Abstract
Our understanding of genetic influences on the response of lipids to specific interventions is limited. In this study, we sought to elucidate effects of rare genetic variants on lipid response to a high-fat meal challenge and fenofibrate (FFB) therapy in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) cohort using an exome-wide sequencing-based association study. Our results showed that the rare coding variants in ITGA7, SIPA1L2, and CEP72 are significantly associated with fasting LDL cholesterol response to FFB (P = 1.24E-07), triglyceride postprandial area under the increase (AUI) (P = 2.31E-06), and triglyceride postprandial AUI response to FFB (P = 1.88E-06), respectively. We sought to replicate the association for SIPA1L2 in the Heredity and Phenotype Intervention (HAPI) Heart Study, which included a high-fat meal challenge but not FFB treatment. The associated rare variants in GOLDN were not observed in the HAPI Heart study, and thus the gene-based result was not replicated. For functional validation, we found that gene transcript level of SIPA1L2 is associated with triglyceride postprandial AUI (P < 0.05) in GOLDN. Our study suggests unique genetic mechanisms contributing to the lipid response to the high-fat meal challenge and FFB therapy.
Keywords: whole-exome sequencing, rare variant, high-density lipoprotein, low-density lipoprotein, triglyceride, cholesterol, genetics, epidemiology, postprandial lipemia
Dyslipidemia, defined as abnormal levels of lipids and/or lipoproteins in the blood (1), is a critical modifiable risk factor for chronic diseases, accounting for almost half of the population attributable risk for adverse cardiovascular events (2). Circulating lipid levels are influenced by both environment (e.g., diet, smoking, and prescription drugs) and genetic variation; twin studies estimate the genetic contribution to explain ∼60% of phenotypic variation (3). Until recently (4), findings from genome-wide association studies have accounted for about 12% of blood lipid variance. The proportion of unexplained variance (∼48%) could be lessened with the inclusion of rare variant analyses, illustrating the important role of the low-frequency polymorphisms in the genetic architecture of lipid traits (5). These rare variants, which were not covered by previous genome-wide association studies but contribute to lipid traits, could be located in coding regions, as well as introns, untranslated regions, and intergene regions.
To date, such high-resolution genetic investigations of lipids have focused only on fasting phenotypes, while knowledge of genetic determinants of lipid response to interventions remains limited. The Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) study cohort provides a unique opportunity to study the effects of rare genetic variants on response to two interventions, a high-fat meal (postprandial lipemia; PPL) and fenofibrate (FFB) treatment, due to its carefully controlled intervention by standardizing the environmental perturbation. The high-fat meal intervention provides a unique and impactful way to study dyslipidemia in a dynamic test, not only because humans spend most of their waking hours in the postprandial state, but also because of the high fat content of Western diets. Furthermore, an elevated postprandial lipid response, followed by delayed clearance, has been shown to predict future risk of cardiovascular disease (6). Additionally, the second GOLDN intervention(the 3 week treatment with micronized FFB)creates an opportunity for pharmacogenomic discovery and a deeper understanding of individual variation to lipid-lowering drugs (7). Prior studies in GOLDN have scanned common variants to identify multiple promising determinants of response to both interventions (8–10); however, they were conducted prior to the availability of high-resolution genomic data. To augment the discoveries from the Genome-Wide Association Study (GWAS), account for further missing heritability, and identify additional functional loci contributing to variation in lipid response to a high-fat meal and treatment with FFB, we performed an exome-wide sequencing study in 894 European Americans from the GOLDN study. We sought to replicate the PPL result using the Heredity and Phenotype Intervention (HAPI) Heart Study, which included a high-fat meal challenge but not FFB treatment. In addition, we sought to validate the associations for our findings using DNA methylation and RNA-sequencing (RNA-Seq) data previously collected in GOLDN.
METHODS
Study population
GOLDN (clinicaltrials.gov NCT00083369) was designed to characterize genetic factors that determine response of lipids to two environmental interventions: a 3 week FFB treatment and a high-fat meal challenge (8, 9, 11). Only Caucasian families with at least two siblings were included. Participants were asked to discontinue any lipid-lowering agents (pharmaceuticals or nutraceuticals) for at least 4 weeks prior to the initial visit. GOLDN recruited and sequenced 894 subjects from 186 families recruited at two centers (Minneapolis, MN, and Salt Lake City, UT). Of the 894 subjects, 810 participants participated in the high-fat meal intervention. After that, 797 GOLDN participants received daily treatment with 160 mg micronized FFB for 3 weeks and were followed for treatment response. Finally, 715 participants participated in another high-fat meal intervention to investigate lipid response to the high-fat meal while treated with FFB. The population size indicates that we have statistical power ranging from 0.5 for h2locus= 0.02 to 1.00 for h2locus= 0.05 or above. Our study has been approved by the committee for the protection of human subjects in the University of Texas Health Science Center at Houston and the other institutions, and it abides by the Declaration of Helsinki principles. Informed consent was obtained from the human subjects.
Clinical measurements
Clinical lipids including HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), and triglycerides (TGs) were measured in this study. TG was measured by using the glycerol-blanked enzymatic method on the Roche COBAS FARA centrifugal analyzer (Roche Diagnostics Corporation) (10). HDL-C was measured by using the same procedure as TG after precipitation of non-HDL cholesterol with magnesium/dextran. LDL-C was measured by using a homogeneous direct method (LDL Direct Liquid Select™ Cholesterol Reagent; Equal Diagnostics) on a Hitachi 911 Automatic Analyzer. The FFB treatment and the high-fat meal intervention in GOLDN have been described previously (10, 12). During the high-fat meal intervention, blood was collected three times for pre- and post-FFB treatment, respectively: draw 1 (fasting) immediately prior to the meal (0 h), draw 2 approximately 3.5 h after the meal, and draw 3 approximately 6 h after the meal. We excluded 14 individuals in pre-FFB and 20 individuals in post-FFB from our postprandial analyses who did not follow the protocol schedule, which resulted in large drawtime deviations (>1 h). HDL- and LDL-C were only assessed at draw 1 both before and after FFB treatment. TG was assessed at each blood draw, which allowed an analysis of response to the high-fat meal and FFB, and change in high-fat meal response with FFB.
Library preparation
Genomic DNA from peripheral blood nucleated cells was extracted by using QIAmp 96 DNA Blood Kits (Qiagen, Hilden, Germany). The integrity and yield of native genomic DNA was verified by a PicoGreen assay for quantitation (Invitrogen, Life Technologies, Carlsbad, CA) and run on a 0.8% agarose gel for quality control (QC). Illumina paired-end small-fragment libraries were constructed according to the manufacturer’s recommendations (Illumina Inc., San Diego, CA) with the following exceptions: 1) 500–1,000 ng of native genomic DNA was fragmented by using the Covaris E220 DNA Sonicator (Covaris, Inc., Woburn, MA) to a size range between 100 and 400 bp; 2) Illumina adaptor-ligated library fragments were amplified in four 50 µl PCR reactions for 18 cycles; and 3) solid-phase reversible immobilization bead clean-up was used for enzymatic purification throughout the library process, as well as for final library size selection targeting 300 to 500 bp fragments.
Exome capture and sequencing
Libraries were pooled precapture and hybridized to NimbleGen SeqCap EZ Human VCRome Library kits (Roche NimbleGen, Madison, WI) according to the manufacturer’s protocol. The concentration of each captured library was determined through KAPA quantitative PCR (Kapa Biosystems, Inc., Woburn, MA) according to the manufacturer’s protocol to produce cluster counts appropriate for the Illumina HiSeq 2000 platform. The libraries were run on a HiSeq 2000 V3 2×101-bp sequencing run according to manufacturer recommendations.
Sequence alignment
Illumina sequencing data in FASTQ format were aligned to the GRCh37-lite reference sequence by using BWA (13) (version 0.5.9) with the parameters: -t 4 -q 5. Duplicates were marked in each Binary Alignment/Map (BAM) file by using Picard (http://broadinstitute.github.io/picard/) (version 1.46). If a sample was sequenced across multiple lanes, the aligned BAM files were merged by using Picard (version 1.46).
Variant calling, variant call file creation, and annotation
Each BAM file was sorted with potential PCR duplications removed by using SAMtools (version 1.2.1) (14, 15). Subject-level single nucleotide variants (SNVs) were called by using the Atlas-SNP2 application with a variant call file (VCF) created for each subject (16). Furthermore, for each chromosome, a population-level VCF was created by using the SAMtools mpileup/BCFtools (13, 14), where the SNVs at all the discovered sites were called and backfilled in multiple samples by using the original BAMs from all the GOLDN subjects. Finally, a combined project-level VCF was created through merging all the 25 population-level VCF files (chromosomes 1–22, X, Y, and mitochondrial) by using the GATK-CatVariants (version 3.5) (17–19). Only biallelic mutations were kept after filtered by using VCFtools (15). For the genotype-level QC, genotypes with read depth <20 or genotyping quality <30 were excluded. For the variant-level QC, the mutations were filtered out if their missing rate was >5%. The project-level VCF was further annotated by using ANNOVAR (20) according to hg19 genome assembly/dbSNP (version 138). Four classes of functional variants (splicing, nonsynonymous, stop-loss, and stop-gain) on chromosomes 1–22 were used for association tests (21).
Sample-level QC
Target region breadth by depth for each sample was evaluated by using RefCov (http://gmt.genome.wustl.edu/packages/refcov/) v0.3 with a Browser Extensible Data file of target regions provided by the exome kit manufacturer. We required that >70% of target bases were covered at >20x; samples below that threshold received additional (top-up) sequencing.
To confirm sample purity and identity, we compared high-density SNP array genotypes (9) (Illumina OmniExpress) to the SNV calls obtained from the sequencing data by using SAMtools (15) version r963, and required >90% genotype concordance to pass a sample. No samples were excluded due to low concordance.
Phenotype definition
All the lipid values were natural log-transformed to achieve normality of residuals. Baseline was defined as the value at 0 h (i.e., fasting) at the pre-FFB visit. Fasting-level response to FFB was defined as the difference of log-transformed lipid plasma concentrations at draw 1 between post- and pre-FFB. Three TG pre-FFB postprandial phenotypes were assessed, including uptake, clearance, and area under the increase (AUI). TG values at each blood draw were natural log-transformed first. Uptake and clearance were defined as the slope of the line of TG response from draw 1 to draw 2 and from draw 2 to draw 3, respectively. Next, AUI was calculated with the transformed value by using the trapezoid rule. Because draw time deviations from protocol could result in procedural errors in AUI measurement, TG values at 6 h were estimated by using the values and draw times of draws 2 and 3 by linear extrapolation. AUI was calculated with the measured values at draws 1 and 2, and the estimated value at 6 h to mitigate the effect caused by draw time deviation. The postprandial phenotype response to FFB were the change of PPL phenotype before and after FFB treatment. These phenotypes included uptake response to FFB, clearance response to FFB, and AUI response to FFB.
Statistical analysis
Genetic associations were assessed by using linear mixed models using RAREMETALWORKER and RAREMETAL (version 4.13.6) (22, 23). We considered a minimal model and a fully adjusted model. In the minimal model, all the associations were adjusted for sex, age, age2, age3, and recruiting center as fixed effects (24, 25). For the FFB analyses, an additional variable measuring the number of pills taken per day (to adjust for compliance) was included as a covariate. In addition, a kinship coefficient considered as a random effect was used to adjust for family relatedness. In the full model, apart from those covariates included in minimal models, additional related lipid levels were included as covariates. For the fasting-level response to FFB and pre-FFB postprandial AUI and uptake, respective baseline levels were included as covariates. For the pre-FFB postprandial clearance phenotype, draw 2 lipid level was used as a covariate. For the three postprandial lipid level response to FFB, their corresponding pre-FFB treatment level and fasting-level response to FFB were included as covariates (supplemental Table S1).
For gene-based analyses, sequence kernel association test (SKAT), simple burden test, Madsen and Browning weighted burden test (MB), and variable threshold test were utilized with multiple frequency thresholds for variant inclusion (MAF < 5% and 1%) (26–28).
Bonferroni corrections were used for both single-variant and gene-based associations. The significant signals were filtered out if they were driven by variants carried by less than three individuals or one single family.
Replication
We sought to replicate our associations for TG postprandial phenotypes using the HAPI Heart Study (29), in which postprandial TG levels were measured at 0, 1, 2, 3, 4, and 6 h after 770 Old Order Amish participants underwent a high-fat feeding intervention identical to the one used in GOLDN. More study procedure details can be found in previous reports (10, 29). HAPI Heart Study participants were genotyped as part of the Trans-Omics for Precision Medicine effort using whole-genome sequencing methods. TG postprandial phenotypes were defined in the HAPI Heart Study similarly as described above in GOLDN but calculated with measured values at 0, 3, and 6 h. Demographic and clinical characteristics of the HAPI Heart Study are listed in supplemental Table S2.
The association of GOLDN top hits in the replication cohort was tested by using RAREMETALWORKER with an identical model to GOLDN. Next, we also performed a joint meta-analysis of GOLDN top hits across all participating cohorts using RAREMETAL.
Functional validation
We sought to validate the associations for our findings using DNA methylation and RNA-Seq data previously collected in GOLDN. CpG site methylation was quantified by using the Illumina (San Diego, CA) Infinium Human Methylation450 Beadchip with 991 participants as described previously (24). The CpG sites within the genes containing significantly associated variants and the intergenic CpG sites near these genes were examined to test whether their methylation levels were associated with lipid levels or not by using linear mixed models.
For transcriptional profiling, 100 unrelated GOLDN participants were selected from the extremes of the BMI distribution. RNA was extracted from buffy coats by using the TRIzol method (ThermoFisher Scientific, Waltham, MA), and the quality was evaluated by using Bioanalyzer (Agilent Technologies, Santa Clara, CA). After sequencing and alignment, we fitted linear mixed models to test for associations between gene transcript level and lipid phenotypes.
RESULTS
Demographic and clinical characteristics
Demographic and clinical characteristics of the study subjects are listed in Table 1 and supplemental Table S2. From 186 families, we included 435 males and 459 females from the GOLDN study. Levels of TG, LDL-C, and HDL-C after FFB treatment were significantly different from pretreatment levels (P ≤ 0.05) (Table 1). As described before (25), TG levels increased significantly during the first 3.5 h and decreased significantly from 3.5 to 6 h after both high-fat meals (pre- and post-FFB) (P ≤ 0.05). Moreover, the post-FFB clearance was significantly different than the pre-FFB clearance (P ≤ 0.05).
TABLE 1.
Clinical characteristics of samples in GOLDN
| Phenotype | Pre-FFB | Post-FFB | ||
| Mean | SD | Mean | SD | |
| BMI, kg/m2 | 28.5 | 5.6 | — | — |
| Glucose, mg/dl | 101.51 | 18.74 | 99.44 | 19.05 |
| Systolic blood pressure, mmHg | 116.08 | 16.82 | — | — |
| Diastolic blood pressure, mmHg | 68.57 | 9.6 | — | — |
| HDL, 0 h, log(mg/dl) | 3.81 | 0.27 | 3.86 | 0.26 |
| LDL, 0 h, log(mg/dl) | 4.78 | 0.27 | 4.60 | 0.31 |
| TG, 0 h, log(mg/dl) | 4.75 | 0.59 | 4.36 | 0.52 |
| TG, 3.5 h, log(mg/dl) | 5.38 | 0.57 | 4.99 | 0.55 |
| TG, 6 h, log(mg/dl) | 5.23 | 0.70 | 4.80 | 0.61 |
| TG uptake, log(mg/(dl*h)) | 0.17 | 0.08 | 0.17 | 0.09 |
| TG clearance, log(mg/(dl*h)) | −0.06 | 0.13 | −0.08 | 0.13 |
| TG AUI, mg*h/dl | 6.71E-04 | 3.37E-04 | 6.65E-04 | 3.16E-04 |
The values were log-transformed.
Exome sequencing
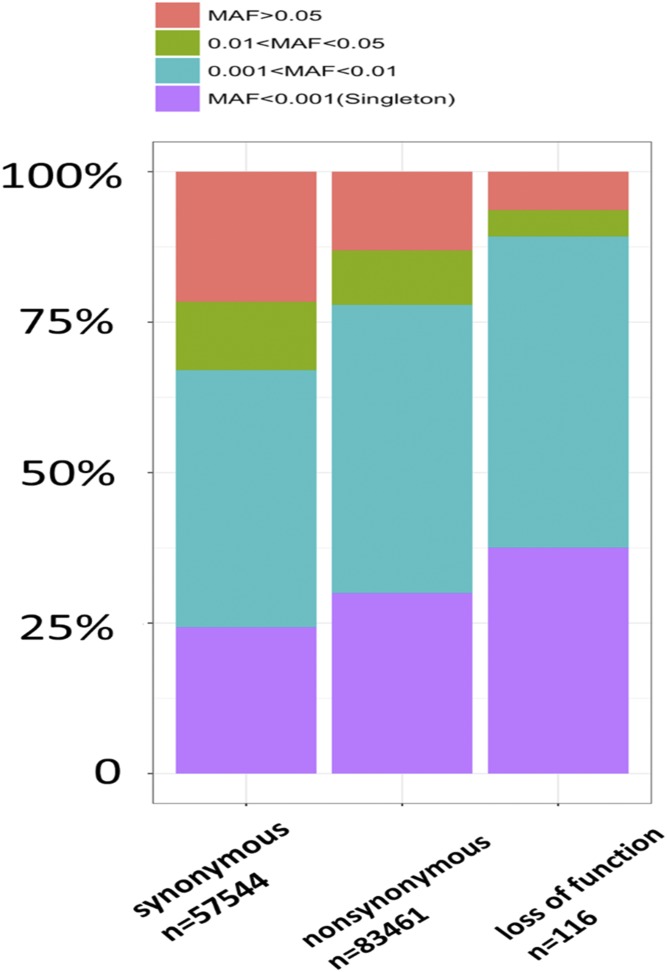
On average, 61,235,513 (SD = 11,606,103) reads per sample were generated, and 98.9% reads were mapped to the reference genome. In total, 968,507 variants (124,661 exonic variants) were called. The average read depth for each sample across the 968,507 variants ranged from 25.5 to 111.2 (mean = 52.3). After filtering out variants with missing rate > 0.05, genotype quality < 30, and read depth < 20, a total of 83,577 functional variants were kept, located within exonic regions of 16,333 genes on autosomes. Of those, 74,216 rare variants with minor allele frequency (MAF) < 0.05 were located in 14,795 genes, while 66,605 rare variants with MAF < 0.01 were located in 14,521 genes. Transitions/transversions ratio of SNPs that passed QC was 2.61, and the heterozygous/homozygous ratio was 1.74. A small percentage (2.8%) of these 83,577 variants were included in our previous GWAS (8), for which the genotype concordance was 98.8%. Across the autosomes, each participant had an average of 1 splicing, 4 stop-loss, 23 stop-gain, and 3,154 nonsynonymous biallelic variants. Rare variants (MAF < 0.05) were more enriched in variants of greater functional impact (Fig. 1), which may indicate purifying selection of deleterious alleles (30, 31).
Fig. 1.
Fraction of SNPs with different MAF range within different categories. The categories include synonymous, nonsynonymous, and loss of function (i.e., splicing, stopgain, and stoploss). The y axis indicates the fraction of SNPs with a different MAF range.
Association tests
By using the Bonferroni correction for multiple testing, P < 5.98E-07 (0.05/83,577) was selected as the exome-wide significance threshold for the single-variant association test, and no single variant was significant by using minimal or full models.
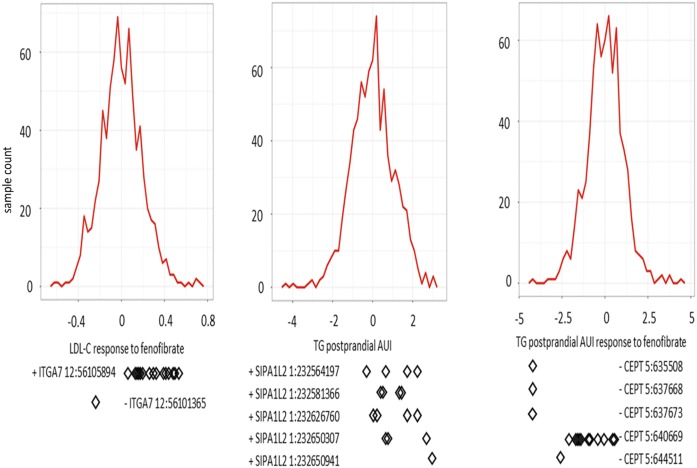
For the gene-based burden test, the Bonferroni corrected significance thresholds were defined as P < 3.38E-06 (0.05/14,795) and 3.44E-06 (0.05/14,521) with MAF thresholds of 0.05 and 0.01, respectively. By using full models, the rare coding variants in three genes, ITGA7, SIPA1L2, and CEP72, were significantly associated with LDL-C fasting level response to FFB, pre-FFB postprandial TG AUI, and postprandial TG AUI response to FFB, respectively (Table 2, Fig. 2). In consideration of the minimally adjusted model, only the rare coding variants in ITGA7 and CEP72 were still significantly associated with the same traits, while the rare coding variants in SIPA1L2 (P = 5.58E-06) did not reach the statistical significance threshold. Below, we present results based on the MAF threshold of 0.01 for the full model unless otherwise noted. The severities of rare variant effects, predicted by PolyPhen2 (32) or SIFT (33), are listed in Table 2.
TABLE 2.
Significant genes and their variants for plasma lipid level according to gene-based test using full model
| Trait | Gene ID | Significant gene P-value | Method | Rare variant # | Rare Variant P < 0.1 | |||||
| Variant ID | Variant P | Variant effect direction | Rare allele carrier # | Mutation effect | Allele frequency in ExAC | |||||
| LDL-C fasting level response to FFB | ITGA7 | 1.24E-07 | SKAT | 12 | 12:56101365:G:T | 5.13E-02 | — | 1 | F->L probably damaging | 1.7E-05 |
| 12:56105894:G:A | 1.74E-07 | + | 15 | T->I possibly damaging | 0.01 | |||||
| Postprandial TG AUI | SIPA1L2 | 2.31E-06 | MB | 22 | 1:232564197:A:G | 7.00E-02 | + | 4 | M->T damaging | NA |
| 1:232581366:C:T | 9.76E-02 | + | 4 | D->N possibly damaging | 4.8E-03 | |||||
| 1:232626760:C:T | 6.83E-02 | + | 4 | A->T tolerated | NA | |||||
| 1:232650307:C:G | 4.24E-02 | + | 3 | G->A tolerated | 1.5E-05 | |||||
| 1:232650941:T:C | 4.21E-03 | + | 1 | T->A benign | 8.4E-04 | |||||
| Postprandial TG AUI response to FFB | CEP72 | 1.88E-06 | MB | 7 | 5:635508:C:T | 5.85E-05 | — | 1 | P->L probably damaging | 4.5E-04 |
| 2.01E-06 | Burden | 5:637668:A:G | 5.85E-05 | — | 1 | K->R possibly damaging | 2.3E-04 | |||
| 5:637673:G:A | 5.85E-05 | — | 1 | D->N benign | 2.3E-04 | |||||
| 5:640669:C:T | 3.40E-02 | — | 11 | R->W possibly damaging | 1.5E-03 | |||||
| 5:644511:C:T | 1.97E-02 | — | 1 | T->I benign | NA | |||||
Rare variant #: the number of rare variant loci within the candidate genes; rare allele carrier #: the number of samples who carry the rare variant; variant P: P value for the null hypothesis of the association test between the rare variant and its corresponding trait. The variants were carried by different families if rare allele carrier number is greater than 1.
Fig. 2.
Distribution of residuals for regression of phenotypes on their covariates. The diamonds indicate corresponding residual values of rare variant carriers. The x axis is the Z score for residuals. The y axis is the sample counts.
Associations with rare variants from known lipid genes from previous GOLDN studies
We also sought to replicate known candidate genes, including APOA5 (34), APOE (11), and APOC3 (35), which were reported to carry variants significantly associated with one or more lipid class in GOLDN (for response phenotypes), as well as fasting lipids in other studies (21). The variants identified in those genes and their P values are listed in supplemental Tables S3 and S4. Overall, we report that APOA5 and APOE, but not APOC3, showed associations with one or more GOLDN lipid traits in single-variant or gene-based tests. We note that some known mutations in these three genes were not identified in our study or were filtered out during QC. For example, a known missense SNP (rs7412) in APOE was filtered out because of low reading depth.
Replication results
The significant gene SIPA1L2 for TG postprandial AUI was tested for association in the HAPI Heart Study. In total, seven exonic functional SNPs were identified in this gene within the HAPI Heart Study, one of which was shared with GOLDN (supplemental Tables S5 and S6). None of the seven SNPs in HAPI Heart Study was significant (supplemental Table S5). The association of SIPA1L2 with TG postprandial AUI in the HAPI Heart Study was not significant (P = 0.97) according to a gene-based test, and the effect direction was different from that in GOLDN. The FFB treatment protocol was not a part of the HAPI Heart study, and thus we could not attempt replication for ITGA7 or CEP72.
Functional validation results
We examined whether differential methylation at cytosine–guanine dinucleotides (CpGs) within and neighboring our top gene findings were associated with our lipid traits. After Bonferroni correction for multiple testing, no methylation of CpG sites within or neighboring those three genes was associated with lipid intervention traits.
We also examined the gene transcript level of candidate genes using RNA-Seq data of GOLDN. The gene transcript level of SIPA1L2 was significantly associated with TG postprandial AUI (P ≤ 0.05), and the gene transcript level of ITGA7 was borderline associated with LDL-C fasting level response to FFB (P = 0.07).
DISCUSSION
High-resolution genetic investigations of lipids have focused only on fasting phenotypes. In this study, we sought to elucidate effects of rare genetic variants on lipid response to a high-fat meal and FFB treatment using an exome-wide sequencing-based association study for the first time. Here, we present preliminary evidence of genetic determinants of lipid response to two interventions. Those genes include ITGA7, SIPA1L2, and CEP72. The SIPA1L2 gene transcript level was associated with the TG postprandial response. In addition, we identified associations related with lipid response to two interventions in candidate genes whose common variants were found to be associated with baseline lipid phenotypes in previous studies.
We identified three genes with rare coding-sequence mutations whose carriers exhibited clinically relevant phenotypic differences compared with noncarriers. Previous studies offer insights into the observed associations. For example, common polymorphisms in ITGA2 (an ITGA7 homolog) were associated with coronary atherosclerosis in a candidate gene association study of the Chinese Han population (36). Variants in CEP164 and CEP68, which are homologs of CEP72, were reported to be associated with TG and LDL-C fasting levels (5, 21, 37). Although no study has indicated that SIPA1L2 is related to lipid level so far, the gene transcript level of SIPA1L2 was significantly associated with TG postprandial AUI (P ≤ 0.05) in the GOLDN RNA-Seq study. Overall, rare variants in these genes warrant further study in relation to PPL and FFB treatment aimed at curbing elevated fasting and postprandial lipid levels.
Previous GOLDN candidate gene studies have interrogated variants in APOA5, APOE, and APOC3 (11, 34, 35) with respect to lipid traits. Although the functions of these genes in controlling lipid fasting levels are well known, we lack the knowledge of the effects of their mutations on lipid level response to interventions. Lai et al. (34) studied associations between an APOA5 variant (rs3135506, chr11:116662407) and baseline TG and TG response to FFB, as well as postprandial TG. Our study replicated their findings (supplemental Table S3), where APOA5 rare variant carriers exhibited significantly higher baseline TG and lower baseline HDL-C levels (P ≤ 0.05) compared with noncarriers. After 3 weeks of FFB treatment, reduction of TG fasting level in APOA5 rare variant carriers was significantly larger than that in noncarriers (P ≤ 0.05), and the HDL-C response was also higher (34). Our study adds to that study by identifying that rs3135506 in APOA5 was also associated with TG postprandial clearance slope, a trait not previously analyzed. The effects of two APOE variants (rs7412 and rs429358) on TG levels has also been examined in GOLDN (11). Our study further showed rs429358 in APOE to be associated with baseline LDL-C and LDL-C response to FFB. Liu et al. (35) reported that two intronic variants in APOC3 were associated with enhanced TG response to FFB treatment. Our study did not cover those variants, and we did not uncover any exonic APOC3 variations with allele count greater than two (supplemental Tables S3 and S4). rs76353203, a stop-gain variant in APOC3, was reported to be associated with decreased baseline TG, increased baseline HDL-C, and decreased baseline LDL-C in the Amish population (38). In our study, we observed consistent directions of this association, but the variant was carried by only one sample, and the association was not significant. Overall, these results continue to add to the body of literature on these important lipid candidate genes.
This study has several limitations. The efficiency of capture probes varies considerably for exome sequencing (supplemental Fig. S1). Thus, the read depths for partial SNPs were low, so that they were excluded by the QC procedure. For example, rs7412, which was known to be associated with lipid levels, was excluded due to low read depth. In addition, participants from the HAPI Heart Study were recruited from the Amish community of Lancaster County, PA, who are descendants of about 200 original founding individuals (facilitating population drift and potential enrichment for rare variants) (29), while the GOLDN cohort was recruited from families of general European ancestry. Thus, external replication of rare variants was likely hindered by the private nature of such polymorphisms to individual populations. Also, cohorts with pharmaceutical intervention phenotypes for lipid traits and exome sequencing are rare, so proper replication of the pharmacogenetic effects is difficult. Moreover, genetic variants in introns, UTR regions, and intergene regions were not examined in our study, which could contribute to the unexplained variance.
CONCLUSION
In our study, novel rare variants in three genes, ITGA7, SIPA1L2, and CEP72, associated with lipid response to a high-fat meal and/or a 3-week FFB treatment were found. Moreover, the gene transcript level of SIPA1L2 was associated with the postprandial TG AUI. We identified new intervention trait associations within two known candidate genes (APOA5 and APOE). In conclusion, we found novel genetic variants that contribute to lipid intervention traits and revealed potential underlying molecular mechanisms, which may inform biomarkers of disease risk and treatment targets.
Supplementary Material
Acknowledgments
We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
Footnotes
Abbreviations:
- AUI
- area under the increase
- BAM
- binary alignment/map
- FFB
- fenofibrate
- GOLDN
- Genetics of Lipid Lowering Drugs and Diet Network
- HAPI
- Heredity and Phenotype Intervention
- HDL-C
- HDL cholesterol
- LDL-C
- LDL cholesterol
- MAF
- minor allele frequency
- MB
- Madsen and Browning weighted burden test
- PPL
- postprandial lipemia
- QC
- quality control
- RNA-Seq
- RNA sequencing
- SNV
- single nucleotide variant
- TG
- triglyceride
- VCF
- variant call file
The work on the GOLDN study has been funded by National Institutes of Health Grants U01HL072524 and R01HL091357. The HAPI Heart Study was supported by National Institutes of Health Grants U01 HL072515 and P30 DK072488. Whole-genome sequencing (WGS) of Amish subjects was provided by the Trans-Omics for Precision Medicine (TOPMed) program through the National Heart, Lung, and Blood Institute. WGS for TOPMed Genetics of Cardiometabolic Health in the Amish (phs000956.v2.p1) was performed at the Broad Institute of MIT and Harvard (3R01HL121007-01S1). Centralized read mapping and genotype calling, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1). Phenotype harmonization, data management, sample-identity QC, and general study coordination were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1). Analysis of the HAPI Heart Study data was supported by P30 DK072488. X.G. and D.Z. are partially supported by National Institute of Food and Agriculture Agriculture and Food Research Initiative Competitive Grant 2015-67015-22975 and US Department of Agriculture Aquaculture Research Program Competitive Grant 2014-70007-22395. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Fodor G. 2010. Primary prevention of CVD: treating dyslipidemia. BMJ Clin. Evid. 2010: 0215. [PMC free article] [PubMed] [Google Scholar]
- 2.Arsenault B. J., Boekholdt S. M., and Kastelein J. J.. 2011. Lipid parameters for measuring risk of cardiovascular disease. Nat. Rev. Cardiol. 8: 197–206. [DOI] [PubMed] [Google Scholar]
- 3.Beekman M., Heijmans B. T., Martin N. G., Pedersen N. L., Whitfield J. B., DeFaire U., van Baal G. C. M., Snieder H., Vogler G. P., and Slagboom P. E.. 2002. Heritabilities of apolipoprotein and lipid levels in three countries. Twin Res. 5: 87–97. [DOI] [PubMed] [Google Scholar]
- 4.Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., Mora S., et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surakka I., Horikoshi M., Mägi R., Sarin A-P., Mahajan A., Lagou V., Marullo L., Ferreira T., Miraglio B., and Timonen S.. 2015. The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 47: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borén J., Matikainen N., Adiels M., and Taskinen M-R.. 2014. Postprandial hypertriglyceridemia as a coronary risk factor. Clin. Chim. Acta. 431: 131–142. [DOI] [PubMed] [Google Scholar]
- 7.Glasser S. P., Wojczynski M. K., Kabagambe E. K., Tsai M. Y., Ordovas J. M., Straka R. J., and Arnett D. K.. 2010. Comparison of postprandial responses to a high-fat meal in hypertriglyceridemic men and women before and after treatment with fenofibrate in the Genetics and Lipid Lowering Drugs and Diet Network (GOLDN) Study. Srx Pharmacol. 2010: 485146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irvin M. R., Rotroff D. M., Aslibekyan S., Zhi D., Hidalgo B., Motsinger-Reif A., Marvel S., Srinivasasainagendra V., Claas S. A., and Buse J. B.. 2016. A genome-wide study of lipid response to fenofibrate in Caucasians: a combined analysis of the GOLDN and ACCORD studies. Pharmacogenet. Genomics. 26: 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aslibekyan S., Goodarzi M. O., Frazier-Wood A. C., Yan X., Irvin M. R., Kim E., Tiwari H. K., Guo X., Straka R. J., and Taylor K. D.. 2012. Variants identified in a GWAS meta-analysis for blood lipids are associated with the lipid response to fenofibrate. PLoS One. 7: e48663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojczynski M. K., Parnell L. D., Pollin T. I., Lai C. Q., Feitosa M. F., O’Connell J. R., Frazier-Wood A. C., Gibson Q., Aslibekyan S., and Ryan K. A.. 2015. Genome-wide association study of triglyceride response to a high-fat meal among participants of the NHLBI Genetics of Lipid Lowering Drugs and Diet Network (GOLDN). Metabolism. 64: 1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irvin M. R., Kabagambe E. K., Tiwari H. K., Parnell L. D., Straka R. J., Tsai M., Ordovas J. M., and Arnett D. K.. 2010. Apolipoprotein E polymorphisms and postprandial triglyceridemia before and after fenofibrate treatment in the Genetics of Lipid Lowering and Diet Network (GOLDN) Study. Circ Cardiovasc Genet. 3: 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Ordovas J. M., Gao G., Province M., Straka R. J., Tsai M. Y., Lai C-Q., Zhang K., Borecki I., and Hixson J. E.. 2008. The SCARB1 gene is associated with lipid response to dietary and pharmacological interventions. J. Hum. Genet. 53: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., and Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., and Sherry S. T.. 2011. The variant call format and VCFtools. Bioinformatics. 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Challis D., Yu J., Evani U. S., Jackson A. R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R. A., and Yu F.. 2012. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., and Daly M.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., Del Angel G., Rivas M. A., and Hanna M.. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auwera G. A., Carneiro M. O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., and Thibault J.. 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 11: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., and Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange L. A., Hu Y., Zhang H., Xue C., Schmidt E. M., Tang Z-Z., Bizon C., Lange E. M., Smith J. D., and Turner E. H.. 2014. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am. J. Hum. Genet. 94: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng S., Liu D., Zhan X., Wing M. K., and Abecasis G. R.. 2014. RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics. 30: 2828–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D. J., Peloso G. M., Zhan X., Holmen O. L., Zawistowski M., Feng S., Nikpay M., Auer P. L., Goel A., and Zhang H.. 2014. Meta-analysis of gene-level tests for rare variant association. Nat. Genet. 46: 200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irvin M. R., Zhi D., Joehanes R., Mendelson M., Aslibekyan S., Claas S. A., Thibeault K. S., Patel N., Day K., and Jones L. W.. 2014. Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid Lowering Drugs and Diet Network study. Circulation 130: 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frazier-Wood A. C., Ordovas J., Straka R., Hixson J., Borecki I., Tiwari H., and Arnett D.. 2013. The PPAR alpha gene is associated with triglyceride, low-density cholesterol and inflammation marker response to fenofibrate intervention: the GOLDN study. Pharmacogenomics J. 13: 312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu M. C., Lee S., Cai T., Li Y., Boehnke M., and Lin X.. 2011. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89: 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price A. L., Kryukov G. V., de Bakker P. I., Purcell S. M., Staples J., Wei L-J., and Sunyaev S. R.. 2010. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madsen B. E., and Browning S. R.. 2009. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell B. D., McArdle P. F., Shen H., Rampersaud E., Pollin T. I., Bielak L. F., Jaquish C., Douglas J. A., Roy-Gagnon M-H., and Sack P.. 2008. The genetic response to short-term interventions affecting cardiovascular function: rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart Study. Am. Heart J. 155: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auer P. L., Reiner A. P., Wang G., Kang H. M., Abecasis G. R., Altshuler D., Bamshad M. J., Nickerson D. A., Tracy R. P., and Rich S. S.. 2016. Guidelines for large-scale sequence-based complex trait association studies: lessons learned from the NHLBI Exome Sequencing Project. Am. J. Hum. Genet. 99: 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tennessen J. A., Bigham A. W., O’Connor T. D., Fu W., Kenny E. E., Gravel S., McGee S., Do R., Liu X., and Jun G.. 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 337: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramensky V., Bork P., and Sunyaev S.. 2002. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 30: 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng P. C., and Henikoff S.. 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai C-Q., Arnett D. K., Corella D., Straka R. J., Tsai M. Y., Peacock J. M., Adiconis X., Parnell L. D., Hixson J. E., and Province M. A.. 2007. fenofibrate effect on triglyceride and postprandial response of apolipoprotein a5 variants: the GOLDN study. Arterioscler. Thromb. Vasc. Biol. 27: 1417–1425. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Ordovas J. M., Gao G., Province M., Straka R. J., Tsai M. Y., Lai C-Q., Zhang K., Borecki I., and Hixson J. E.. 2009. Pharmacogenetic association of the APOA1/C3/A4/A5 gene cluster and lipid responses to fenofibrate: the genetics of lipid-lowering drugs and diet network study. Pharmacogenet. Genomics. 19: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Fu W., Xie F., Wang Y., Chu X., Wang H., Shen M., Sun W., Lei R., and Yang L.. 2010. Common polymorphisms in ITGA2, PON1 and THBS2 are associated with coronary atherosclerosis in a candidate gene association study of the Chinese Han population. J. Hum. Genet. 55: 490–494. [DOI] [PubMed] [Google Scholar]
- 37.Helgadottir A., Gretarsdottir S., Thorleifsson G., Hjartarson E., Sigurdsson A., Magnusdottir A., Jonasdottir A., Kristjansson H., Sulem P., and Oddsson A.. 2016. Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nat. Genet. 48: 634–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu W., Cheng Y-C., Chen K., Wang H., Gerhard G. S., Still C. D., Chu X., Yang R., Parihar A., and O’Connell J. R.. 2015. Evidence for several independent genetic variants affecting lipoprotein (a) cholesterol levels. Hum. Mol. Genet. 24: 2390–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.