measured by densitometry. I: Representative electron microscopy micrographs of BEAS2b cells following ASM inhibition with imipramine (50 μM; 6 h). Note several small intracytoplasmic bodies in control cells (white arrow) compared with multiple large intracytoplasmic bodies with heterogeneous density in ASM-inhibited cells (black arrows). The graphs in B, D, F, and H show individual data points from independent experiments, mean ± SEM; ***P ≤ 0.001 (ANOVA; Tukey’s post hoc test).
ASM inhibition induces autophagy with lysosomal degradative potential
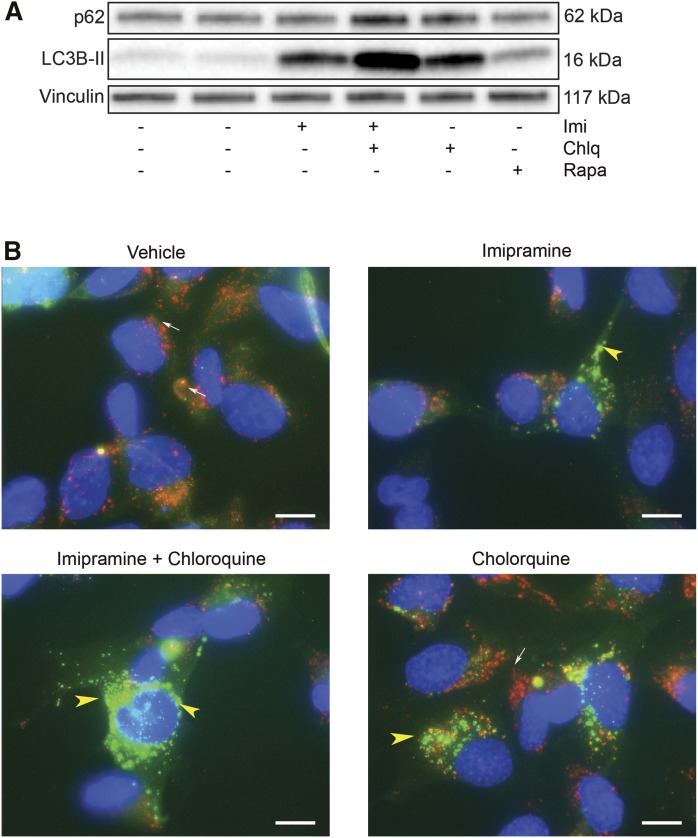
To clarify whether the increased LC3B-II abundance following ASM inhibition is the result of increased production of autophagosomes versus that of reduced clearance of autophagosomes produced at a basal rate, experiments were performed in HPAECs in the presence or absence of chloroquine, which increases lysosomal pH, diminishes the activity of acidic proteases, reduces autophagosome-lysosome fusion, and decelerates the degradation of LC3B-II associated with autophagosomes (autophagic flux). Compared with cells treated with imipramine alone, those treated with both imipramine and chloroquine exhibited further increases in LC3B-II levels as well as p62 levels (Fig. 3A). This result suggests that autophagosome turnover is present in ASM-inhibited cells and that LC3B-II accumulation is more likely due to increased autophagosome formation rather than a significant blockage in autophagic flux (17).
Fig. 3.
Effect of ASM inhibition on lysosomal degradative potential. A: Representative Western blot of p62, LC3B-II, and vinculin (loading control) following treatment with the ASM inhibitor, imipramine (50 μM, 4 h) and low-dose chloroquine (Chlq, 5 μM, 4 h) or rapamycin (Rapa, 1 μM, 4 h). B: Representative immunofluorescence images of bronchial epithelial cells (BEAS2b cells) expressing eGFP-LC3B (green) immunostained with LAMP1 antibody (red; white arrows) following treatment with imipramine (50 μM, 4 h) and/or chloroquine (50 μM, 4 h). Note the increased green punctae in imipramine-treated cells (indicating increased autophagosomes) and relatively low red signal (indicating nonfused lysosomes); intense green punctae in imipramine-chloroquine cotreated cells (yellow arrowhead, indicating increased undigested autophagosomes); increased red immunofluorescence (white arrow) along with green punctae (yellow arrowhead), indicating autophagosomes that do not fuse with lysosomes in chloroquine-treated cells.
As a complementary approach to determine lysosomal degradation of autophagy following ASM inhibition, we used BEAS2b cells stably expressing eGFP-LC3B (Fig. 3B). The colocalization of eGFP-LC3B (green) and LAMP1 (red), a marker of endosomes and lysosomes, was assessed by immunofluorescence microscopy following ASM inhibition in the presence of chloroquine. Vehicle-treated cells exhibited cytosolic diffuse green immunofluorescence with minimal colocalization with LAMP1-positive (red) endolysosomal vesicles (Fig. 3B). Imipramine treatment increased the number of punctate eGFP-containing autophagic vesicles that colocalized with LAMP1, suggesting autophagolysosomal fusion (Fig. 3B). Cells cotreated with imipramine and chloroquine had more intracellular eGFP punctate vesicles, suggesting nonfused autophagosomes (Fig. 3B) when compared with imipramine treatment alone (Fig. 3B). Together, these results suggest that ASM inhibition induces autophagy with degradative potential.
ASM inhibition leads to decreased Sph levels
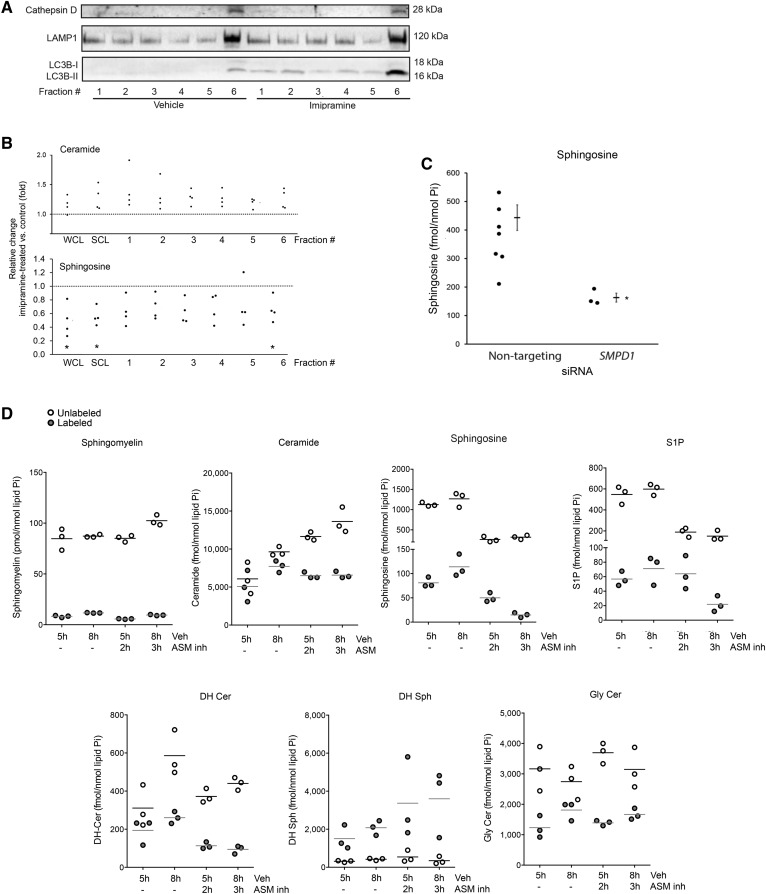
Because the sphingolipid metabolism is highly dynamic and organelle specific, we next determined levels of sphingolipid metabolites following ASM inhibition in whole cells and in lysosome-enriched fractions obtained by isopycnic separation. To analyze proximal changes in sphingolipids following ASM inhibition, we selected to measure these at the earliest time point (30 min) of detectable LYNUS alterations, such as decreased TFEB phosphorylation. Following subcellular fractionation, we noted that fraction six (in order of increasing density) contained the mature forms of both cathepsin D and LAMP1, indicating lysosome enrichment (Fig. 4A). Fraction six also contained LC3B-II (Fig. 4A) as well as the nucleus marker, histone h3, and the mitochondrial marker, voltage-dependent anion channel (VDAC) (not shown). Following imipramine treatment, neither whole cell lysates nor the lysosome-enriched subcellular fraction exhibited decreased ceramides (Fig. 4B, C); instead, ceramides were modesty increased on account of less abundant species (supplemental Fig. S4). Similarly, inhibition of ASM with imipramine did not significantly change sphingomyelins within this timeframe (supplemental Fig. S4) or for up to 24 h (data not shown). The most remarkable change in sphingolipid species following pharmacological ASM inhibition was a significant decrease in relative Sph levels (Fig. 4B). As a complementary approach, we transfected HPAECs with nontargeting pool 1, nontargeting pool 2, or SMPD1 siRNA (1–3.33 μM; 48–72 h) and noted that, similar to imipramine, ASM knockdown significantly reduced Sph concentrations compared with control conditions (Fig. 4C).
Fig. 4.
Effect of ASM inhibition on sphingolipids. A: Representative immunoblots of markers of lysosome (cathepsin D) and of endomembranes (LAMP1 and the autophagy marker, LC3B-II) in the indicated subcellular fractions (fractions 1–6, obtained by isopycnic separation) of HPAECs treated with imipramine (50 μM, 30 min) or control vehicle. Note the increased abundance of autophagosomal marker LC3B-II in all imipramine-treated fractions. B: Relative levels of ceramide and Sph in HPAECs exposed to the ASM inhibitor, imipramine, compared with cells exposed to vehicle, measured by MS/MS. Each data point represents an independent experiment. Data are expressed as mean ± SEM; *P ≤ 0.05 imipramine versus control. C: Absolute Sph levels in HPAECs transfected with nontargeting pool 1, nontargeting pool 2, or SMPD1 siRNA (1–3.33 μM; 48–72 h). Each data point represents an independent experiment. Data are expressed as mean ± SEM; *P ≤ 0.05. Cer, ceramide. D: Levels of U-[13C,15N]serine-labeled and unlabeled sphingolipids measured by MS/MS following treatment with vehicle (Veh) or imipramine (ASM inh) for the indicated time. The horizontal bar represents the mean.
To further study how ASM inhibition caused Sph depletion, we performed pulse labeling of de novo synthesized sphingolipids with U-[13C,15N]serine. Cells were allowed to incorporate the label for 5 or 8 h, of which the last 2 or 3 h, respectively, were in the presence of vehicle or imipramine. The quantitation of labeled and nonlabeled sphingolipid levels by MS/MS established that ASM inhibition had modest effects on sphingomyelin levels and, indeed, did not reduce, but rather increased, ceramide levels at the expense of markedly decreasing Sph and S1P production (Fig. 4D). Because sphingolipid metabolites, such as dihydroceramide, DHSph, and glycosylceramides, did not significantly change following imipramine incubation (Fig. 4D), these data indicate that inhibition of ASM was intimately linked to inhibition of acid ceramidase. It should be also noted that it was mostly the levels of short chain ceramides (14:0; 16:0, and 18:0) that were increased following imipramine (supplemental Fig. S4A), which was confirmed in pulse labeled experiments where 16:0 ceramide species, but not 24:1 (supplemental Fig. S4B) or 24:0 (data not shown), were increased as a result of imipramine treatment, suggesting that the effect of the metabolic block at the ASM/acid ceramidase level is linked to the preferential accumulation of newly formed short-chain ceramide molecules.
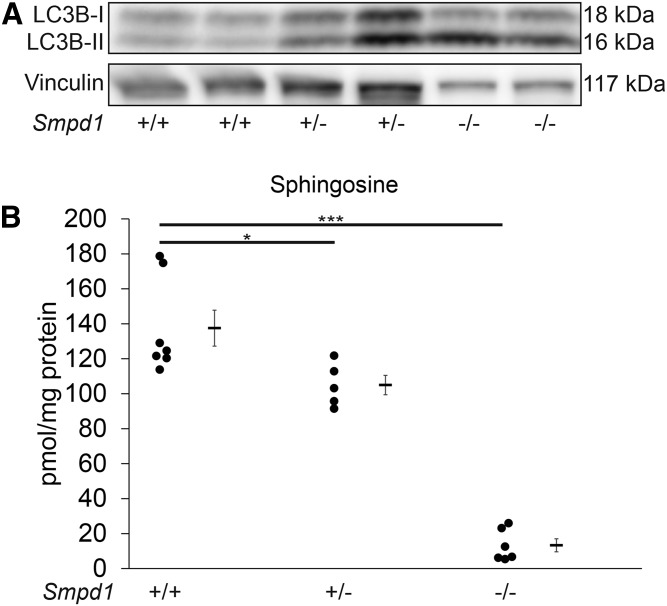
ASM inhibition increases LC3B-II levels and decreases Sph levels in vivo
To investigate whether the marked effects of ASM inhibition on autophagy are recapitulated in vivo, we measured for LC3B-II by Western blot in whole lung lysates of Smpd1+/− mice, known to exhibit 50% reductions in ASM activity compared with wild-type mice (18). Smpd1+/− lungs had an increased abundance of LC3B-II compared with Smpd1+/+ littermates (Fig. 5A). Sph levels were significantly lower in the lungs of Smpd1+/− mice compared with Smpd1+/+ mice, indicating that Sph abundance depends on ASM (Fig. 5B) and that the elevation in LC3B-II following ASM inhibition is associated with decreased Sph both in vitro and in vivo.
Fig. 5.
Effect of ASM inhibition on lung autophagy and Sph levels in vivo. A, B: Representative immunoblot of LC3B-I and -II and vinculin (loading control) in whole lung lysates of mice expressing baseline levels (Smpd1+/+) or deficient (Smpd1+/− and Smpd1−/−) in ASM (each lane represents a separate mouse lung lysate of each genotype). B: Sph levels in whole lungs of Smpd1+/+, Smpd1+/−, and Smpd1−/−) mice. Graph shows individual data from distinct animals, means, and SEM. *P ≤ 0.05, ***P ≤ 0.001 (ANOVA; Dunnett’s post hoc test).
DISCUSSION
Our study demonstrates, for the first time, an important homeostatic functional relationship between ASM and LYNUS activity, indicating that a baseline ASM activity is required for mTOR signaling and autophagy inhibition. Our data indicate that, upon ASM inhibition, mTOR fails to phosphorylate P70-S6k and TFEB becomes dephosphorylated and translocates to the nucleus, events associated with overall increased TFEB abundance and signs of increased autophagy with degradative potential. Although the precise mechanism underlying the functional connection between ASM-LYNUS remains to be elucidated, our data suggest that it may be linked to the critical role of ASM in sustaining Sph levels in the lysosome. These new findings, that implicate basal ASM activity in fundamental lysosome functions, complement a large body of knowledge relating ASM hyperactivation to lysosome (dys)function during multiple conditions.
ASM inhibition reduced the phosphorylation of mTOR at Ser 2448, which typically occurs through Akt pathway signaling and is required for mTOR catalytic activity. Because the phosphorylation of Akt was unaffected following ASM inhibition, the effect is likely to occur downstream of Akt. The phosphorylation of mTOR at Ser 2448 is also dependent, through a feedback loop, on the phosphorylation (activation) of its downstream target, P70-S6k (19), which was markedly decreased by ASM inhibition. It is therefore likely that ASM activity participates in the feedback signaling between P70-S6k and mTOR Ser 2448 phosphorylation. How this occurs remains elusive. We hypothesized that these effects may be ascribed to signaling or structural effects of sphingolipids metabolized by ASM. Because we noted increased, rather than decreased, ceramide upon ASM inhibition, this may inhibit mTOR activity via the SET-PP2A signaling axis (20), but the measured increases were rather modest. However, because lysosomal ASM-generated ceramides are spatially constrained within lysosomes, they may contribute to lysosomal membrane biophysical properties, such as stiffness. These membrane perturbations may affect docking and signaling by interfering with the insertion of farnesylated complex anchors or by rearranging microdomains. Nevertheless, because Sph was the most marked and consistently affected sphingolipid following ASM inhibition, it is likely that the decreased Sph content in the lysosome may have significantly altered the lysosomal calcium efflux, ultimately affecting TFEB phosphorylation through calcineurin (21). Alternatively, subsequent reductions in S1P levels may be implicated, although increased, rather than decreased, intracellular S1P has been associated with triggering autophagy (22). The pro-autophagic effect of S1P production and the importance of the subcellular localization of S1P accumulation have recently been described in neuronal cells (23–25). This body of work suggests that the autophagy triggered by ASM inhibition, which was associated with decreased S1P levels, occurs via a different mechanism than by S1P overproduction.
The consistent decrease in Sph upon ASM inhibition has been previously reported and suggests either reduced substrate (ceramide) for acid ceramidase to produce Sph or indicates that inactivation of ASM, which is known to form a physical complex with acid ceramidase, directly inhibits the ceramidase activity (26). We could not document decreased substrate (ceramide) availability for acid ceramidase and our pulse-labeling experiments indicate that ASM inhibition rapidly causes marked inhibition of acid ceramidase activity, as well. Because Sph readily crosses lysosomal membranes, reduced Sph, which could generate ceramide via recycling in the endoplasmic reticulum, may compensatorily stimulate de novo ceramide synthesis or reduce ceramide utilization for complex sphingolipid synthesis (27) (see schematic in supplemental Fig. S5). Pulse-labeling analyses did suggest modest stimulation of de novo synthesis of several ceramide species (C14:0 to C18:0), of which the most abundant was the palmitoyl (C16:0) ceramide. These effects may explain why ceramide levels were not found to be decreased for up to 72 h following ASM inhibition and implicate decreased Sph as a driver of autophagy, rather than increases in previously described autophagic drivers, S1P or ceramide. Furthermore, the subcellular localization of lower levels of Sph may be important, because an inhibitor of de novo sphingolipid production in the endoplasmic reticulum did not increase LC3B-II levels (28); whereas, we show here that inhibition of Sph production in the lysosome effectively triggered lysosomal autophagy. However, the sphingolipid metabolism is notoriously reactive to perturbations, and we recognize that we have not analyzed the kinetics of all sphingolipid species’ changes following ASM inhibition. Another limitation of our study is the lack of direct correlation of the magnitude of the SMPD1 mRNA decrease following knockdown with the marked enzymatic activity inhibition that followed this approach, especially in the absence of documented ASM protein levels, which are known to have a relatively short half-life of only 5 h (16).
Our results, using chloroquine, immunofluorescence of LC3B-II and lysosome markers, and electron microscopy, indicate that ASM inhibition triggered lysosomal autophagy with preserved flux, which is defined as degradation of autophagosomes within lysosomes. Furthermore, the finding that LC3B-II was absent in the less dense subcellular fractions of control cells, but present in all fractions of ASM-inhibited cells, indicates the presence of elongating phagophores (29), which suggests rapid induction of autophagy following ASM inhibition. Electron microscopy images not only demonstrated the increased number of structures suggestive of autophagic vesicles following ASM inhibition, but also showed that the content of these vesicles was more heterogeneous, a feature previously described to reflect various stages of cargo degradation in autophagolysosomes (30). This finding, in the context of the rest of our other data, may indicate that autophagic induction and pleiotropic fusion may occur simultaneously following ASM inhibition. The autophagic activity following ASM inhibition was associated with lysosomal integrity, as appreciated by immunofluorescence with LAMP1 antibodies, acridine orange staining (data not shown), and electron microscopy. Autophagy was also associated with preserved cell viability, as assessed by annexin V and propidium iodide staining and thymidine incorporation (data not shown). Our results, obtained in primary cells, immortalized macrophages, or lung epithelial cell lines, differ from those reported in a cancer cell line that overexpressed eGFP-LC3, which showed that desmethylclomipramine, a metabolite of clomipramine that structurally resembles imipramine, blocked autophagic flux (31). Such a discrepancy may be related to differences in baseline ASM or mTOR activities among different cell types, rather than the use of a different pharmacological inhibitor. Our results do not contradict the report that autophagosomes may move more slowly in the absence of ASM due to decreased calcium efflux from the lysosome (32), because our data indicate that ASM inhibition induces autophagy and that the autophagic flux is present, and we cannot dispute that such flux may occur at slower rates than in other conditions of autophagy.
In conclusion, our study demonstrates that lysosomal ASM is an important determinant of homeostatic mTOR signaling and LYNUS function, which suggests that this sphingolipid metabolic enzyme is an important regulator of autophagy. The clinical implications of these results relate to the widespread use of antidepressants in various clinical settings, and their potential to affect conditions where initiation of autophagy with degradative potential is desirable.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Ronald Wek, Zhong-Yin Zhang, Xiao-Ming Yin, Charlie Dong, and Peter Roach for lending their knowledge and expertise to this project. The authors thank Caroline Miller for her help with electron microscopy experiments and Daniela N. Petrusca, Karina S. Serban, Kevin Ni, Danting Cao, and Jacob Saliba for technical assistance.
Footnotes
Abbreviations:
- ASM
- acid sphingomyelinase
- BEAS2b cell
- immortalized bronchial epithelial cell
- C17:0-Cer
- N-heptadecanoyl-sphingosine
- C17-Sph
- 17-carbon analog of sphingosine
- C17-S1P
- 17-carbon analog of sphingosine-1-phosphate
- DH
- dihydro
- HLMVEC
- primary human lung microvascular endothelial cell
- HPAEC
- primary human pulmonary artery endothelial cell
- HUVEC
- primary human umbilical vein endothelial cell
- LAMP1
- lysosome-associated membrane protein 1
- LC3B
- microtubule-associated protein 1 light chain 3β
- LYNUS
- lysosomal nutrient-sensing complex
- MRM
- multiple reaction monitoring
- mTOR
- mammalian target of rapamycin
- P70-S6k
- P70-S6 kinase
- qPCR
- quantitative PCR
- SMPD1
- sphingomyelin phosphodiesterase 1
- Sph
- sphingosine
- S1P
- sphingosine-1-phosphate
- TFEB
- transcription factor EB
This work was supported by National Heart, Lung, and Blood Institute Grants RO1HL077328 (I.P.) and F31HL126459 (M.J.J.) and National Institute on Drug Abuse Grant R21DA029249 (I.P.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Patschan S., Chen J., Polotskaia A., Mendelev N., Cheng J., Patschan D., and Goligorsky M. S.. 2008. Lipid mediators of autophagy in stress-induced premature senescence of endothelial cells. Am. J. Physiol. Heart Circ. Physiol. 294: H1119–H1129. [DOI] [PubMed] [Google Scholar]
- 2.Lee J. K., Jin H. K., Park M. H., Kim B. R., Lee P. H., Nakauchi H., Carter J. E., He X., Schuchman E. H., and Bae J. S.. 2014. Acid sphingomyelinase modulates the autophagic process by controlling lysosomal biogenesis in Alzheimer’s disease. J. Exp. Med. 211: 1551–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuchman E. H. 2010. Acid sphingomyelinase, cell membranes and human disease: lessons from Niemann-Pick disease. FEBS Lett. 584: 1895–1900. [DOI] [PubMed] [Google Scholar]
- 4.Settembre C., Fraldi A., Medina D. L., and Ballabio A.. 2013. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14: 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z. H., Kim H. P., Sciurba F. C., Lee S. J., Feghali-Bostwick C., Stolz D. B., Dhir R., Landreneau R. J., Schuchert M. J., Yousem S. A., et al. . 2008. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One. 3: e3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu C., Alterman M., and Dobrowsky R. T.. 2005. Ceramide displaces cholesterol from lipid rafts and decreases the association of the cholesterol binding protein caveolin-1. J. Lipid Res. 46: 1678–1691. [DOI] [PubMed] [Google Scholar]
- 7.Chatelut M., Leruth M., Harzer K., Dagan A., Marchesini S., Gatt S., Salvayre R., Courtoy P., and Levade T.. 1998. Natural ceramide is unable to escape the lysosome, in contrast to a fluorescent analogue. FEBS Lett. 426: 102–106. [DOI] [PubMed] [Google Scholar]
- 8.Höglinger D., Haberkant P., Aguilera-Romero A., Riezman H., Porter F. D., Platt F. M., Galione A., and Schultz C.. 2015. Intracellular sphingosine releases calcium from lysosomes. eLife. 4: e10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Settembre C., De Cegli R., Mansueto G., Saha P. K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T. J., et al. . 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guenther G. G., Peralta E. R., Rosales K. R., Wong S. Y., Siskind L. J., and Edinger A. L.. 2008. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc. Natl. Acad. Sci. USA. 105: 17402–17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeidan Y. H., and Hannun Y. A.. 2007. Activation of acid sphingomyelinase by protein kinase Cdelta-mediated phosphorylation. J. Biol. Chem. 282: 11549–11561. [DOI] [PubMed] [Google Scholar]
- 12.Jackson W. T., Giddings T. H. Jr., Taylor M. P., Mulinyawe S., Rabinovitch M., Kopito R. R., and Kirkegaard K.. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berdyshev E. V., Gorshkova I. A., Usatyuk P., Zhao Y., Saatian B., Hubbard W., and Natarajan V.. 2006. De novo biosynthesis of dihydrosphingosine-1-phosphate by sphingosine kinase 1 in mammalian cells. Cell. Signal. 18: 1779–1792. [DOI] [PubMed] [Google Scholar]
- 14.Berdyshev E. V., Gorshkova I. A., Garcia J. G., Natarajan V., and Hubbard W. C.. 2005. Quantitative analysis of sphingoid base-1-phosphates as bisacetylated derivatives by liquid chromatography-tandem mass spectrometry. Anal. Biochem. 339: 129–136. [DOI] [PubMed] [Google Scholar]
- 15.Clarke N. G., and Dawson R. M.. 1981. Alkaline O leads to N-transacylation. A new method for the quantitative deacylation of phospholipids. Biochem. J. 195: 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurwitz R., Ferlinz K., and Sandhoff K.. 1994. The tricyclic antidepressant desipramine causes proteolytic degradation of lysosomal sphingomyelinase in human fibroblasts. Biol. Chem. Hoppe Seyler. 375: 447–450. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N., and Yoshimori T.. 2007. How to interpret LC3 immunoblotting. Autophagy. 3: 542–545. [DOI] [PubMed] [Google Scholar]
- 18.Horinouchi K., Erlich S., Perl D. P., Ferlinz K., Bisgaier C. L., Sandhoff K., Desnick R. J., Stewart C. L., and Schuchman E. H.. 1995. Acid sphingomyelinase deficient mice: a model of types A and B Niemann-Pick disease. Nat. Genet. 10: 288–293. [DOI] [PubMed] [Google Scholar]
- 19.Chiang G. G., and Abraham R. T.. 2005. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J. Biol. Chem. 280: 25485–25490. [DOI] [PubMed] [Google Scholar]
- 20.Delgoffe G. M. 2016. PP2A’s restraint of mTOR is critical for T(reg) cell activity. Nat. Immunol. 17: 478–479. [DOI] [PubMed] [Google Scholar]
- 21.Medina D. L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., et al. . 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lépine S., Allegood J. C., Park M., Dent P., Milstien S., and Spiegel S.. 2011. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ. 18: 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du C., Ren Y., Yao F., Duan J., Zhao H., Du Y., Xiao X., Duan H., and Shi Y.. 2017. Sphingosine kinase 1 protects renal tubular epithelial cells from renal fibrosis via induction of autophagy. Int. J. Biochem. Cell Biol. 90: 17–28. [DOI] [PubMed] [Google Scholar]
- 24.Mitroi D. N., Karunakaran I., Graler M., Saba J. D., Ehninger D., Ledesma M. D., and van Echten-Deckert G.. 2017. SGPL1 (sphingosine phosphate lyase 1) modulates neuronal autophagy via phosphatidylethanolamine production. Autophagy. 13: 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moruno Manchon J. F., Uzor N. E., Finkbeiner S., and Tsvetkov A. S.. 2016. SPHK1/sphingosine kinase 1-mediated autophagy differs between neurons and SH-SY5Y neuroblastoma cells. Autophagy. 12: 1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J. H., and Schuchman E. H.. 2006. Acid ceramidase and human disease. Biochim. Biophys. Acta. 1758: 2133–2138. [DOI] [PubMed] [Google Scholar]
- 27.Riboni L., Prinetti A., Bassi R., Viani P., and Tettamanti G.. 1998. The effects of exogenous sphingosine on Neuro2a cells are strictly related to the overall capacity of cells to metabolize sphingosine. J. Biochem. 124: 900–904. [DOI] [PubMed] [Google Scholar]
- 28.Mizumura K., Justice M. J., Schweitzer K. S., Krishnan S., Bronova I., Berdyshev E. V., Hubbard W. C., Pewzner-Jung Y., Futerman A. H., Choi A. M. K., et al. . Sphingolipid regulation of lung epithelial cell mitophagy and necroptosis during cigarette smoke exposure. FASEB J. Epub ahead of print. January 5, 2018; doi:10.1096/fj.201700571R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molino D., Zemirli N., Codogno P., and Morel E.. 2017. The Journey of the autophagosome through mammalian cell organelles and membranes. J. Mol. Biol. 429: 497–514. [DOI] [PubMed] [Google Scholar]
- 30.Ylä-Anttila P., Vihinen H., Jokitalo E., and Eskelinen E. L.. 2009. Monitoring autophagy by electron microscopy in mammalian cells. Methods Enzymol. 452: 143–164. [DOI] [PubMed] [Google Scholar]
- 31.Rossi M., Munarriz E. R., Bartesaghi S., Milanese M., Dinsdale D., Guerra-Martin M. A., Bampton E. T., Glynn P., Bonanno G., Knight R. A., et al. . 2009. Desmethylclomipramine induces the accumulation of autophagy markers by blocking autophagic flux. J. Cell Sci. 122: 3330–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Xu M., Pitzer A. L., Xia M., Boini K. M., Li P. L., and Zhang Y.. 2014. Control of autophagy maturation by acid sphingomyelinase in mouse coronary arterial smooth muscle cells: protective role in atherosclerosis. J. Mol. Med. (Berl.). 92: 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.