Abstract
Lipoxygenase (LOX)-catalyzed oxidation of the essential fatty acid, linoleate, represents a vital step in construction of the mammalian epidermal permeability barrier. Analysis of epidermal lipids indicates that linoleate is converted to a trihydroxy derivative by hydrolysis of an epoxy-hydroxy precursor. We evaluated different epoxide hydrolase (EH) enzymes in the hydrolysis of skin-relevant fatty acid epoxides and compared the products to those of acid-catalyzed hydrolysis. In the absence of enzyme, exposure to pH 5 or pH 6 at 37°C for 30 min hydrolyzed fatty acid allylic epoxyalcohols to four trihydroxy products. By contrast, human soluble EH [sEH (EPHX2)] and human or murine epoxide hydrolase-3 [EH3 (EPHX3)] hydrolyzed cis or trans allylic epoxides to single diastereomers, identical to the major isomers detected in epidermis. Microsomal EH [mEH (EPHX1)] was inactive with these substrates. At low substrate concentrations (<10 μM), EPHX2 hydrolyzed 14,15-epoxyeicosatrienoic acid (EET) at twice the rate of the epidermal epoxyalcohol, 9R,10R-trans-epoxy-11E-13R-hydroxy-octadecenoic acid, whereas human or murine EPHX3 hydrolyzed the allylic epoxyalcohol at 31-fold and 39-fold higher rates, respectively. These data implicate the activities of EPHX2 and EPHX3 in production of the linoleate triols detected as end products of the 12R-LOX pathway in the epidermis and implicate their functioning in formation of the mammalian water permeability barrier.
Keywords: linoleic acid; lipoxygenase; trihydroxy-linoleate; 14,15-epoxyeicosatrienoic acid; epidermis; ichthyosis
Among the gene products required for formation of the mammalian epidermal water barrier are the two lipoxygenase (LOX) enzymes, 12R- LOX and epidermal LOX3 (eLOX3) (1, 2). Inactivating mutations in either enzyme leads to trans-epidermal water loss and the fish skin symptoms of congenital ichthyosis in afflicted families (3) and to neonatal lethality in mice (2). The 12R-LOX/eLOX3 substrate in the epidermis is the essential fatty acid, linoleate, which is esterified to the omega-hydroxyl of the very long chain fatty acid component of the unique epidermal acylceramides [esterified omega-hydroxyacyl-sphingosine (Cer-EOS)] (4, 5). Products of the actions of 12R-LOX and eLOX3 are detectable in the outer epidermis as epoxy-hydroxy (epoxyalcohol) derivatives of the ceramide-linoleate ester (6). Oxidation of the linoleate moiety is proposed to facilitate hydrolysis of the ester bond, freeing the omega hydroxyl of the ceramide for coupling to cross-linked proteins, a vital step in sealing the water barrier (6).
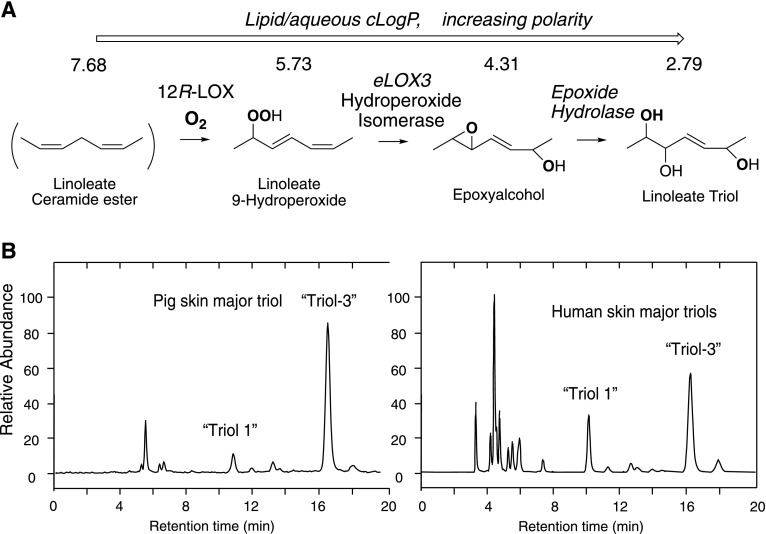
Recent evidence suggests that this oxidative pathway in the outer epidermis is extended by transformation of the epoxyalcohol formed by the action of 12R-LOX and eLOX3 to a trihydroxy derivative (Fig. 1A) (7). For illustrative purposes (the relevance of which will be evident later with the presentation of Results), the pattern of linoleate triols previously reported to be esterified in Cer-EOS of pig and human epidermis is shown in Fig. 1B (7). It is a central tenet of the hypothesis governing the involvement of the LOX pathway in epidermal barrier construction that oxidation of the linoleate ester has a disruptive effect on the lipid environment and that this facilitates esterase-catalyzed removal of the (oxidized) linoleate (6). Thus, implicating the linoleate triol in the pathway supports and extends the hypothesis because the triols have the high polarity needed for disruption of the lipid environment, facilitating cleavage of the oxidized Cer-EOS for covalent coupling of the free omega-hydroxyacyl-sphingosine (Cer-OS) to protein. The relative polarities of linoleate, its 9-hydroperoxide, the epoxyalcohol, and a linoleate triol are shown on Fig. 1A as the trend toward a reduced logP partition coefficient, with formation of the triol making a major impact.
Fig. 1.
The proposed 12R-LOX pathway in the mammalian epidermis and formation of linoleate triols. A: The 12R-LOX/eLOX3 pathway of the mammalian epidermis and proposed involvement of EH forming linoleate triol. The stepwise transformations increase the polarity of the unionized fatty acid moiety (indicated by the decreased lipid/aqueous partition coefficients, cLogP calculated by ChemDraw). B: Previously reported patterns of linoleate triols esterified in Cer-EOS as detected by LC-MS of pig and human epidermis (7). Structures of triol-1 and -3 are the same as illustrated later in Fig. 5A of the Results.
The above considerations bring in the potential involvement of one or more epoxide hydrolases (EHs) in producing the distinctive patterns of linoleate triols previously observed in pig and human epidermis and illustrated in Fig. 1B (7). The two well-studied enzymes in mammalian biology are microsomal EH [mEH (EPHX1)] and soluble EH [sEH (EPHX2)] (8), and in this work we evaluate both for their ability to hydrolyze the relevant fatty acid epoxides. The EH gene family also includes EH3, EH4, and paternally expressed gene 1/mesoderm specific transcript (peg1/MEST) (9). The peg1/MEST is an unlikely candidate; it is an imprinted gene with roles in development (10) and not so far shown to have EH activity. EH4 is similarly unstudied as an EH; the main site of its gene expression is the CNS (11), and more recently it is attributed a role in sebocyte lipid droplet biogenesis (12). The third additional candidate is EH3, which is known to hydrolyze fatty acid cis-epoxyeicosatrienoic acids (EETs) [which have proposed roles in epidermal differentiation (13)], and EH3 is highly expressed in the skin (14); moreover, the highest expression of EH3 mRNA is in the outer epidermis where the water permeability barrier is formed (15). From comparison of expression patterns of candidate genes with disease-related gene clusters, EPHX3 is also recognized as a potential ichthyosis gene (16). For all of these reasons, EH3 was selected along with mEH and sEH for evaluation of their catalytic activity with epidermis-related fatty acid epoxides.
EXPERIMENTAL PROCEDURES
Materials
Linoleic acid, methyl linoleate, and linoelaidic acid were purchased from Nu-Chek Prep Inc. (Elysian, MN). The 9R-hydroperoxy-octadecadienoic acid (HPODE) was prepared using Anabaena 9R-LOX (6). Epoxyalcohol and triol derivatives of 9-hydroperoxy-linoleic acid were prepared as previously described (17). Pentafluorobenzyl (PFB) bromide, p-toluene sulfonic acid, pyridinium p-toluenesulfonate, diisopropylethylamine, 2,2-dimethoxypropane (DMP), and trimethylamine were obtained from Sigma-Aldrich (St. Louis, MO). The p-toluene sulfinic acid was obtained by acidification of aqueous solutions of the commercially available sodium salt with HCl (18). The mixture of eight deuterated linoleate triols was prepared by autoxidation of [2H4]linoleic acid, as previously described (7). The [2H8]14,15-dihydroxy-eicosatrienoic acid was prepared by m-chloroperoxybenzoic acid (mCPBA) treatment of d8-arachidonic acid, isolation of the 14,15-EET, and acid hydrolysis to the d8-14,15-diol. Two synthetic trihydroxy derivatives of linoleic acid, 9R,10R,13R-trihydroxy-11E-octadecenoic acid (triol-1) and the 9R,10S,13R isomer (triol-3), were prepared by total synthesis using methodology to be described elsewhere.
Derivatization procedures
PFB esters were prepared by dissolving the d4 standards or triol analyte in 20 μl of acetonitrile, 20 μl of PFB bromide in acetonitrile (1:19, v/v), and 20 μl of diisopropylethylamine in acetonitrile (1:9, v/v). The solution was incubated at room temperature under argon for 30 min and then evaporated to dryness under nitrogen. Acetonide (DMP) derivatives of the triols were prepared by treatment of the fatty acid PFB ester with 20 μl of 1 mM pyridinium p-toluenesulfonate in acetone/DMP (1:1 by volume) for 30 min at room temperature. Samples were then taken to dryness under a stream of nitrogen and redissolved in straight-phase (SP)-HPLC solvent (hexane/IPA, 100:1, v/v) for subsequent HPLC-UV or LC-MS analysis.
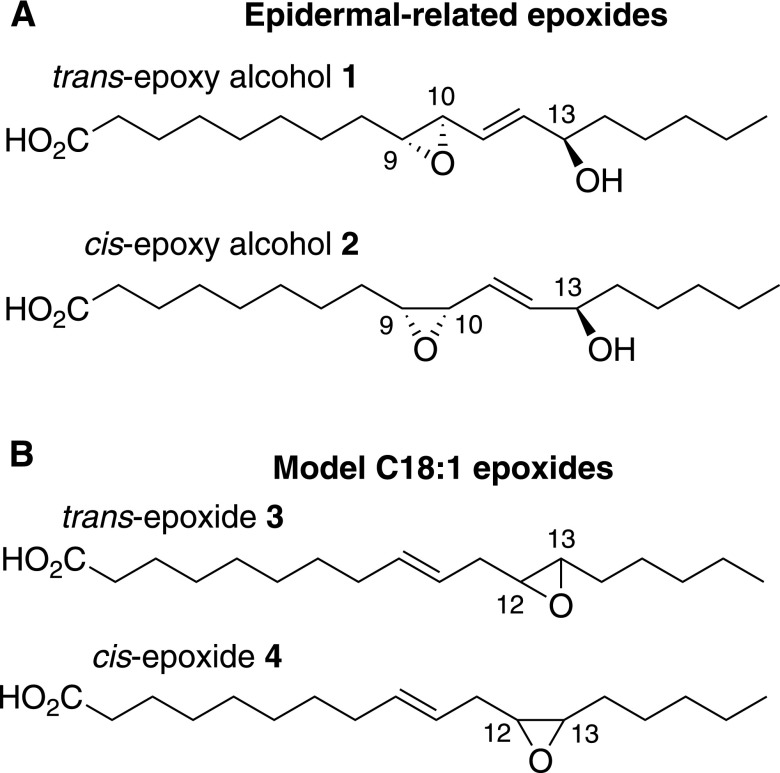
Preparation of 9R,10R-trans-epoxy-13R-hydroxy-octadeca-11E-enoic acid (1) and 9R,10S-cis-epoxy-13R-hydroxy-octadeca-11E-enoic acid (2)
These allylic epoxides were isolated following hematin treatment of 9R-HPODE as described (17) (Fig. 2A). Briefly, 9R-HPODE was prepared in 5–10 mg quantities using recombinant Anabaena 9R-LOX (19) and converted by hematin treatment to a mixture of products that were separated by straight-phase (SP)-HPLC. The 9R,10R-trans-epoxy-13R-hydroxy-octadeca-11E-enoic acid (trans-epoxyalcohol 1) was recovered in about 10% yield and the more minor 9R,10S-cis-epoxy-13R-hydroxy-octadeca-11E-enoic acid (cis-epoxyalcohol 2) in ∼3% yield. The products were quantified by conversion of an aliquot of approximately 50 μg to the PFB ester and, after SP-HPLC purification, quantified using the UV chromophore at 263 nm in ethanol (ε = 570 M−1 cm−1, unpublished observations); although this could lead to a slight underestimate of the free acid concentration due to incomplete recovery, the method was considered accurate to within 10%.
Fig. 2.
Structures of the linoleate-derived trans and cis allylic epoxyalcohols 1 and 2 and the model cis and trans fatty acid epoxides 3 and 4. The epidermal-related epoxides are trans-epoxyalcohol 1 and cis-epoxyalcohol 2. The model C18:1 epoxides are trans-epoxide 3 and cis-epoxide 4.
Preparation of 12,13-trans-epoxy-octadeca-9E-enoic acid (trans-epoxide 3)
Linoelaidic acid (20 mg), the 9E,12E geometric isomer of linoleic acid, was epoxidized using equimolar mCPBA in dichloromethane for 30 min at room temperature (Fig. 2B). The two major products, the 12,13- and 9,10-trans-epoxides, were separated on SP-HPLC using a semi-preparative Beckman 5 μm Ultrasphere silica column (250 × 10 mm) with a solvent of hexane/IPA/glacial acetic acid (100/0.4/0.02) and a flow rate of 5.0 ml/min; the peaks were detected by UV monitoring at 205 nm and eluted at 25.8 min (12,13-trans-epoxides) and 31.8 min (9,10-trans-epoxides). The products were quantified by weighing.
Preparation of 12,13-cis-epoxy-octadeca-9E-enoic acid (cis-epoxide 4)
This epoxide was prepared by initial isomerization of methyl linoleate (20 mg) to a mixture of the nonconjugated 9E,12Z and 9Z,12E isomers (18), which were isolated by silver ion chromatography (Fig. 2B). Following epoxidation of the 9E,12Z and 9Z,12E isomers with mCPBA, the products were separated by SP-HPLC using a Thomson silica column (5 μm, 4.6 × 250 mm), a solvent of hexane/IPA (100:0.2), and a flow rate of 0.5 ml/min. The mixture chromatographed as three peaks in 1:3:2 ratio in order of elution. From preliminary studies using a model compound, 6E,9Z-pentadecadiene (20), we found that mCPBA favored epoxidation of the cis double bond by 2:1 over trans. From this, and knowing that on SP-HPLC 12,13-epoxy-linoleates elute before the 9,10-epoxides (21), we could interpret the 1:3:2 ratio as an early-eluting 12,13-trans-epoxide (area of 1), a mixture of 12,13-cis-epoxide (area of 2) and 9,10-trans-epoxide (area of 1), and lastly a peak of the 9,10-cis-epoxide (area of 2). This was later confirmed by NMR. The mixture of 12,13-cis-epoxy-9E and 9,10-trans-epoxy-12Z eluted at 13.1 min and was resolved by silver ion chromatography using a Waters 5 μm, 4.6 × 250 mm column in the Ag+ form with a solvent of hexane/IPA (100:1) and a flow rate of 0.5 ml/min. The first eluting peak, 12,13-cis-epoxy-octadeca-9E-enoate at ∼9 min retention time was collected and alkali-hydrolyzed to the free acid. It was quantified by HPLC peak area (205 nm) in comparison to trans-epoxide 3, yield 175 μg. NMR in C6D6 showed, δ ppm: 5.52, 1H, ddd, J9,10 = 15.4 Hz, H9; 5.46, 1H, ddd, J9,10 = 15.4 Hz, H10; 2.87, 1H, dt, J12,13 = 4.1 Hz, H12; 2.75, 1H, dt, J12,13 = 4.2 Hz, H13; 2.27, 1H, dd, H11a; 2.11, 1H, ddd, H11b; 2.04, 1H, t, H2; 1.55–1.05, 18H, m, H3, 4, 5, 6, 7, 14, 15, 16, 17; 0.85, 3H, t, H18.
Analysis of the pH sensitivity of allylic epoxides
The trans-epoxyalcohol 1 and cis-epoxyalcohol 2 (2 μl in ethanol, ∼1 μg) were incubated in 48 μl of pH 5, pH 6, pH 7, and pH 8 0.1 M phosphate buffers for times up to 1 h at 37°C. After incubation at 37°C, 50 μl of cold acetonitrile were added and the samples were placed on ice and analyzed in turn (10% aliquots, lowest pH first) by reversed-phase (RP)-HPLC-UV using a Kinetex 2.6 μm C18 column (100 × 3 mm) with a 0.5 μm PEEK precolumn microfilter, a solvent of CH3CN/water/HAc (45/55/0.01), and flow rate of 0.4 ml/min with UV monitoring at 205 nm. To suppress ionization and achieve consistent retention times of the pH 7 and pH 8 samples, 2 μl of 10-fold diluted glacial acetic acid were added immediately prior to injection.
The nonenzymatic hydrolysis products from the pH 5 and pH 6 conditions were collected using a Waters Symmetry C18 column, 4.6 × 250 mm, a solvent of CH3CN/water/HAc (50/50/0.01), and a flow rate of 1 ml/min, giving a retention time of ∼4.5–4.8 min and ∼4.2–4.4 min for the triol products from trans-epoxyalcohol 1 and cis-epoxyalcohol 2, respectively (detected at 205 nm). The products were converted to the PFB ester DMP derivative and analyzed by atmospheric pressure chemical ionization-LC-MS using a TSQ Vantage instrument (Thermo Scientific) with a Waters Alliance 2690 HPLC system, and a Kinetex 100A 2.6 μm HILIC column (100 × 2.1 mm) with a solvent of hexane/IPA (100:1, v/v) and a flow rate of 0.4 ml/min. The atmospheric pressure chemical ionization vaporizer temperature was set to 300°C and the electrospray ionization probe temperature was set to 150°C with selected ion monitoring of negative ions at m/z 369 for the unlabeled triol derivatives and m/z 373 for a mixture of eight d4-triol standards prepared as described (7).
Preparation of the human EH3, sEH, and mEH
A full-length sequence of human EH3 (GenBank accession BC132960) was PCR-amplified from a template plasmid obtained from the Integrated Molecular Analysis of Genomes and their Expression (IMAGE) Consortium (Clone identification number 40146982) through Open Biosystems. The PCR reactions were performed using KOD Hot Start polymerase (EMD Millipore) with primers EH3BglATG (5′-GAAGATCTATGCCGGAGCTGGTGGTGACCG-3′) and EH3stopEco (5′-CGGAATTCCTAGTCCAGCAGGTCTTGCAAGAAGGC-3′) following the manufacturer’s recommended procedures. The use of primers EH3BglATG and EH3stopEco placed BglII and EcoRI sites at the 5′ and 3′ ends of the coding sequence, respectively. The amplicon (1.1 kbp) was inserted into the cloning vector, pCR-Blunt II-TOPO (Invitrogen), and sequenced in both directions in order to confirm the authenticity of the sequence. Subsequently, the 1.1 kbp-long insert was excised by digestion with BglII and EcoRI, and then ligated using T4 DNA ligase (New England BioLabs) into the BglII and EcoRI sites of the baculovirus transfer vector, pAcUW21, in order to generate the recombinant baculovirus, Ac-hEH3, that expresses the human EH3, following published procedures for pAcUW21-hEH3 (22). The human sEH and mEH were cloned in similar baculoviruses previously (23, 24). Human sEH, mEH, and EH3 were produced in a baculovirus expression system as previously described (23). Human sEH was purified by affinity chromatography (25), mEH was partially purified by chromatography (26), and a microsomal cell extract was used as a source of human EH3. The human sEH, mEH, and EH3 preparations were 97, 80, and 4% pure, respectively, as judged by SDS-PAGE and scanning densitometry. The activity of the purified enzymes was tested using radioactive surrogate substrates (27). The human sEH and human EH3 activities were tested using [3H]trans-diphenyl-propene oxide as substrate, and found to be 7,100 and 48 nmol·min−1·mg−1, respectively. The human mEH activity was measured with [3H]cis-stilbene oxide, and found to be 490 nmol·min−1 ·mg−1.
Mouse EH3 protein expression
Three different constructs of murine EH3 sequence (see Results) that we refer to as full-length, N-truncated, and N-truncated+7aa-insert, were ordered with an additional N-terminal (His)6 tag from Genewiz (South Plainfield, NJ); the full-length sequence was codon-optimized for expression in Sf9 cells, whereas the truncated constructs were ordered with the natural sequences. The three constructs were first tested in Escherichia coli in a pET plasmid only to see no enzyme activity, so expression was tested in Sf9 cells in the pVL1393 vector (PharMingen, San Diego, CA). Expression was carried out using the AB Vector Proeasy™ baculovirus expression system. Cell lysates were tested for EH activity and, paradoxically, the codon-optimized full-length cDNA failed to express with enzyme activity; therefore, further experiments were carried out with the N-truncated proteins. Although lysates of the N-truncated constructs had activity, no band was evident on SDS-PAGE and the activity failed to solubilize using a number of detergents, so cell lysates were used in our experiments.
Enzymatic activity and pH activity profile with model cis and trans epoxy standards
Enzymatic hydrolysis of trans-epoxide 3 and cis-epoxide 4 (∼0.75 μg) were compared with two different EHs, human sEH (purified protein:100 ng) and human EH3, used as cell lysates containing 54 μg of protein, respectively. Incubations were conducted for 60 min at 37°C in a 0.5 ml Eppendorf tube containing 12 μl of enzyme lysate and 1 μl of epoxyalcohol substrate in ethanol (0.75 μg) made up to a total volume of 50 μl with phosphate buffer (0.1 mM, pH 8.0). The pH activity profiles using the model trans-epoxide 3 (50 μM) were conducted in 50 μl aliquots of 0.1 M phosphate buffers at pH 5, 6, 7, or 8 for 30 min at 37°C with human sEH and human EH3. Reactions were terminated after 60 min by addition of an equal volume of acetonitrile. Then the samples were centrifuged for 10 min at 16,000 g, the supernatant was injected into RP-HPLC using a Kinetex 2.6 μm C18 column (100 × 3 mm) with a 0.5 μm PEEK precolumn microfilter, a solvent of CH3CN/water/HAc (45/55/0.01), and flow rate of 0.4 ml/min, giving a retention time of 2.2 and 2.3 min for diol products from trans-epoxide 3 and cis-epoxide 4 (detected at 205 nm). Immediately prior to HPLC of the samples, approximately 2 μl of 10-fold dilute glacial acetic acid were added to suppress ionization and achieve consistent retention times, mixed, and the aliquot injected immediately.
Incubations of allylic epoxyalcohols with EHs
Enzymatic hydrolysis of trans-epoxyalcohol 1 and cis-epoxyalcohol 2 (∼1 μg) was examined with five different EHs, human sEH (purified protein:100 ng), human EH3, N-truncated murine EH3, and N-truncated+7aa-insert murine EH3 (used as cell lysates containing 54, 74, and 68 μg of protein, respectively), and human mEH (purified protein:100 ng). Incubations were conducted for 60 min at 37°C in a 0.5 ml Eppendorf tube containing 12 μl of enzyme lysate, 1 μg of epoxyalcohol substrate in ethanol (1 μl) made up to a total volume of 50 μl with Tris buffer (10 mM, pH 8.0) containing NaCl (100 mM), EDTA (1 mM), and gelatin (0.1% w/v). The reaction was terminated after 60 min by addition of an equal volume of acetonitrile. Then the samples were taken to dryness under a stream of nitrogen and redissolved with 100 μl CH3CN/water/HAc (60/40/0.01). After centrifugation for 10 min at 16,000 g, the supernatant was injected into RP-HPLC with a Waters Symmetry C18 column, 4.6 × 250 mm, a solvent of CH3CN/water/HAc (60/40/0.01), and flow rate of 0.75 ml/min, giving retention times of 4.0 and 3.7 min for triol products from RRR trans- and RSR cis-epoxyalcohol; the peaks (205 nm) were collected manually and subsequently derivatized for LC-MS analysis as described above.
RESULTS
Acid-catalyzed hydrolysis of linoleate allylic epoxyalcohols
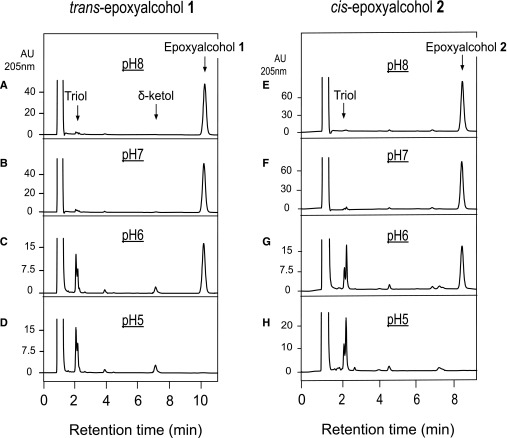
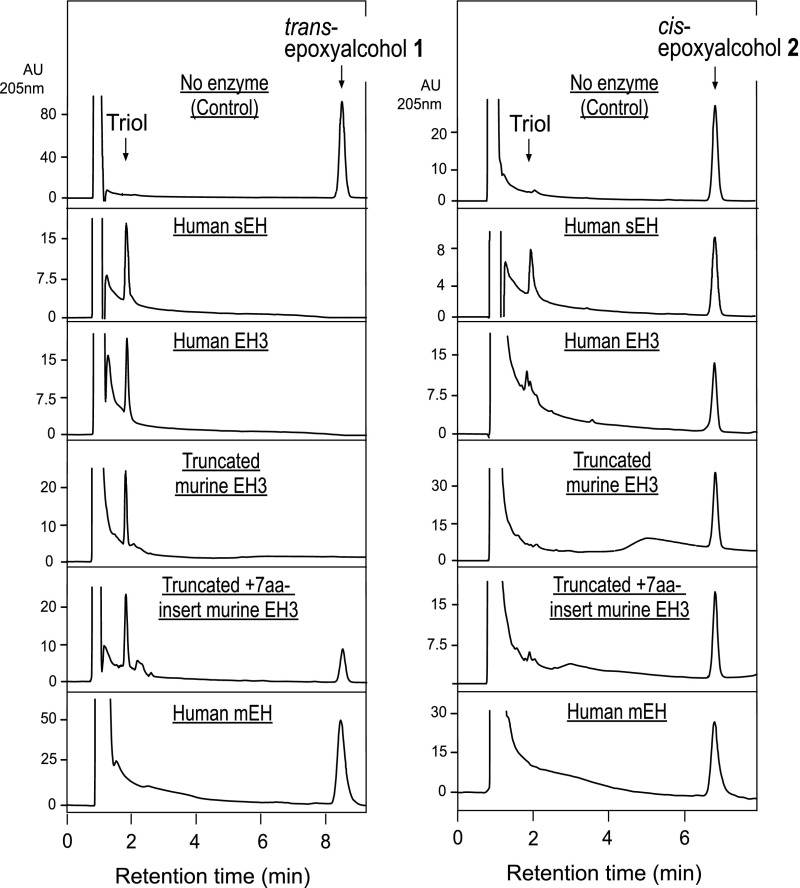
Prior to assessing the characteristics of enzymatic hydrolysis of skin-related linoleate epoxyalcohols, it is important to describe their sensitivity to nonenzymatic hydrolysis. The pH of the mammalian outer epidermis is slightly acidic [the pH for human skin lies in the broad range 4.0–7.0 (28, 29)] and the epoxyalcohols derived via 12R-LOX and eLOX3 catalysis include an allylic epoxide structure that should be acid sensitive. Therefore, we carefully assessed the effects of a mildly acidic environment on the hydrolytic degradation of epidermal epoxyalcohols. Incubated in pH 8 or pH 7 buffer, the skin-related trans-epoxyalcohol 1 was not hydrolyzed over the course of 30 min at 37°C (Fig. 3A, B), whereas nonenzymatic hydrolysis was about 70% complete at pH 6 (Fig. 3C) and full hydrolysis occurred after 30 min in pH 5 buffer (Fig. 3D). The cis-epoxyalcohol 2 showed essentially the same pH sensitivity, being fully hydrolyzed after 30 min at 37°C in pH 5 buffer (Fig. 3E–H).
Fig. 3.
RP-HPLC analysis of the pH sensitivity of epoxyalcohols 1 and 2 exposed to pH 5–8. The trans-epoxyalcohol 1 and cis-epoxyalcohol 2 (30 μM) were incubated in 50 μl aliquots of 0.1 M phosphate buffers at pH 5, 6, 7, or 8 for 30 min at 37°C. The samples were then injected directly on RP-HPLC. The HPLC analysis used a Kinetex 2.6 μ 100 × 3 mm column with an isocratic solvent acetonitrile/water/glacial acetic acid (45/55/0.01, by volume) at a flow rate of 0.4 ml/min with UV detection at 205 nm. Retention times are illustrated for the starting material (epoxyalcohol), the triol hydrolysis products, and a δ-ketol formed during acid-catalyzed rearrangement (17).
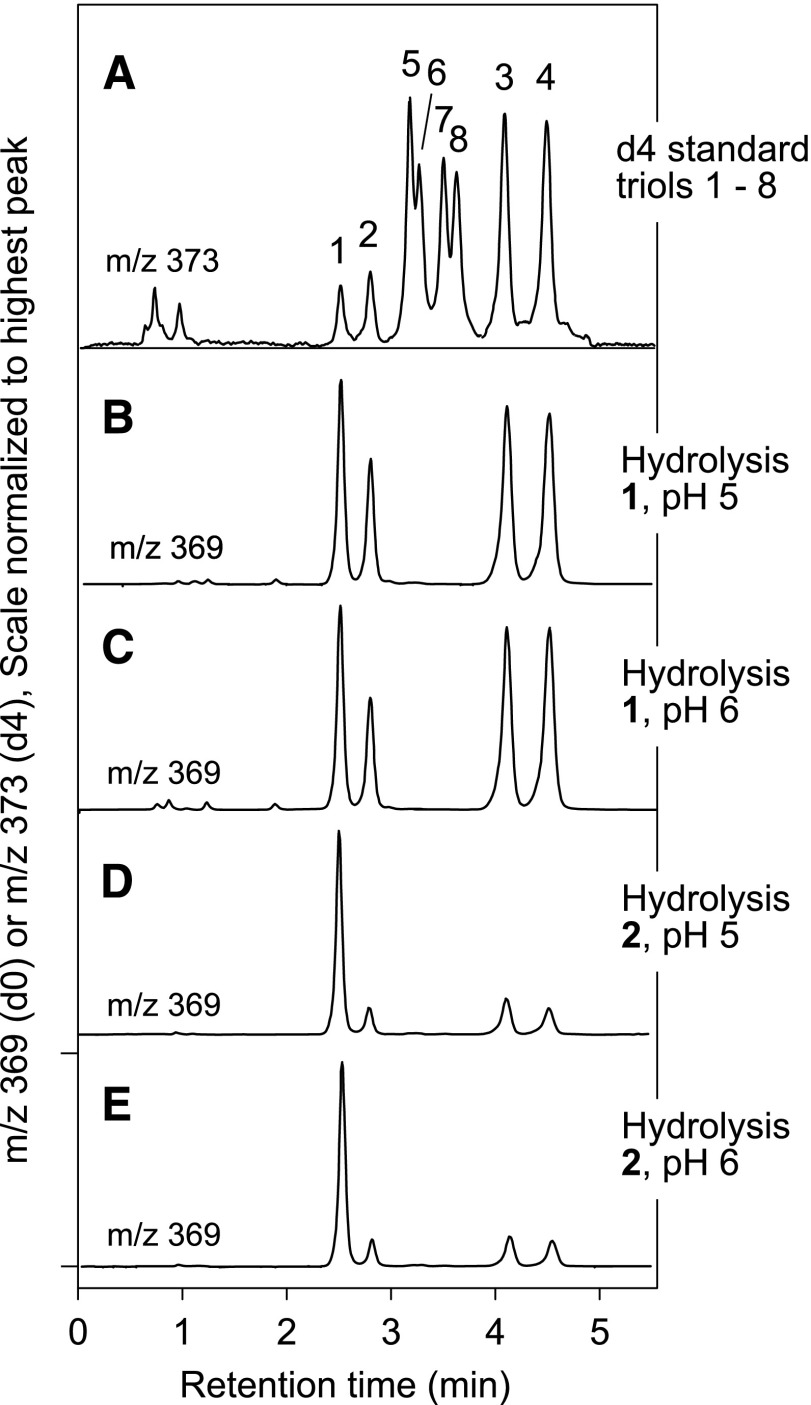
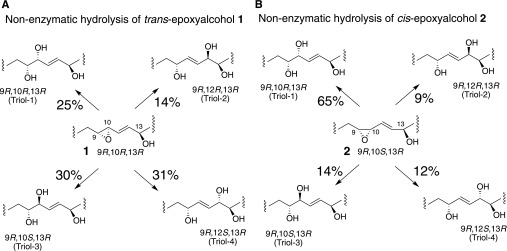
The individual triol products of these nonenzymatic hydrolyses were identified by LC-MS of the PFB ester DMP derivative, a method that allows assignment of eight separate triol isomers, illustrated in Fig. 4A (7) (and the same HPLC method illustrated in Fig. 1B of the Introduction). Nonenzymatic hydrolysis of the trans-epoxyalcohol 1 at pH 6 or pH 5 gave four triol products (designated here as triols 1–4), as expected from bidirectional attack of water at both C-10 and C-12 (Fig. 4B, C; Fig. 5A); in three separate experiments, the relative proportions of triols 1–4 averaged 25:14:30:31, respectively (Fig. 5A), similar to the result using 0.1% aqueous glacial acetic acid, which averaged 28:18:26:28. In contrast to the four prominent triols formed from the trans epoxide, the corresponding cis-epoxyalcohol 2 gave one prominent hydrolysis product in 65% relative abundance along with three minor triols (proportions 65:9:14:12, respectively, Fig. 4D, E and Fig. 5B), similar to the proportions using 0.1% aqueous glacial acetic acid (63:9:14:14).
Fig. 4.
Identification of the triol products of nonenzymatic hydrolysis of epoxyalcohols 1 and 2. Triol isomers were separated on LC-MS by SP-HPLC of the 1,2-acetonide PFB ester derivative and detected as the M – PFB ion at m/z 369 (7). The top chromatogram illustrates separation of eight triol isomers (A). B, C: Show that trans-epoxyalcohol 1 is hydrolyzed at pH 5 or pH 6 to a mixture of triols 1, 2, 3 and 4. D, E: The cis-epoxyalcohol 2 is hydrolyzed at pH 5 or pH 6 to a mixture of four triols, predominantly 1, and also including triols 2, 3, and 4. The HPLC analysis used a Phenomenex HILIC column (100 × 2 mm) with a solvent system of hexane/IPA (100:1, v/v) at a flow rate of 0.5 ml/min.
Fig. 5.
Structures of the triol products of nonenzymatic hydrolysis of epoxyalcohols 1 and 2. The percentage of each triol product of nonenzymatic hydrolysis is shown next to the arrows.
Preparation of mouse EH3
Analysis of the EPHX3 sequences in GenBank shows that rodent EH3 is expressed with an additional 57 amino acids on the N terminus and that mouse EH3 is found in a second isoform that contains a seven amino acid insert after position 138 (Fig. 6). We elected to express one construct containing the full-length sequence including the seven amino acid insert, and two variants (with or without the seven amino acid insert) with the rodent-specific 57 amino acid N-terminal sequence removed (Fig. 6). Heterologous expression of these constructs in several strains of E. coli gave a strong band at the expected molecular mobility on SDS-PAGE, but the proteins were insoluble and completely lacking in catalytic activity. Expression of active enzyme in Sf9 insect cells was successful with both of the truncated forms of the enzyme, whereas the full-length construct exhibited no activity. Although activity was readily measurable for the N-terminally shortened constructs, the expression level was insufficient to visualize the EH3 protein on SDS-PAGE. Lysates of EH3-expressing Sf9 cells were used for all experiments. Similar findings to the above were reported for human EH3 expression in E. coli and Sf9 cells (14) and, similarly, we used Sf9 cell lysates for assessment of human EH3 activity.
Fig. 6.
Amino acid sequences of EH3 (EPHX3). On top is the sequence of human EH3. Below are three constructs of mouse EH3 prepared for testing in the current study, each with a N-terminal (His)6 tag: i) truncated mouse EH3 with the rodent-specific 57 amino acid N-terminus removed; ii) truncated mouse EH3 containing a seven amino acid insert (VSVPPVK) at position 138; and iii) full-length mouse EH3. Arrowheads (with numbering of human EH3 residues) indicate the α/β-hydrolase catalytic triad (D173, D307, H337) and the two tyrosines (Y220, Y281) important for initial activation of the oxirane ring (14).
Enzymatic hydrolysis of model linoleate epoxides
Both sEH and EH3 are known to efficiently hydrolyze the arachidonate-derived cis-EETs and sEH is known also to metabolize trans-EETs (30). We were interested in comparing the hydrolase activity with cis and trans epoxides of otherwise identical structure, and as we did not have the trans-EET analogs available, we prepared model linoleate epoxides 3 and 4 for these experiments. These non-allylic epoxides are chemically stable at pH 5 or 6, which proved to be useful for comparing the hydrolase activities of sEH and EH3 at the slightly acidic pH values typical of human epidermis.
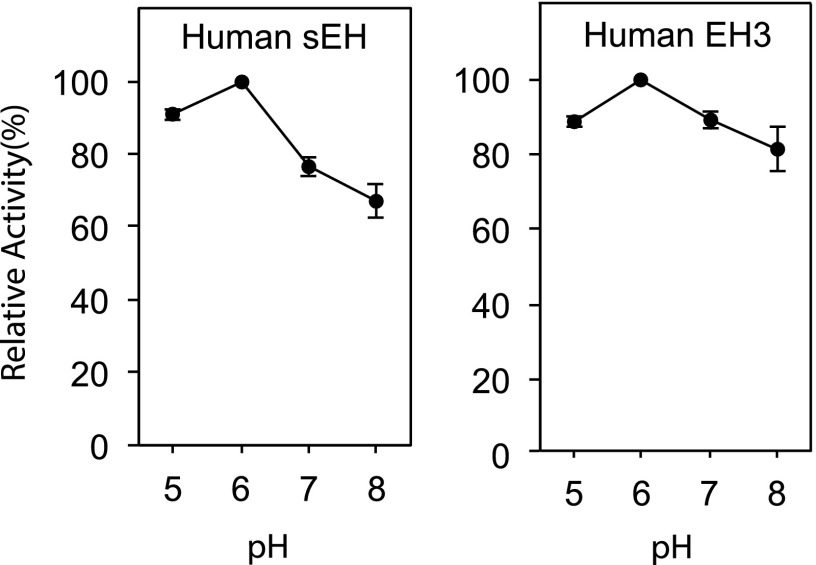
As an initial screen for EH activity comparing trans-epoxide 3 with cis-epoxide 4, the substrates were incubated for 1 h at 37°C with human sEH, human EH3, and one of the mouse EH3 constructs. The concentrations of enzymes used were those known to metabolize EETs. Under these conditions, recombinant sEH (100 ng/ml) partially hydrolyzed both the trans- and cis-epoxides, showing a preference for the trans epoxide 3 (Fig. 7B), similar to reported properties with trans- and cis-EETs (30). Human EH3 and mouse EH3 showed similar activities, hydrolyzing both epoxides 3 and 4, with a preference for trans-epoxide 3 (Fig. 7C, D). Using trans-epoxide 4 as substrate, the pH profiles of activity of sEH and EH3 were compared at pH 5–8 (Fig. 8).
Fig. 7.
Comparison of the enzymatic hydrolysis of model cis and trans fatty acid epoxides 3 and 4. The trans-epoxide 3 and the cis-epoxide 4 were incubated in 50 μl aliquots of 0.1 M phosphate buffers at pH 8 with human sEH, human EH3, and truncated mouse EH3 containing the seven amino acid insert (see Fig. 6). The samples were then injected directly on RP-HPLC; the samples were slightly acidified by addition of 2 μl of 10-fold dilute glacial acetic acid immediately before injection on column. The HPLC analysis used a Kinetex 2.6 μ 100 × 3 mm column with an isocratic solvent, acetonitrile/water/glacial acetic acid (60/40/0.01, by volume), at a flow rate of 0.4 ml/min with UV detection at 205 nm. Retention times are illustrated for the starting material (model fatty acid epoxide) and the diol hydrolysis products.
Fig. 8.
pH profile of EH activities with trans-epoxide 3. The model trans-epoxide 3 (50 μM) was incubated in 50 μl aliquots of 0.1 M phosphate buffers at pH, 5, 6, 7, or 8 for 30 min at 37°C with human sEH and human EH3. The samples were then injected directly on RP-HPLC; the pH 7 and 8 samples were slightly acidified before injection on column. Chromatographic conditions were the same as in Fig. 7. The data points represent n = 3, ± range.
Enzymatic hydrolysis of allylic linoleate epoxides
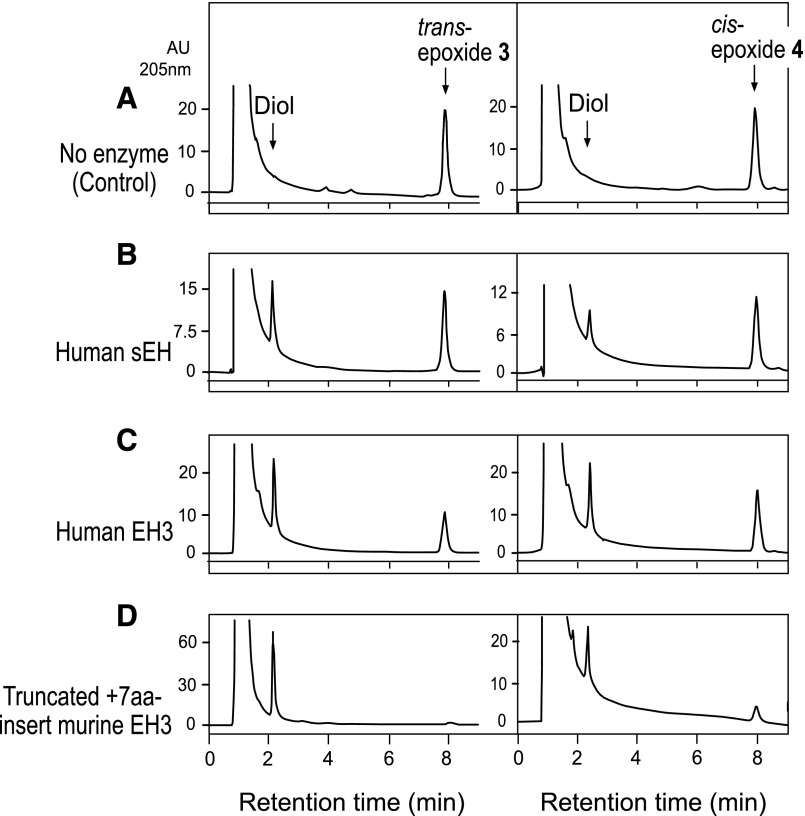
The trans-epoxyalcohol 1 and cis-epoxyalcohol 2 were tested with human sEH, human EH3, murine EH3 (both the N-truncated form and the N-truncated+7aa-insert). Hydrolysis products of the epoxyalcohol, triol, were collected and derivatized to PFB ester, then to DMP for further analysis. Figure 9 compares the RP-HPLC profiles of linoleate allylic epoxyalcohol substrate and the more polar hydrolysis products. Under the conditions used, trans-epoxyalcohol 1 was completely hydrolyzed using human sEH, human EH3, and N-truncated murine EH3 and mostly hydrolyzed using the N-truncated+7aa-insert murine EH3. The cis-epoxyalcohol 2 was hardly hydrolyzed using human EH3, N-truncated murine EH3, and the N-truncated+7aa-insert murine EH3, and ∼30% hydrolysis was observed using human sEH. Human mEH did not hydrolyze trans-epoxyalcohol 1 or cis-epoxyalcohol 2 (Fig. 9).
Fig. 9.
RP-HPLC analysis of the hydrolysis of epoxyalcohols 1 and 2 by EHs. The trans-epoxyalcohol 1 and the cis-epoxyalcohol 2 were incubated in 50 μl aliquots of Tris buffer (10 mM, pH 8.0) with human sEH, human EH3, two N-terminally truncated constructs of mouse EH3 (with and without the seven amino acid insert, Fig. 6), and human mEH. Chromatographic conditions were the same as in Fig. 3. Retention times are illustrated for the starting material (epoxyalcohol) and the triol hydrolysis products.
Identification of the enzymatic hydrolysis products of linoleate allylic epoxyalcohols
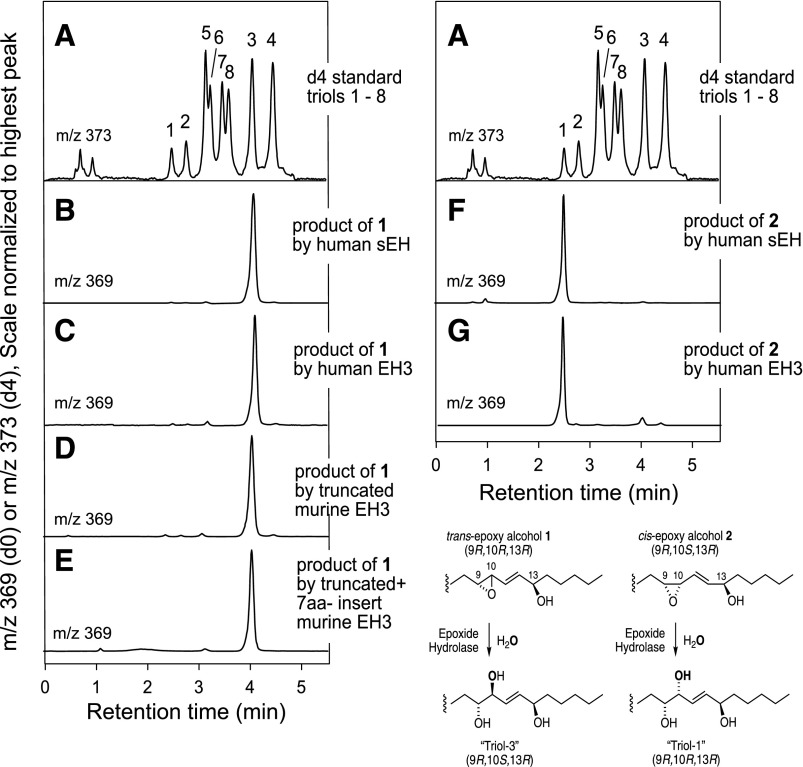
Applying the same LC-MS assay used to characterize the mixtures of triols formed nonenzymatically, human sEH, human EH3, and murine EH3 (both the N-truncated form and the N-truncated+7aa-insert) were shown to convert trans-epoxyalcohol 1 exclusively to triol-3 (Fig. 10A–E). Likewise, the hydrolysis of cis-epoxyalcohol 2 by human sEH and human EH3 exclusively gave triol-1 (Fig. 10F, G).
Fig. 10.
Identification of the triol products of enzymatic hydrolysis of epoxyalcohols 1 and 2. Triol isomers were separated on LC-MS by SP-HPLC of the 1,2-acetonide PFB ester derivative and detected as the M – PFB ion at m/z 369 (7). The top chromatogram illustrates separation of eight triol isomers (A). B–E: Show that human sEH, human EH3, and two constructs of mouse EH3 each hydrolyze epoxyalcohol 1 exclusively to triol 3. F, G: Show that human sEH and human EH3 hydrolyze epoxyalcohol 2 exclusively to triol 1. The HPLC analysis used a Phenomenex HILIC column (100 × 2 mm) with a solvent system of hexane/IPA (100:1, v/v) at a flow rate of 0.4 ml/min.
Comparison of the rates of EET and linoleate allylic epoxide hydrolysis by sEH and EH3
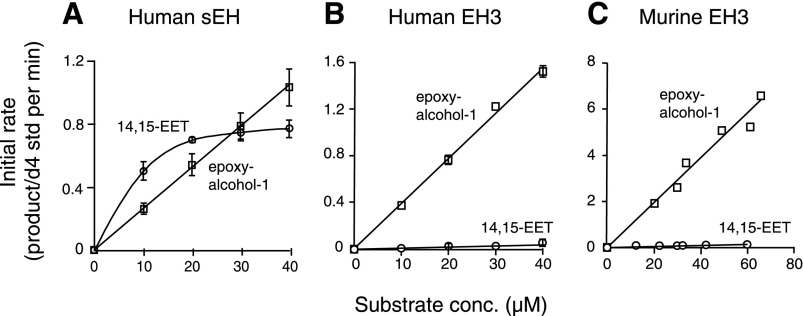
Both sEH and EH3 are known to metabolize EET-type epoxides (14). In the case of the well-studied sEH, the reports include the metabolism of both cis- and trans-EETs (30) and the hydrolysis of the linoleate 9,10- and 12,13-cis-epoxides (31). Here, we compared the rates of hydrolysis of 14,15-EET and trans-epoxyalcohol 1 under identical incubation conditions with sEH and EH3. The hydrolysis products were quantified by LC-MS using deuterated internal standards prepared for the study and the results are illustrated in Fig. 11. Using the rate versus substrate concentration data (Fig. 11), the turnover rates were calculated at low substrate concentration (1 μM) to observe first-order behavior. Human sEH showed 2.1-fold higher activity in the hydrolysis of cis-14,15-EET over trans-epoxyalcohol 1 (Fig. 11A). By contrast, human and murine EH3 showed 31-fold and 39-fold differences, respectively, with the preference reversed in favor of trans-epoxyalcohol 1 hydrolysis over cis-14,15-EET (Fig. 11B, C).
Fig. 11.
Comparison of the rates of hydrolysis of trans-epoxyalcohol 1 and 14,15-EET. The initial rates of transformation of trans-epoxyalcohol 1 (open squares) and 14,15-EET (open circles) are plotted against concentration of the epoxide on incubation with human sEH (A), human EH3 (B), and truncated mouse EH3 (C). Incubations were conducted in 50 μl aliquots of 0.1 M phosphate buffer at pH 8 at 37°C and terminated by addition of 50 μl acetonitrile and deuterated internal standards (d4-triol or d8-diol). The samples were analyzed using a Kinetex 2.6 μ C18 column (100 × 3 mm) with an isocratic solvent of acetonitrile/15 mM ammonium acetate pH 8.5 (25/75, by volume for triol analysis and 35/65, by volume for diol) at a flow rate of 0.4 ml/min, with LC-MS detection at m/z 329 (d0) and 333 (d4) for the triol product from trans-epoxyalcohol 1 and m/z 337 (do) and 345 (d8) for the diol product from 14.15-EET. The graph was obtained by using KaleidaGraph 4.0 (Synergy Software, Reading, PA); A, B: Data are expressed as mean ± SEM, n = 4. C: Data points represent the average of two or three determinations from two independent experiments.
DISCUSSION
EHs implicated in epidermal water barrier formation
The aim of this study was to investigate the catalytic activities of EH enzymes potentially involved in formation of the epidermal permeability barrier. So far, no EH has been tested with substrates directly relevant to the LOX epidermal permeability pathway. Herein, we demonstrate that the EHs, sEH and EH3, enzymatically hydrolyze trans-epoxyalcohol 1 (skin-related allylic epoxide) to RSR triol-3, the most abundant triol isomer in human and porcine epidermis (7) (see Fig. 1B), and the cis-epoxyalcohol 2 is hydrolyzed to triol-1. This is consistent with the mechanism whereby EH inserts oxygen from water (via the active site aspartate) with reversal of configuration (Fig. 10). Our results are compatible with a significant role of the EHs, EH3 and sEH, in the epidermal LOX pathway. There is compelling experimental and analytical support for a role of an EH in skin barrier formation; although on the basis of the lack of a skin phenotype in the single knockouts of sEH and EH3 already reported (32, 33), there is functional redundancy of EHs in the epidermis. Soluble EH is well-expressed in skin (34), and transcriptome analysis of human granular keratinocytes identified EH3 as highly expressed in the outermost cells of human epidermis (20-fold over basal cells) (15). Also, coexpression analysis of mouse and human disease-related gene clusters identified EH3 as a predicted ichthyosis gene (P value 10−14) (16). In fact, on the basis of the tissue expression of mouse EH3, highest in skin, lung, and upper gastrointestinal tract, Decker et al. concluded that “Based on this observation, it is tempting to speculate on a potential role of EH3 in barrier formation, because the above tissues represent the major contact surfaces with the outside” (Ref. 14; p. 2042).
Analysis of the relative rates of hydrolysis of 14,15-EET and trans-epoxyalcohol 1 at very low substrate concentrations (the initial slopes of the rate versus substrate concentration lines reflecting the ratio of kcat/Km) showed that sEH hydrolyzes each substrate within the same order of magnitude (2-fold more efficiently in favor of 14,15-EET). By contrast, both human and mouse EH3 greatly favor the trans-epoxyalcohol 1, by factors of 31- and 39-fold, respectively. Also, Fig. 9 shows that EH3 has a great preference for the trans-allylic epoxyalcohol 1 over the cis analog 2, further supporting its potential relevance to the hydrolase activity in the 12R-LOX/eLOX3 pathway.
Reported nonenzymatic hydrolysis of other fatty acid epoxides
Although to the best of our knowledge the acid instability of the arachidonic acid-derived EETs or the equivalent linoleate analogs has not been studied in detail, one of the chemical procedures for their hydrolysis to diols involves treatment with 70% perchloric acid (35), i.e., the epoxides are moderately stable and strongly acidic conditions or exceptionally long incubation times are required for their complete hydrolysis. An exception is 5,6-EET, in which the free ionized carboxyl at C-1 facilitates hydrolysis and 5,6-EET is chemically unstable at pH 8.0 and 25°C and complete hydrolysis was observed within 7.5 h (36). At the extreme end of the spectrum of instability of fatty acid epoxides are leukotriene A4, with its epoxide moiety allylic to a conjugated triene and with an estimated half-life of 3.6 s at pH 7.4 and 25°C (37), and the fatty acid allene oxides, with a double bond impinging directly on the epoxide and a reported half-life of 30–40 s at pH 7.4 and 0°C (38, 39).
Among the fatty acid epoxyalcohols [including the hepoxilins (40)], it is recognized that the hepoxilin B analogs with the alcohol group between the epoxide and a double bond are stable for short periods at pH 3 (41), whereas the hepoxilin A types, which are analogs of the epoxyalcohols 1 and 2 studied here, can survive only briefly at pH 3 (1). Herein, we refine this understanding by showing the hydrolysis of 1 and 2 at pH 5 (complete within 30 min at 37°C) and pH 6, which gives about 50% hydrolysis under the same conditions. Most significantly, the pattern of nonenzymatic hydrolysis products of the epoxyalcohol of the LOX pathway in the epidermis (epoxyalcohol 1) gives four triols in 14, 25, 30, and 30% relative abundance (Figs. 4, 5). By contrast, the actions of sEH and EH3 give a single main triol, the same one that is prominent in pig and human epidermis (see Fig. 1). This evidence greatly strengthens the argument that EH(s) participate in the production of the linoleate triols detected in the mammalian epidermis. Some doubt remains on the enzymatic or nonenzymatic origin of triol-1 in the epidermis. Under very mild acidic conditions (pH 5, pH 6), the cis-epoxyalcohol 2 is hydrolyzed nonenzymatically predominantly to triol-1 (and exclusively to triol-1 by the action of EH enzymes), leaving open the basis of its synthesis in the skin.
CONCLUSIONS
The current studies, along with earlier findings, are consistent with a role for EH in the 12R-LOX/eLOX pathway in the epidermis. The next steps needed to test the postulated involvement in the formation of esterified linoleate triols and in epidermal barrier function under in vivo conditions will include the comparison of single or multiple EPHX knockout mice. This will define the functioning of specific EHs in the production of esterified linoleate triols and their significance to the epidermal permeability barrier, and contribute to development of an integrated model of epidermal barrier formation. Notably, loss of the covalently bound ceramides, as occurs with 12R-LOX deficiency, is similarly a feature of the gene inactivation of eLOX3 (42), P450 CYP4F22 (that omega-hydroxylates Cer to give Cer-OS) (43), PNPLA1 (that couples linoleate to Cer-OS) (44, 45), and CGI-58 (ABHD5, that facilitates the formation of CER-EOS) (46). Taken together, the importance of this work is for the basic understanding of a vital process for mammalian survival [“that permits terrestrial life” (Ref. 47; p. 231)], and for understanding the disease-related pathology and symptomology associated not only with the relatively rare cases of ARCI (48) but also symptomatic of such common skin conditions as atopic dermatitis, a huge clinical issue (49, 50).
Footnotes
Abbreviations:
- Cer-EOS
- esterified omega-hydroxyacyl-sphingosine
- Cer-OS
- omega-hydroxyacyl-sphingosine
- cis-epoxide 4
- 12,13-cis-epoxy-octadeca-9E-enoic acid
- cis-epoxyalcohol 2
- 9R,10S-cis-epoxy-13R-hydroxy-octadeca-11E-enoic acid
- DCM
- dichloromethane
- DMP
- 2,2-dimethoxypropane
- EET
- epoxyeicosatrienoic acid
- EH
- epoxide hydrolase
- EH3
- epoxide hydrolase-3 (EPHX3)
- eLOX3
- epidermal lipoxygenase-3
- HPODE
- hydroperoxy-octadecadienoic acid
- LOX
- lipoxygenase
- mCPBA
- m-chloroperoxybenzoic acid
- mEH
- microsomal epoxide hydrolase (EPHX1)
- PFB
- pentafluorobenzyl
- RP-HPLC
- reversed-phase-HPLC
- sEH
- soluble epoxide hydrolase (EPHX2)
- SP-HPLC
- straight-phase-HPLC
- trans-epoxide 3
- 12,13-trans-epoxy-octadeca-9E-enoic acid trans-epoxyalcohol 1, 9R,10R-trans-epoxy-13R-hydroxy-octadeca-11E-enoic acid
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR-51968 (A.R.B.), National Institutes of Health Shared Resource Grant P30 CA068485, National Institute of General Medical Sciences Grant GM-115722 (G.A.S.), and National Institute of Environmental Health Sciences Grants R01-ES002710 and P42 ES04699 (B.D.H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Muñoz-Garcia A., Thomas C. P., Keeney D. S., Zheng Y., and Brash A. R.. 2014. The importance of the lipoxygenase-hepoxilin pathway in the mammalian epidermal barrier. Biochim. Biophys. Acta. 1841: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krieg P., and Furstenberger G.. 2014. The role of lipoxygenases in epidermis. Biochim. Biophys. Acta. 1841: 390–400. [DOI] [PubMed] [Google Scholar]
- 3.Elias P. M., Williams M. L., Holleran W. M., Jiang Y. J., and Schmuth M.. 2008. Pathogenesis of permeability barrier abnormalities in the ichthyoses: inherited disorders of lipid metabolism. J. Lipid Res. 49: 697–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wertz P. W., and Downing D. T.. 1983. Acylglucosylceramides of pig epidermis: structure determination. J. Lipid Res. 24: 753–758. [PubMed] [Google Scholar]
- 5.Bowser P. A., Nugteren D. H., White R. J., Houtsmuller U. M., and Prottey C.. 1985. Identification, isolation and characterization of epidermal lipids containing linoleic acid. Biochim. Biophys. Acta. 834: 419–428. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Y., Yin H., Boeglin W. E., Elias P. M., Crumrine D., Beier D. R., and Brash A. R.. 2011. Lipoxygenases mediate the effect of essential fatty acid in skin barrier formation: a proposed role in releasing omega-hydroxyceramide for construction of the corneocyte lipid envelope. J. Biol. Chem. 286: 24046–24056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiba T., Thomas C. P., Calcutt M. W., Boeglin W. E., O’Donnell V. B., and Brash A. R.. 2016. The precise structures and stereochemistry of trihydroxy-linoleates esterified in human and porcine epidermis and their significance in skin barrier function: implication of an epoxide hydrolase in the transformations of linoleate. J. Biol. Chem. 291: 14540–14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DuTeaux S. B., Newman J. W., Morisseau C., Fairbairn E. A., Jelks K., Hammock B. D., and Miller M. G.. 2004. Epoxide hydrolases in the rat epididymis: possible roles in xenobiotic and endogenous fatty acid metabolism. Toxicol. Sci. 78: 187–195. [DOI] [PubMed] [Google Scholar]
- 9.Decker M., Arand M., and Cronin A.. 2009. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch. Toxicol. 83: 297–318. [DOI] [PubMed] [Google Scholar]
- 10.Lefebvre L., Viville S., Barton S. C., Ishino F., Keverne E. B., and Surani M. A.. 1998. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat. Genet. 20: 163–169. [DOI] [PubMed] [Google Scholar]
- 11.Lord C. C., Thomas G., and Brown J. M.. 2013. Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim. Biophys. Acta. 1831: 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahlhoff M., Frohlich T., Arnold G. J., Muller U., Leonhardt H., Zouboulis C. C., and Schneider M. R.. 2015. Characterization of the sebocyte lipid droplet proteome reveals novel potential regulators of sebaceous lipogenesis. Exp. Cell Res. 332: 146–155. [DOI] [PubMed] [Google Scholar]
- 13.Ladd P. A., Du L., Capdevila J. H., Mernaugh R., and Keeney D. S.. 2003. Epoxyeicosatrienoic acids activate transglutaminases in situ and induce cornification of epidermal keratinocytes. J. Biol. Chem. 278: 35184–35192. [DOI] [PubMed] [Google Scholar]
- 14.Decker M., Adamska M., Cronin A., Di Giallonardo F., Burgener J., Marowsky A., Falck J. R., Morisseau C., Hammock B. D., Gruzdev A., et al. 2012. EH3 (ABHD9): the first member of a new epoxide hydrolase family with high activity for fatty acid epoxides. J. Lipid Res. 53: 2038–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toulza E., Mattiuzzo N. R., Galliano M. F., Jonca N., Dossat C., Jacob D., de Daruvar A., Wincker P., Serre G., and Guerrin M.. 2007. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. 8: R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ala U., Piro R. M., Grassi E., Damasco C., Silengo L., Oti M., Provero P., and Di Cunto F.. 2008. Prediction of human disease genes by human-mouse conserved coexpression analysis. PLOS Comput. Biol. 4: e1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas C. P., Boeglin W. E., Garcia-Diaz Y., O’Donnell V. B., and Brash A. R.. 2013. Steric analysis of epoxyalcohol and trihydroxy derivatives of 9-hydroperoxy-linoleic acid from hematin and enzymatic synthesis. Chem. Phys. Lipids. 167–168: 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson T. W., and Strassburger P.. 1976. Sulfinic acid catalyzed isomerization of olefins. J. Org. Chem. 41: 791–793. [Google Scholar]
- 19.Zheng Y., Boeglin W. E., Schneider C., and Brash A. R.. 2008. A 49-kDa mini-lipoxygenase from Anabaena sp. PCC 7120 retains catalytically complete functionality. J. Biol. Chem. 283: 5138–5147. [DOI] [PubMed] [Google Scholar]
- 20.Tallman K. A., Roschek B. Jr., and Porter N. A.. 2004. Factors influencing the autoxidation of fatty acids: effect of olefin geometry of the nonconjugated diene. J. Am. Chem. Soc. 126: 9240–9247. [DOI] [PubMed] [Google Scholar]
- 21.Gao B., Boeglin W. E., Zheng Y., Schneider C., and Brash A. R.. 2009. Evidence for an ionic intermediate in the transformation of fatty acid hydroperoxide by a catalase-related allene oxide synthase from the Cyanobacterium Acaryochloris marina. J. Biol. Chem. 284: 22087–22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weyer U., Knight S., and Possee R. D.. 1990. Analysis of very late gene expression by Autographa californica nuclear polyhedrosis virus and the further development of multiple expression vectors. J. Gen. Virol. 71: 1525–1534. [DOI] [PubMed] [Google Scholar]
- 23.Beetham J. K., Tian T. G., and Hammock B. D.. 1993. cDNA cloning and expression of a soluble epoxide hydrolase from human liver. Arch. Biochem. Biophys. 305: 197–201. [DOI] [PubMed] [Google Scholar]
- 24.Morisseau C., Newman J. W., Wheelock C. E., Hill Iii T., Morin D., Buckpitt A. R., and Hammock B. D.. 2008. Development of metabolically stable inhibitors of mammalian microsomal epoxide hydrolase. Chem. Res. Toxicol. 21: 951–957. [DOI] [PubMed] [Google Scholar]
- 25.Wixtrom R. N., Silva M. H., and Hammock B. D.. 1988. Affinity purification of cytosolic epoxide hydrolase using derivatized epoxy-activated Sepharose gels. Anal. Biochem. 169: 71–80. [DOI] [PubMed] [Google Scholar]
- 26.Akatsuka T., Kobayashi N., Ishikawa T., Saito T., Shindo M., Yamauchi M., Kurokohchi K., Miyazawa H., Duan H., Matsunaga T., et al. 2007. Autoantibody response to microsomal epoxide hydrolase in hepatitis C and A. J. Autoimmun. 28: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morisseau C., and Hammock B. D.. 2001. Measurement of soluble epoxide hydrolase (sEH) activity. Curr. Protoc. Toxicol. Chapter 4: Unit 4.23. [DOI] [PubMed] [Google Scholar]
- 28.Lambers H., Piessens S., Bloem A., Pronk H., and Finkel P.. 2006. Natural skin surface pH is on average below 5, which is beneficial for its resident flora. Int. J. Cosmet. Sci. 28: 359–370. [DOI] [PubMed] [Google Scholar]
- 29.Braun-Falco O., and Korting H. C.. 1986. Normal pH value of human skin. [Article in German] Hautarzt. 37: 126–129. [PubMed] [Google Scholar]
- 30.Jiang H., Zhu A. G., Mamczur M., Morisseau C., Hammock B. D., Falck J. R., and McGiff J. C.. 2008. Hydrolysis of cis- and trans-epoxyeicosatrienoic acids by rat red blood cells. J. Pharmacol. Exp. Ther. 326: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moghaddam M. F., Grant D. F., Cheek J. M., Greene J. F., Williamson K. C., and Hammock B. D.. 1997. Bioactivation of leukotoxins to their toxic diols by epoxide hydrolase. Nat. Med. 3: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinal C. J., Miyata M., Tohkin M., Nagata K., Bend J. R., and Gonzalez F. J.. 2000. Targeted disruption of soluble epoxide hydrolase reveals a role in blood pressure regulation. J. Biol. Chem. 275: 40504–40510. [DOI] [PubMed] [Google Scholar]
- 33.Hoopes S. L., Gruzdev A., Edin M. L., Graves J. P., Bradbury J. A., Flake G. P., Lih F. B., DeGraff L. M., and Zeldin D. C.. 2017. Generation and characterization of epoxide hydrolase 3 (EPHX3)-deficient mice. PLoS One. 12: e0175348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enayetallah A. E., French R. A., Thibodeau M. S., and Grant D. F.. 2004. Distribution of soluble epoxide hydrolase and of cytochrome P450 2C8, 2C9, and 2J2 in human tissues. J. Histochem. Cytochem. 52: 447–454. [DOI] [PubMed] [Google Scholar]
- 35.Corey E. J., Niwa H., and Falck J. R.. 1979. Selective epoxidation of eicosa-cis-5,8,11,14-tetraenoic (arachidonic) acid and eicosa-cis-8,11,14-trienoic acid. J. Am. Chem. Soc. 101: 1586–1587. [Google Scholar]
- 36.VanderNoot V. A., and VanRollins M.. 2002. Capillary electrophoresis of cytochrome P-450 epoxygenase metabolites of arachidonic acid. 1. Resolution of regioisomers. Anal. Chem. 74: 5859–5865. [DOI] [PubMed] [Google Scholar]
- 37.Fitzpatrick F. A., Morton D. R., and Wynalda M. A.. 1982. Albumin stabilizes leukotriene A4. J. Biol. Chem. 257: 4680–4683. [PubMed] [Google Scholar]
- 38.Hamberg M. 1987. Mechanism of corn hydroperoxide isomerase: detection of 12,13(S)-oxido-9(Z),11-octadecadienoic acid. Biochim. Biophys. Acta. 920: 76–84. [Google Scholar]
- 39.Hamberg M. 2000. New cyclopentenone fatty acids formed from linoleic and linolenic acids in potato. Lipids. 35: 353–363. [DOI] [PubMed] [Google Scholar]
- 40.Pace-Asciak C. R. 2015. Pathophysiology of the hepoxilins. Biochim. Biophys. Acta. 1851: 383–396. [DOI] [PubMed] [Google Scholar]
- 41.Chawengsub Y., Gauthier K. M., Nithipatikom K., Hammock B. D., Falck J. R., Narsimhaswamy D., and Campbell W. B.. 2009. Identification of 13-hydroxy-14,15-epoxyeicosatrienoic acid as an acid-stable endothelium-derived hyperpolarizing factor in rabbit arteries. J. Biol. Chem. 284: 31280–31290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krieg P., Rosenberger S., de Juanes S., Latzko S., Hou J., Dick A., Kloz U., van der Hoeven F., Hausser I., Esposito I., et al. 2013. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J. Invest. Dermatol. 133: 172–180. [DOI] [PubMed] [Google Scholar]
- 43.Ohno Y., Nakamichi S., Ohkuni A., Kamiyama N., Naoe A., Tsujimura H., Yokose U., Sugiura K., Ishikawa J., Akiyama M., et al. 2015. Essential role of the cytochrome P450 CYP4F22 in the production of acylceramide, the key lipid for skin permeability barrier formation. Proc. Natl. Acad. Sci. USA. 112: 7707–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirabayashi T., Anjo T., Kaneko A., Senoo Y., Shibata A., Takama H., Yokoyama K., Nishito Y., Ono T., Taya C., et al. 2017. PNPLA1 has a crucial role in skin barrier function by directing acylceramide biosynthesis. Nat. Commun. 8: 14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grond S., Eichmann T. O., Dubrac S., Kolb D., Schmuth M., Fischer J., Crumrine D., Elias P. M., Haemmerle G., Zechner R., et al. 2017. PNPLA1 deficiency in mice and humans leads to a defect in the synthesis of omega-O-acylceramides. J. Invest. Dermatol. 137: 394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grond S., Radner F. P. W., Eichmann T. O., Kolb D., Grabner G. F., Wolinski H., Gruber R., Hofer P., Heier C., Schauer S., et al. 2017. Skin barrier development depends on CGI-58 protein expression during late-stage keratinocyte differentiation. J. Invest. Dermatol. 137: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madison K. C. 2003. Barrier function of the skin: “la raison d’être” of the epidermis. J. Invest. Dermatol. 121: 231–241. [DOI] [PubMed] [Google Scholar]
- 48.Eckl K. M., de Juanes S., Kurtenbach J., Natebus M., Lugassy J., Oji V., Traupe H., Preil M. L., Martinez F., Smolle J., et al. 2009. Molecular analysis of 250 patients with autosomal recessive congenital ichthyosis: evidence for mutation hotspots in ALOXE3 and allelic heterogeneity in ALOX12B. J. Invest. Dermatol. 129: 1421–1428. [DOI] [PubMed] [Google Scholar]
- 49.Elias P. M., and Schmuth M.. 2009. Abnormal skin barrier in the etiopathogenesis of atopic dermatitis. Curr. Opin. Allergy Clin. Immunol. 9: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elias P. M., and Wakefield J.. 2014. Mechanisms of abnormal lamellar body secretion and the dysfunctional skin barrier in atopic dermatitis. J. Allergy Clin. Immunol. 134: 781–791.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]