Abstract
Background
KIF18B was identified as a potential oncogene by analysis of The Cancer Genome Atlas database.
Materials and methods
We assessed KIF18B expression and explored its clinical significance in cervical cancer tissues. We have also evaluated the effects of KIF18B on cervical cancer cell proliferation, migration, and invasion both in vitro and in vivo.
Results
Our results show that KIF18B is overexpressed in cervical cancer tissues and is associated with a large primary tumor size, an advanced FIGO stage, and an advanced tumor grade. Knockdown of KIF18B induces cell cycle G1-phase arrest and inhibits the proliferation, migration, and invasion of cervical cancer cells, whereas its overexpression promotes proliferation, migration, and invasion in these cells. Moreover, silencing of KIF18B reduces expression of CyclinD1, β-catenin, C-myc, and p-GSK3β expression.
Conclusion
These data suggest that KIF18B can serve as a novel oncogene that promotes the tumorigenicity of cervical cancer cells by activating Wnt/β-catenin signaling pathway.
Keywords: KIF18B, cervical cancer, CyclinD1, Wnt/β-catenin signaling pathway
Introduction
Cervical cancer is the second-most common malignancy in women worldwide and is often diagnosed at a local and advanced stage.1 Most cases of cervical cancer occur in developing countries, with a trend of younger patients being observed.2–4 Although the 5-year survival rate of early cervical cancer is 85% or higher, the prognosis of locally advanced disease remains unfavorable, with an overall 5-year survival rate of approximately 30%–50%.5 Therefore, it is urgent to identify novel molecular markers of cervical cancer to facilitate a more accurate prediction of clinical outcomes and prescribe effective treatments.
The Wnt/β-catenin pathway plays key roles in embryogenesis, homeostasis, and stem cell regeneration, and pluripotency. In the canonical pathway, Wnt activation disrupts the destruction complex, allowing β-catenin to accumulate and subsequently translocate to the nucleus,6 where it interacts with a transcription factor of the TCF/LEF-1 family; the result is increased expression of oncogenes, such as c-Myc and CyclinD1.7 Regulation of Wnt/β-catenin activity is closely related to the development of various cancers, including lung cancer, breast cancer, liver cancer, colon cancer, and cervical cancer.8–10 Thus, targeting Wnt/β-catenin signaling is a promising strategy for treating cancers, with many drugs currently in clinical trials.
Analysis of The Cancer Genome Atlas (TCGA) database showed that the KIF18B gene is highly expressed in cervical cancer tissue but exhibits low expression in paracancerous tissue. It is also associated positively with a large primary tumor size and an advanced TNM stage. KIF18B is a member of the kinesin family of motor proteins, which is closely related to the pairing and separation of chromosomes in mitosis.11–13 These proteins typically contain two large globular heads that attach to microtubules, a central coiled region, and a light-chain region that connects the protein to the intracellular component to be transferred.14 Kinesins are divided into the following types according to their motor region: the NH3-terminal, middle, and COOH-terminal motor domain types. Vimentin, which is involved in a variety of intracellular processes, such as cell division and proliferation, intracellular transport and brain cell development, and survival,15,16 plays an extremely important role in the formation of the intracellular bipolar spindle. Studies have shown that certain kinesins are closely related to tumorigenesis and development. KIF4 is considered the main tumor trigger, and the absence of this motor protein will lead to aneuploidy in mouse embryonic stem cells15 or tumor formation in nude mice. A previous study showed that KIF4 is associated with a poor prognosis in breast cancer.17 Furthermore, Ishikawa et al18 observed higher KIF2C mRNA expression in 120 cases of colorectal cancer than in adjacent normal tissues. KIF2C/MCAK is also upregulated in proliferative tumors, suggesting that KIF2C is associated with cell hyperproliferation. Shimo et al19 detected 27,648 genes in 81 patients with breast cancer by gene chip analysis and found KIF2C gene expression to be significantly enhanced. Additionally, examination of 10 breast cancer cell lines revealed high KIF2C expression in nine. De et al20 showed that overexpression of KIFC1, KIF1A, KIF5A, and KIFC3 can increase docetaxel chemotherapy resistance in breast cancer cells, and Liao et al21 found that KIF18A is closely related to the overall survival and progression-free survival in hepatic carcinoma patients. Moreover, overexpression of KIF18B indicates a poor prognosis in hepatic carcinoma. To predict whether KIF18B is a cancer driver gene with carcinogenic effects, we in a previous study retrieved only literature on the relationship between KIF18B and cancer, which primarily included bioinformatic analyses.22 However, the function of KIF18B in humans has remained unclear. In the present study, we show that KIF18B is a potential oncogene that promotes cervical cancer cell proliferation and migration in vitro and in vivo. Furthermore, our results indicate that the Wnt/β-catenin pathway may contribute to the carcinogenicity of KIF18B overexpression.
Materials and methods
Cell culture and transfection
Human cervical cancer cell lines C33A, HeLa, Siha, and Caski were obtained from Shanghai Life Science Institute Cell Library (Shanghai, People’s Republic of China). HaCaT cells (an immortalized human papillomavirus [HPV]-negative skin keratinocyte line), which were purchased from Nanjing Kaiji Biotechnology Company, were used as normal controls. C33A, HeLa, and Siha cell lines were cultured in Minimum Essential Medium (MEM) (HyClone; GE Health-care Life Sciences, Logan, UT, USA), and HaCaT and Caski cell lines were cultured in RPMI 1640 media (Kaiji, Nanjing, People’s Republic of China). All media were supplemented with 10% fetal bovine serum (FBS) (HyClone; GE Health-care Life Sciences), penicillin–streptomycin liquid (Thermo Fisher Scientific Inc., Waltham, MA, USA), and 0.25 µg/mL amphotericin B (Ameresco Inc., Framingham, MA, USA), and cells were cultured at 37°C in a humidified atmosphere of 5% CO2. Transfection was performed using Lipofectamine RNAi MAX, a small interfering RNA (siRNA) transfection protocol (Thermo Fisher Scientific). We used nonsense RNAi (nsRNA) as a negative control for KIF18B siRNA. Transfection efficiency was detected by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) and western blotting. We designed three siRNAs using the following sequences: siRNA-1 for KIF18B, 5′-GCUCAUCAACGUCCUCAAUTT-3′ (sense), 5′-AUUGAGGACGUUGAUGAGCTT-3′ (antisense); siR-NA-2 for KIF18B, 5′-GCUACCAGGAGGUGUAUAATT-3′ (sense), 5′-UUAUACACCUCCUGGUAGCTT-3′ (antisense); siRNA-3 for KIF18B, 5′-CCAGUUUCC AUGAAUGCAUTT-3′ (sense), 5′-AUGCAUUCAUGGA AACUGGTT-3′ (antisense). The negative control sequences for the nonsense siRNA are as follows: 5′-UUCUC CGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGAC ACGUUCGGAGAATT-3′ (antisense). The human KIF18B-targeting small hairpin RNA (shRNA) sequence was designed based on siRNA-1, siRNA-2, and nsRNA. We generated recombinant lentiviral particles, and cells were transfected with KIF18B or negative control recombinant lentivirus (shRNA-KIF18B or shRNA-NC, respectively). A Genechem-KIF18B plasmid expressing the full-length human KIF18B protein was purchased from Genechem, and an empty plasmid was used as a negative control. KIF18B cDNA was cloned into the pcDNA3.1 vector (GENEray Biotechnology) to construct overexpression plasmids, and the empty vector was used as a negative control. shRNA to knockdown KIF18B was generated according to the siRNA sequence (GENEray Biotechnology). Transfection efficiency was measured by qRT-PCR and western blotting.
RNA extraction, quantitative real-time reverse transcription polymerase chain reaction
Cervical cancer cells were harvested and collected, and TRI-zol reagent (Thermo Fisher Scientific) was used to extract total RNA. A NanoDrop 2000 (Thermo Fisher Scientific Inc, USA) was used to measure the quantity of RNA. For qRT-PCR, a reverse transcription kit (Takara, cat: RR036A) was employed to reverse transcribe 1,000 ng total RNA to cDNA in a final volume 20 µL. qRT-PCR was performed with SYBR Select Master Mix (Applied Biosystems, Cat: 4472908). The primers used for KIF18B, p21, p27, CyclinD1, CyclinE, and β-actin are listed in Table 1. QuantStudio™ 6 Flex Real-Time PCR System was used to collect qRT-PCR data; the qRT-PCR reaction included an initial denaturation step at 95°C for 10 minutes, followed by 40 cycles at 92°C for 15 seconds, and 60°C for 1 minute. Each sample was carried out in triplicate, and relative expression was calculated and normalized using the 2−ΔΔCt method relative to β-actin.
Table 1.
Primers used in qRT-PCR analysis
| Gene | Primer sequence |
|---|---|
| KIF18B | Forward: GCTGCAAGTAGTGGTACGGG |
| Reverse: CCTCAGGGTTAAACACCAGCA | |
| p21 | Forward: AGACCATGTGGACCTGTCACTGG |
| Reverse: GTTTGGAGTGGTAGAAATCTGTC | |
| p27 | Forward: TGCAACCGATTCTTCTCAA |
| Reverse: CAAGCAGTGATGTATCTGATAAACAAG | |
| CyclinD1 | Forward: GCTGCGAAGTGGAAACCATC |
| Reverse: CCTCCTTCTGCACACATTTGAA | |
| CyclinE | Forward: ACTCAACGTGCAAGCCTCG |
| Reverse: GCTCAAGAAAGTGCTGATCCC | |
| β-actin | Forward: TGACGTGGACATCCGCAAAG |
| Reverse: CTGGAAGGTGGACAGCG |
Abbreviation: qRT-PCR, quantitative real-time reverse transcription polymerase chain reaction.
Western blotting
Cells were harvested and processed in lysis buffer on ice (KeyGEN, Nanjing, People’s Republic of China); a BCA Kit (KeyGEN) was utilized to quantitate protein concentrations. Equal amounts of protein were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes. The membranes were blocked in 2% bovine serum albumin (BSA) in Tris-buffered saline/Tween 20 (TBS-T) for 1 hour and subsequently incubated overnight (4°C) with antibodies against KIF18B (Biorbyt, orb184615, 1:500), p21 (CST, 2947, 1:1,000), p27 (CST, 3688, 1:1,000), CyclinD1 (CST, 2978, 1:1,000), CyclinE1 (CST, 4136, 1:1,000), C-myc (Santa Cruz, sc-764, 1:500), β-catenin (Abcam, ab6302, 1:1,500), GSK3β (CST, 12456, 1:1,000), phospho-GSK3β (CST, 5558,1:1,000), or β-actin (CST, 8H10D10, 1:1,000). After washing with TBS-T, the membrane was incubated with a goat antirabbit or a goat antimouse horseradish peroxidase-conjugated secondary antibody (1:10,000; Abcam) at room temperature for 2 hours. The blots were visualized by enhanced chemiluminescence detection (Thermo Fisher Scientific Inc). All experiments were independently repeated at least three times.
Cell proliferation assays and colony-formation assay
Cell proliferation was measured using Cell Counting Kit-8 (CCK-8) (KeyGEN). Cells were plated in 96-well plates at a density of 2,000 cells in 100 µL per well, and absorbance was measured at 450 nm using an ELx-800 universal microplate reader. Each experiment was repeated independently in quadruplicate. For colony-formation assays, a total of 100 transfected cells in a fresh six-well plates were maintained in serum-free medium containing 10% FBS; the medium was replaced every 3 or 4 days. After 2 weeks, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The visible colonies were counted; each experiment was repeated three times.
Wound-healing assay
Cell migratory ability was detected by using a wound-healing assay. Cells were seeded in 6-well plates and cultured in complete medium until near confluence (80%–90%). The culture medium was discarded, and a sterile, 100-µL pipette tip was used to generate a wound. The cells were washed two times and incubated in serum-free medium for 24 hours, after which images were captured.
Cell migration and invasion assays
Transwell chambers were used to assess the migration and invasion properties of tumor cells. HeLa and Siha cells (5×104 cells) in 100 µL of serum-free DMEM were seeded onto cell inserts and placed in wells filled with 600 µL of DMEM supplemented with 10% FBS. After 24 hours of incubation at 37°C and 5% CO2, the nonmigrating cells present on the upper surface of the filter were removed. The membranes were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet. The number of migrating cells from four random fields was calculated under a microscope. Each sample was assayed in triplicate. A similar system with matrigel-coated membranes was used for assessing invasion.
Flow cytometry analysis
Flow cytometry was used to detect the cell cycle distribution and apoptosis. For the former, cells were harvested and fixed with 70% ethanol at 20°C for 24 hours and subsequently stained with 50 µg/mL propidium iodide at 37°C in the dark for 10 minutes. The samples were analyzed using a FACSCalibur flow cytometer. The percentage of cells in G0–G1, S, and G2–M phases was counted and compared. For apoptosis analysis, cells were washed and resuspended, and the Annexin V-FITC cell apoptosis detection kit was used according to the manufacturer’s instructions (BD Biosciences). After incubation at room temperature in the dark for 20 minutes, the cells were immediately analyzed by flow cytometry using a FACScan. All samples were detected in triplicate.
In vivo experiments
A total of eight nude mice (ages 4–6 weeks) for the present study were purchased from an accredited animal facility at the Nanjing Medical University School of Medicine. All animal studies were conducted in accordance with National Institutes of Health animal use guidelines and all animal protocols were approved by Nanjing Medical University Animal Care Committee. Briefly, 1.0×106 exponentially growing HeLa or Siha cells transfected with shRNA-KIF18B or shRNA-NC were subcutaneously injected into the axilla region. Tumor volume was measured every 10 days using calipers. Approximately 6 weeks later, the mice were anesthetized with 1% sodium pentobarbital and intraperitoneally injected with luciferin. The tumor weights were measured, and tumors were collected for immunohistochemistry (IHC).
Tissue microarray (TMA) and IHC
TMA was performed to evaluate the clinical utility of KIF18B as a prognostic marker. Briefly, formalin fixed paraffin-embedded archived tissues of 62 cervical cancer samples were arranged in tissue array blocks (Shanghai BioChip Co Ltd., Shanghai, People’s Republic of China). Each spot was accompanied with case information, including sex, age, pathological grade, and clinical stage. The study was conducted in accordance with the provisions of the Ethics Committee of Nanjing Medical University, and written informed consent was obtained from each participant involved. This study was approved by the Ethics Boards of the Cancer Institute of Jiangsu Province. KIF18B staining was scored independently by two observers (including a pathologist) according to the intensity and percentage of positive cells. Staining intensity was scored according to 4 grades: 0 (negative), 1 (weak), 2 (moderate), or 3 (strong). The product (percentage of positive cells and respective intensity scores) was used as the final staining score (a minimum value of 0 and a maximum value of 300).
Statistical analysis
SPSS 20.0 was used for statistical analysis. Data are presented as the mean ± SD from at least three independent experiments. The statistical analysis was performed using Student’s t-test or one-way analysis of variance. A P-value <0.05 was considered statistically significant. Data graphs were generated with GraphPad Prism 5.0 software.
Results
KIF18B is overexpressed in cervical cancer tumor tissues and correlates with more aggressive clinical characteristics
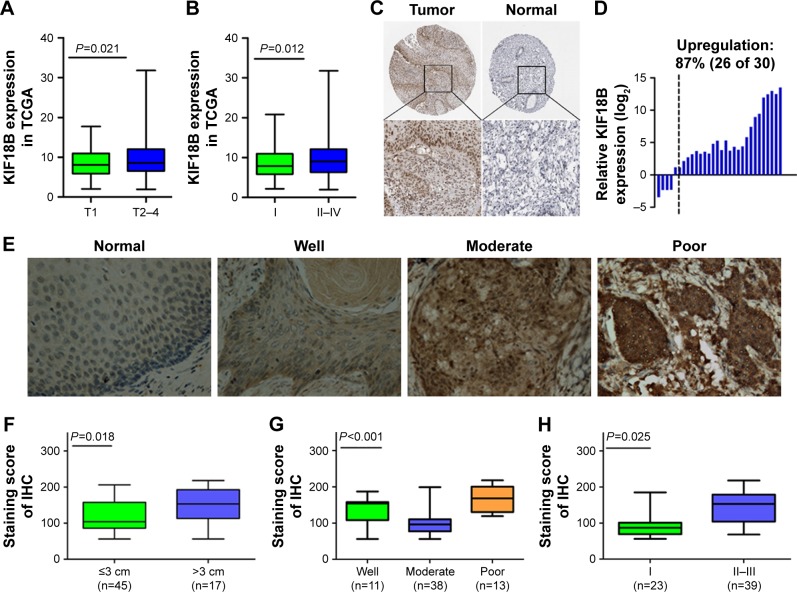
By analyzing the TCGA_CESC_exp_HiSeqV22015-02-24 dataset, cervical cancer tissues showed 3.4-fold hyperexpression of KIF18B compared with normal cervical tissues (P<0.0001), in close association with a large primary tumor size and an advanced FIGO stage (Figure 1A and B). Subsequently, Human Protein Atlas IHC analyses showed that KIF18B was not expressed in normal cervical tissues but was expressed in 11 of 12 (91.7%) cervical cancer tissue samples (Figure 1C). We next detected expression of KIF18B in 30 paired cervical cancer tissue samples (tumor and adjacent normal cervical tissues) using qRT-PCR and found that KIF18B was overexpressed in 87% (26 of 30) of these patients (P<0.001; Figure 1D).
Figure 1.
KIF18B is overexpressed in cervical cancer tumor tissues and correlates with aggressive clinical characteristics.
Notes: (A) KIF18B closely associated with a larger primary tumor size in TCGA database. (B) KIF18B is closely associated with the FIGO stage in TCGA database. (C) Normal cervical tissues do not express KIF18B, but several cervical cancer tissues are positive for KIF18B. (D) KIF18B is overexpressed in 87% (26 of 30) of cervical cancer tissues. (E) Representative images of KIF18B staining in normal cervical tissue and at different histological differentiation levels (from well to poor); magnification ×40. (F–H) KIF18B overexpression is associated with primary tumor size (P=0.018), an advanced tumor grade (P<0.001), and staging (P=0.025) in cervical cancer tissue.
Abbreviations: TCGA, The Cancer Genome Atlas; IHC, immunohistochemistry.
OD-CT-RpUtr03-003 and OD-CT-RpUtr03-006 (purchased from Shanghai Xinchao Biological Company) were used for IHC detection of KIF18B expression in normal cervical tissues and in cervical cancer tissues. As shown in Figure 1E, cervical cancer tissues exhibited hyperexpression of KIF18B compared with normal cervical tissues (Figure 1E), and this overexpression was positively correlated with a large primary tumor size (P=0.018), an advanced tumor grade (P<0.001) and an advanced FIGO stage (P=0.025) (Figure 1F–H). However, no associations between KIF18B expression and age (P=0.934) or lymph node metastasis (P=0.167) were observed (Table 2).
Table 2.
Correlation between KIF18B expression and clinical characteristics in cervical cancer patients (ChIP) (n=62)
| Characteristics | No of patients n=62 | KIF18B-low cases | KIF18B-high cases | P-value |
|---|---|---|---|---|
| Age (years) | ||||
| <50 | 36 | 17 | 19 | 0.934 |
| ≥50 | 26 | 12 | 14 | |
| Differentiation | ||||
| Well (I or I–II) | 11 | 8 | 3 | <0.001 |
| Moderate (II) | 38 | 18 | 20 | |
| Poor (II–III or III) | 13 | 4 | 9 | |
| Diameter of tumor | ||||
| ≤3 cm | 45 | 18 | 27 | 0.018* |
| >3 cm | 17 | 5 | 12 | |
| FIGO stage | ||||
| I | 23 | 15 | 8 | 0.025* |
| II and III | 39 | 14 | 15 | |
| Lymph node metastasis | ||||
| No | 23 | 16 | 7 | 0.167 |
| Yes | 39 | 14 | 25 |
Note:
P<0.05, calculated with Pearson’s χ2 test.
Knockdown of KIF18B inhibits cervical cancer cell proliferation, invasion, and migration in vitro
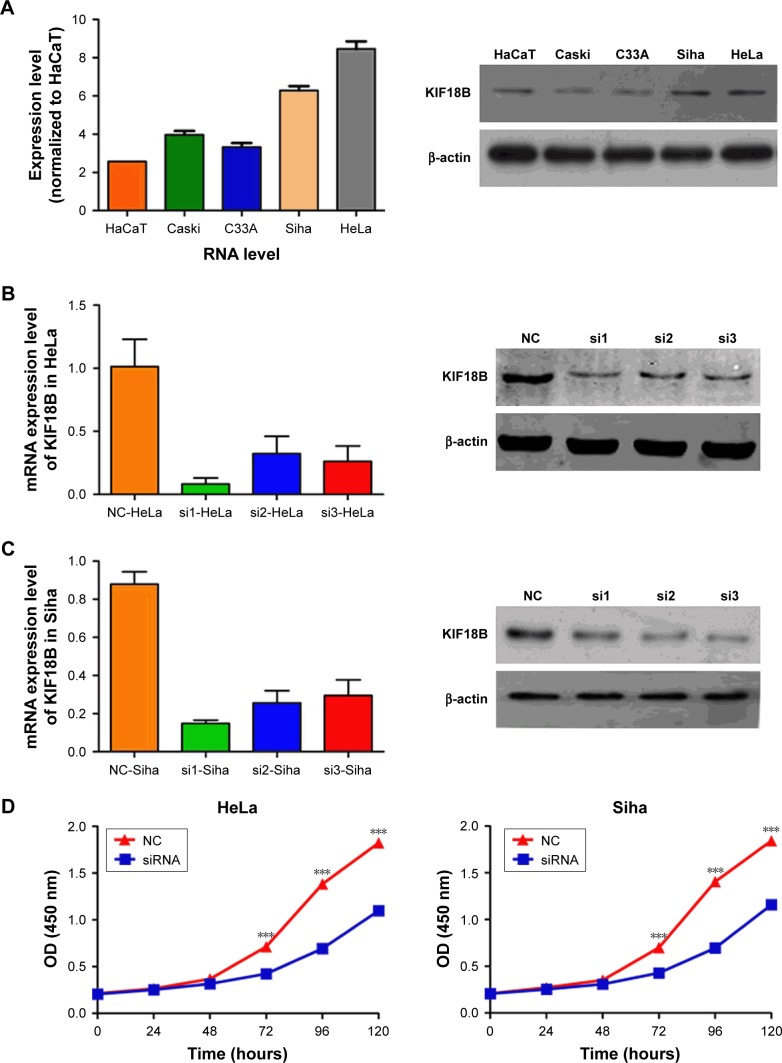
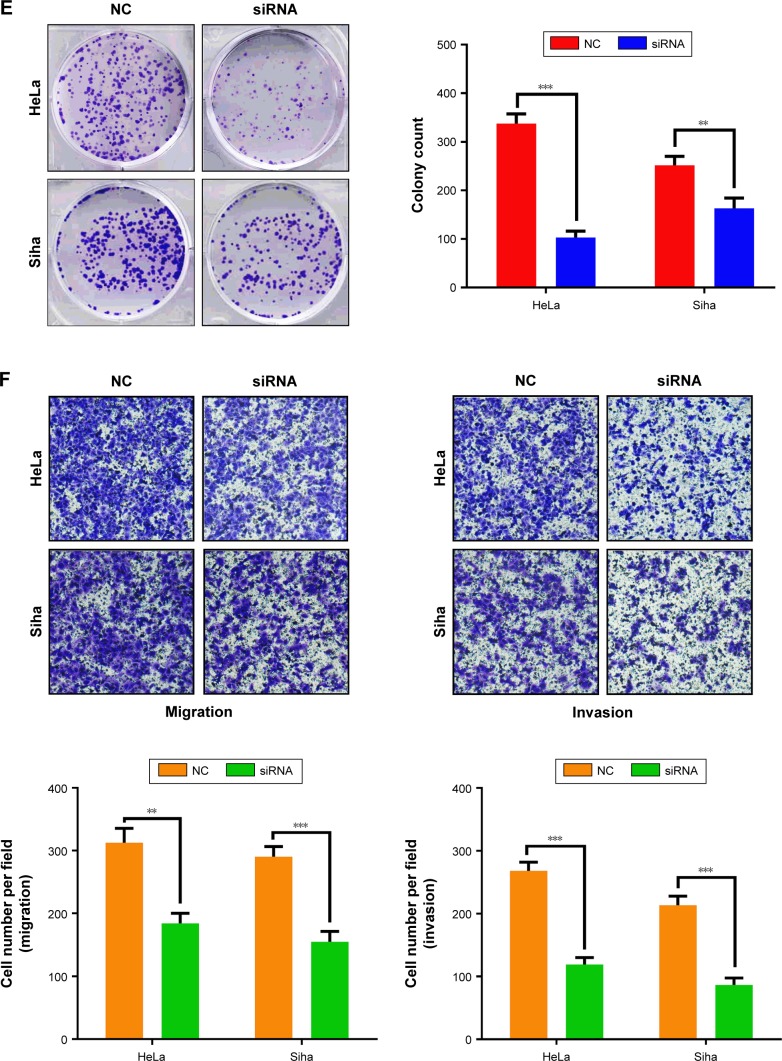
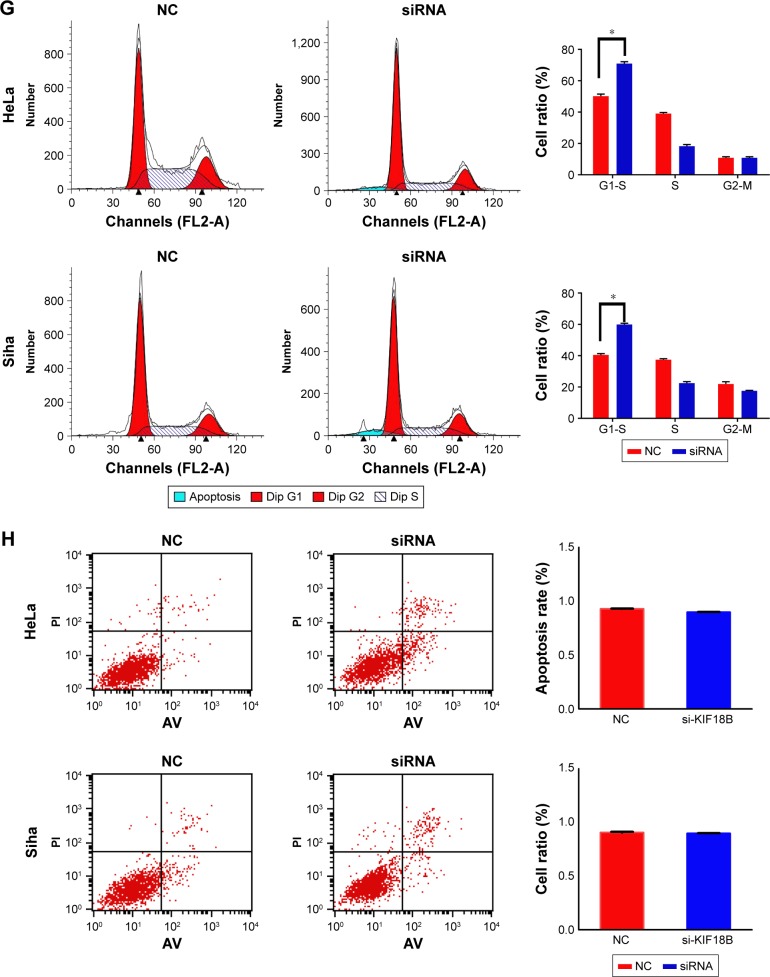
Expression of KIF18B was compared among different cervical cancer cell lines. KIF18B was hyperexpressed in HeLa and Siha cell lines compared with that in cultured human keratinocyte (HaCaT) cells (Figure 2A). To investigate the biological function of KIF18B in vitro, three different sets of siRNAs (siRNA-1, siRNA-2, and siRNA3) were utilized to knock down KIF18B, all of which effectively decreased KIF18B mRNA and protein expression (Figure 2B and C). As shown in Figure 2D, CCK-8 assays revealed that KIF18B knockdown attenuated the proliferation of both HeLa and Siha cells. Moreover, si-KIF18B-transfected cells displayed fewer colonies than did cells transfected with control siRNA (si-NC) (Figure 2E). A Transwell assay also showed that the migration of HeLa and Siha cells was inhibited by siRNA-mediated knockdown of KIF18B (Figure 2F), and a Matrigel invasion assay revealed that si-KIF18B treatment impaired the invasion capacities of HeLa and Siha cells (Figure 2F). Finally, the effect of KIF18B on cell cycle distribution and apoptosis was evaluated by flow cytometry. As shown in Figure 2G, compared with si-NC treatment, si-KIF18B treatment increased the percentage of HeLa and Siha cells in G1 phase. However, no significant difference in apoptosis was found between HeLa and Siha cells treated with si-NC and si-KIF18B (Figure 2H).
Figure 2.
Knockdown of KIF18B inhibits cervical cancer cell proliferation, invasion, and migration in vitro.
Notes: (A) KIF18B mRNA and protein are hyperexpressed in HeLa and Siha cell lines. (B and C) Three specific siRNAs (siRNA-1, siRNA-2, and siRNA-3) were designed and synthesized, with siRNA-1 showing better efficiency. (D) CCK-8 assays showed that knock down of KIF18B inhibited the proliferation of both HeLa and Siha cells. (E) Colony numbers of HeLa and Siha cells transfected with si-KIF18B are fewer than cells transfected with si-NC. (F) Migration and invasion rates of HeLa and Siha cells transfected with si-KIF18B are decreased compared with those in NC cells. Scale bar 100 μm. (G) HeLa and Siha cells transfected with si-KIF18B display more arrest at G1 phase than do cells transfected with si-NC. (H) No significant difference for apoptosis was observed between the si-NC and si-KIF18B groups for both HeLa and Siha cells. *P<0.05, **P<0.01, ***P<0.001. Error bars indicate the SEM.
Abbreviations: siRNA, small interfering RNA; NC, negative control; AV, annexin V.
Overexpression of KIF18B promotes cervical cancer cell proliferation, invasion, and migration in vitro
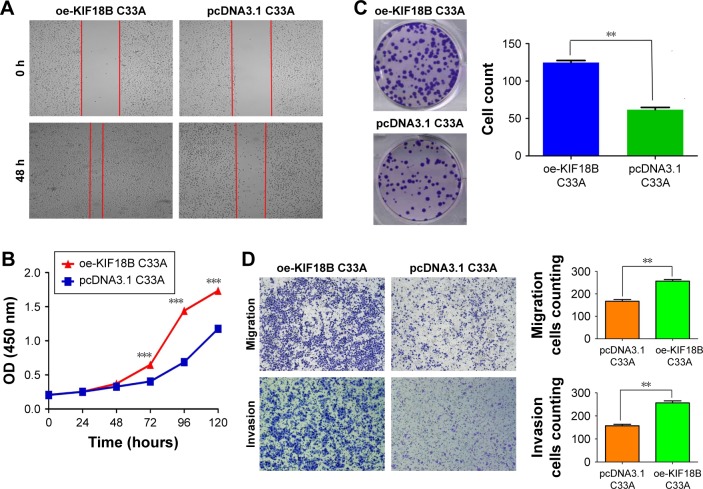
KIF18B expression in the C33A and Caski cell lines was low compared with cultured HaCaT cells (Figure 2A). As shown in Figure 3A, a wound-healing assay showed that the migration of oe-KIF18B C33A cells was increased, and a Transwell assay yielded similar results (Figure 3D). CCK-8 assays revealed that KIF18B overexpression promoted C33A cell proliferation (Figure 3B). In addition, more colonies of oe-KIF18B-transfected cells were observed compared with cells transfected with control pcDNA3.1 (Figure 3C). A Matrigel invasion assay also showed that oe-KIF18B treatment impaired C33A cell invasion (Figure 3D) and overexpression of KIF18B in Caski cell line also can promote cervical cancer cell proliferation, invasion, and migration (Figure S1).
Figure 3.
Overexpression of KIF18B promotes cervical cancer cell proliferation, invasion, and migration in vitro.
Notes: (A) The wound-healing assay showed that the migration of oe-KIF18B C33A cells was increased. (B) CCK-8 assays revealed that overexpression of KIF18B promoted the proliferation of C33A cells. (C) oe-KIF18B-transfected C33A cells had more colonies than did cells transfected with the control, pcDNA3.1. (D) Migration and invasion assays showed that oe-KIF18B treatment impaired the migration and invasion capacities of C33A cells. Scale bar 100 μm. **P<0.01, ***P<0.001. Error bars indicate the SEM.
KIF18B plays a carcinogenic role by activating Wnt/β-catenin signaling
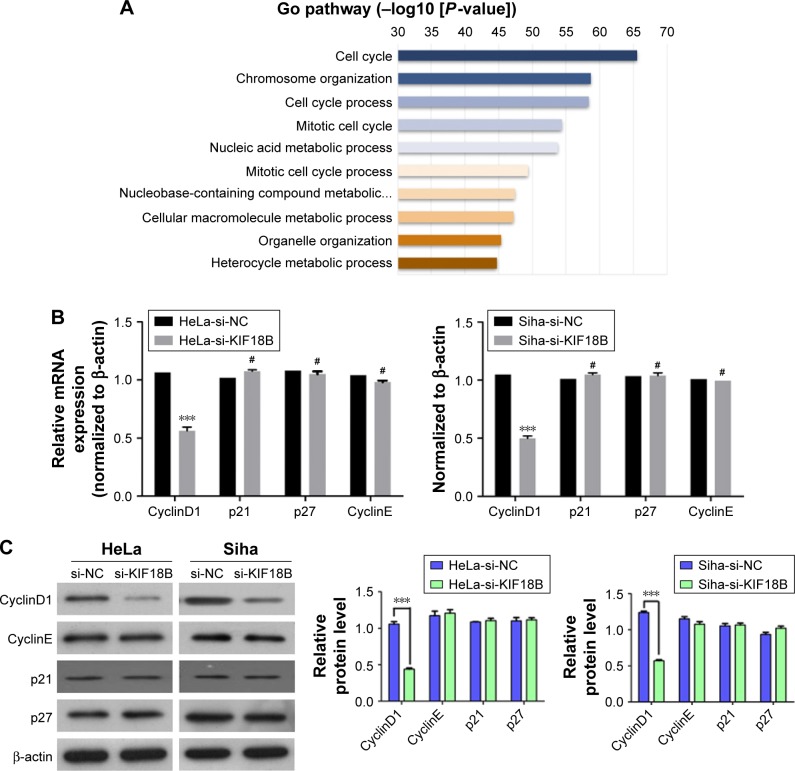
Next, we used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (DAVID Bioinformatics Resources 6.7) to evaluate a list of genes co-expressed with KIF18B that were obtained from cBioPortal data by using both RNA sequencing (RNA-Seq) and microarray results of cervical cancer (TCGA, Provisional). Enrichment of the cell cycle pathway was found for most of the genes co-expressed with KIF18B (Figure 4A). Considering that KIF18B knockdown decreased cancer cell proliferation, migration, and invasion and promoted G1-phase cell cycle arrest, we examined expression of cell cycle-related genes in KIF18B-knockdown cells using qRT-PCR and western blot analyses. Consistent with the enrichment analyses, si-KIF18B treatment decreased both the mRNA and protein levels of CyclinD1 compared with si-NC treatment, whereas expression of p21, p27, and CyclinE was not affected (Figure 4B and C). We then assessed proteins related to the Wnt/β-catenin pathway and found that compared to the si-NC group, the si-KIF18B group showed decreased levels of C-myc, β-catenin, and the phosphorylation of GSK3β, but the total level of GSK3 β has no obvious differences (Figure 4D).
Figure 4.
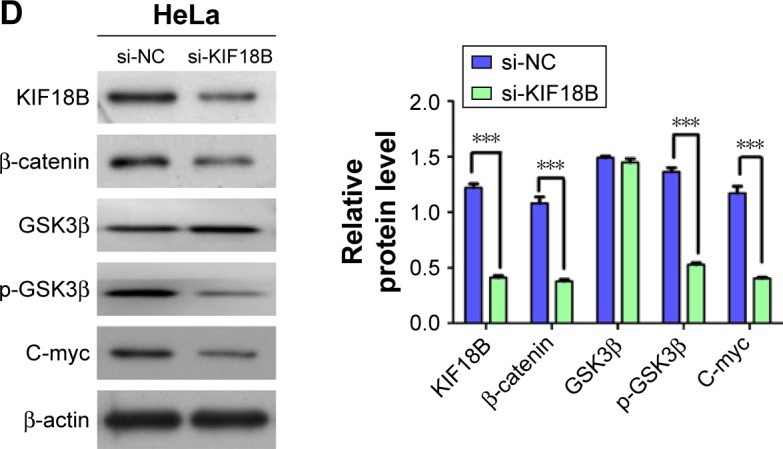
KIF18B plays a carcinogenic role by activating the Wnt/β-catenin signaling pathway.
Notes: (A) Gene Ontology analysis indicates enrichment of cell cycle pathways among genes co-expressed with KIF18B. (B) CyclinD1 mRNA expression was reduced after transfection with si-KIF18B, but the expression of p21, p27, and CyclinE were not altered both in HeLa and Siha cells. (C) CyclinD1 protein expression was decreased after transfection with si-KIF18B, with no difference in the expression of p21, p27, or CyclinE expression both in HeLa and Siha cells. (D) In HeLa cell, compared to si-NC treatment, si-KIF18B treatment resulted in decreased protein levels of C-myc, β-catenin, and p-GSK3β, but the total level of GSK3β has no obvious difference. ***P<0.001, #no significance. Error bars indicate the SEM.
Abbreviation: NC, negative control.
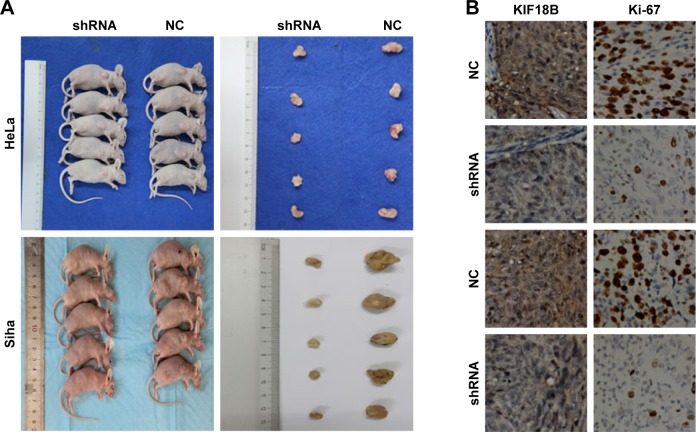
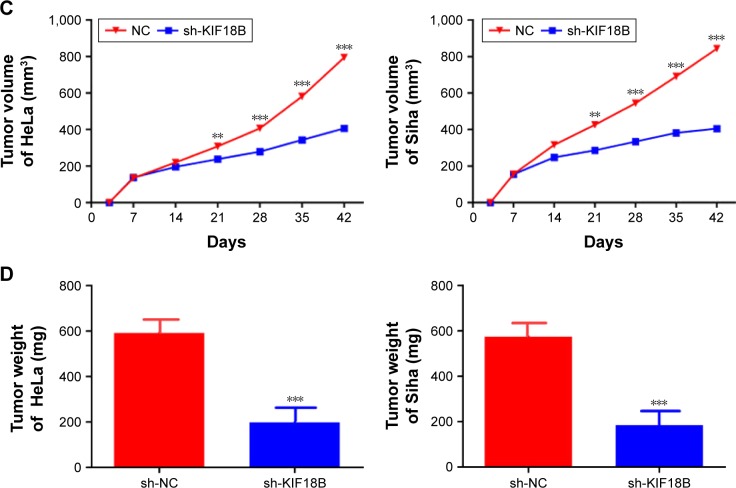
Knockdown of KIF18B suppresses tumor growth in vivo
We have used a nude mouse xenograft model produced using HeLa and Siha cells. When compared with the control group, tumor volumes and tumor weight were smaller in the shKIF18B-treated groups for both cell lines (Figure 5A, C and D). IHC staining revealed weaker Ki67 expression in the sh-NC group than in the sh-KIF18B group (Figure 5B), suggesting that silencing KIF18B inhibits tumor growth in vivo.
Figure 5.
Knockdown of KIF18B suppresses tumor growth in vivo.
Notes: (A) Tumor nodules from mice injected with sh-KIF18B cells are significantly smaller than those from mice injected with NC cells. (B) sh-KIF18B tumors generated from both HeLa and Siha cell lines have less dense Ki-67 staining. Magnification ×40.(C) Compared with the NC group, the sh-KIF18B group showed reduced tumor sizes. (D) Compared with the NC group, the sh-KIF18B group showed reduced tumor weight. **P<0.01, ***P<0.001. Error bars indicate the SEM.
Abbreviations: shRNA, small hairpin RNA; NC, negative control.
Discussion
Kinesins are a class of motor proteins found in all eukaryotic cells, from yeast to humans.23 These motor proteins move along microtubules through hydrolysis of ATP; thus, kinesins are ATPases. Recently, specific kinesin motor proteins have been studied as key proteins that regulate mitotic events and potential targets of therapy.24–26 Lee et al11 initially identified KIF18B as a member of the kinesin family and demonstrated that this protein may play an important role in cell division in a cell cycle-dependent manner. In addition, its homolog KIF18A has been closely associated with the pairing and separation of medullary chromosomes in mitosis.27 Abnormal expression of the KIF18A protein leads to sister chromosome separation abnormalities, subsequently causing cell aneuploidy, and ultimately inducing tumor.28 Several studies have reported that KIF18A is closely related to breast cancer, stomach cancer, and renal cancer.29–31
In addition to previous studies that reported KIF18B upregulation in hepatic carcinoma,22 we provide the first evidence that KIF18B is overexpressed in cervical cancer and is associated with a large tumor size and an advanced TNM stage. KIF18B is also upregulated in several cervical cancer cell lines compared to HaCaT cells. Experiments were performed using CCK8 and colony-formation assays, and we observed that knockdown of KIF18B significantly inhibited cell proliferation and tumorigenesis by arresting cell cycle progression at G1 phase. Transwell and Matrigel assays further demonstrated that KIF18B knockdown decreases cancer cell migration and invasion capacities. We also overexpressed KIF18B in C33A cells, which promoted proliferation and tumorigenesis based on CCK-8 assays and enhanced invasion and migration based on Transwell and Matrigel assays. In vivo tumorigenesis was evaluated by inoculating shRNA-transfected tumor cells into nude mice, and we found that shRNA significantly disrupted the tumorigenic ability of these cells, with lower expression of KIF18B and Ki67 in the shRNA group than in the NC group (P<0.05).
Flow cytometric analysis indicated that si-KIF18B treatment strongly inhibits G1 cell cycle progression, and KEGG analyses produced similar results. We hypothesized that KIF18B promotes the proliferation, migration, and invasion of cervical cancer cells by inducing G1-phase cell cycle progression. We then assessed several G1 phase-related genes to explore the potential mechanism and found that expression of only CyclinD1 was significantly decreased by siRNA-mediated KIF18B knockdown as CyclinD1 is the upstream molecule of β-catenin, and the Wnt/β-catenin pathway play important role in tumor cell migration and invasion. Cell migration and invasion are important aspects of cancer progression. The Wnt/β-catenin pathway plays a key role in cancer cell proliferation, survival, differentiation, and epithelial to mesenchymal transition.32,33 When Wnt ligands bind to transmembrane receptors, Wnt signaling can be initiated,34 and β-catenin aggregates within the nucleus and forms a β-catenin/TCF/LEF transcription complex. There is evidence that many important oncogenes or tumor suppressors regulate the Wnt/β-catenin signaling pathway and thereby regulate the migration and invasion of tumor cells.35,36 In addition, many transcription factor also participate in this regulatory process.37–39 In the present study, we tested the effect of KIF18B knockdown on the Wnt/β-catenin signaling pathway. Compared with the control group, silencing of KIF18B expression decreased the total level of β-catenin, C-myc, and the phosphorylation of GSK3β, but the total level of GSK3β has no obvious differences.
Conclusion
The results of the present study suggest that KIF18B is widely overexpressed in cervical cancer in both cells and tissues and that upregulation of KIF18B correlates with an advanced tumor stage and a large tumor size. KIF18B can promote cervical cancer cell proliferation in vitro and tumor growth in vivo. In addition, silencing of KIF18B suppresses cell cycle progression by inhibiting CyclinD1 expression and decreasing expression of Wnt/β-catenin pathway-related proteins. This finding suggests that KIF18B plays an oncogenic role in cervical cancer and might serve as a novel prognostic biomarker in cervical cancer patients.
Supplementary material
Overexpression of KIF18B promotes Caski cell proliferation, invasion, and migration.
Notes: (A) The wound-healing assay showed that the migration of oe-KIF18B Caski cells was increased. (B) CCK-8 assays revealed that overexpression of KIF18B promoted the proliferation of Caski cells. (C) oe-KIF18B-transfected Caski cells had more colonies than did those transfected with control pcDNA3.1. (D) The migration and invasion assay showed that oe-KIF18B treatment impaired the migration and invasion capacities of Caski cells. Scale bar 100μm. **P<0.01, ***P<0.001. Error bars indicate the SEM.
Abbreviation: CCK-8, Cell Counting kit-8.
Acknowledgments
This work was supported by Six Summit Investigator Grant of Jiangsu Province (2016-WSW-020) and Youth project of Nanjing Medical University Affiliated Cancer Hospital (ZQ201515) for Yaqin Wu and all other authors.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Garland SM, Cuzick J, Domingo EJ, et al. Recommendations for cervical cancer prevention in Asia Pacific. Vaccine. 2008;(Suppl 12):M89–M98. doi: 10.1016/j.vaccine.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Schiffman MH, Brinton LA. The epidemiology of cervical carcinogenesis. Cancer. 1995;76(10 Suppl):1888–1901. doi: 10.1002/1097-0142(19951115)76:10+<1888::aid-cncr2820761305>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 5.Petignat P, Roy M. Diagnosis and management of cervical cancer. BMJ. 2007;335(7623):765–768. doi: 10.1136/bmj.39337.615197.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136(19):3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu N, Kawakami K, Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Dev Biol. 2012;370(1):71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Anastas JN, Moon RT. WNT signaling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Sokol SY. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development. 2011;138(20):4341–4350. doi: 10.1242/dev.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YM, Kim E, Park M, et al. Cell cycle-regulated expression and subcellular localization of a kinesin-8 member human KIF18B. Gene. 2010;466(1–2):16–25. doi: 10.1016/j.gene.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10(1):60–73. doi: 10.1016/s0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- 13.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407(6800):41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 14.Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88(3):1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 15.Hirokawa N, Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Exp Cell Res. 2004;1(1):50–59. doi: 10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Mazumdar M, Lee JH, Sengupta K, Ried T, Rane S, Misteli T. Tumor formation via loss of a molecular motor protein. Curr Biol. 2006;16(15):1559–1564. doi: 10.1016/j.cub.2006.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa K, Kamohara Y, Tanaka F, et al. Mitotic centromere-associated kinesin is a novel marker for prognosis and lymph node metastasis in colorectal cancer. Br J Cancer. 2008;98(11):1824–1829. doi: 10.1038/sj.bjc.6604379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimo A, Tanikawa C, Nishidate T, et al. Involvement of kinesin family member 2C/mitotic centromere-associated kinesin overexpression in mammary carcinogenesis. Cancer Sci. 2008;99(1):62–70. doi: 10.1111/j.1349-7006.2007.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De S, Cipriano R, Jackson MW, Stark GR. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009;69(20):8035–8042. doi: 10.1158/0008-5472.CAN-09-1224. [DOI] [PubMed] [Google Scholar]
- 21.Liao W, Huang G, Liao Y, et al. High KIF18A expression correlates with unfavorable prognosis in primary hepatocellular carcinoma. Oncotarget. 2014;5(21):10271–10279. doi: 10.18632/oncotarget.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itzel T, Scholz P, Maass T, et al. Translating bioinformatics in oncology: guilt-by-profiling analysis and identification of KIF18B and CDCA3 as novel driver genes in carcinogenesis. Bioinformatics. 2015;31(2):216–224. doi: 10.1093/bioinformatics/btu586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miki H, Okada Y, Hirokaw N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15(9):467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Miglarese MR, Carlson RO. Development of new cancer therapeutic agents targeting mitosis. Expert Opin Investig Drugs. 2006;15(11):1411–1425. doi: 10.1517/13543784.15.11.1411. [DOI] [PubMed] [Google Scholar]
- 25.Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28(1–2):197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- 26.Kaestner P, Bastians H. Mitotic drug targets. J Cell Biochem. 2010;111(2):258–265. doi: 10.1002/jcb.22721. [DOI] [PubMed] [Google Scholar]
- 27.Gardner MK, Odde DJ, Bloom K. Kinesin-8 molecular motors: putting the brakes on chromosome oscillations. Trends Cell Biol. 2008;18(7):307–310. doi: 10.1016/j.tcb.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mustaly HM, Ganem NJ. eLS. John Wiley & Sons Ltd; Chichester: [Accessed November 18, 2017]. Mitosis: chromosome segregation and stability. Available from: http://www.els.net. [Google Scholar]
- 29.Kasahara M, Nagahara M, Nakagawa T, et al. Clinicopathological relevance of kinesin family member 18A expression in invasive breast cancer. Oncol Lett. 2016;12(3):1909–1914. doi: 10.3892/ol.2016.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen QI, Cao B, Nan N, et al. Elevated expression of KIF18A enhances cell proliferation and predicts poor survival in human clear cell renal carcinoma. Exp Ther Med. 2016;12(1):377–383. doi: 10.3892/etm.2016.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Yang S, Sun R, Lu M, Wu Y, Li Y. Expression of KIF18A in gastric cancer and its association with prognosis. Zhonghua Wei Chang Wai Ke Za Zhi. 2016;19(5):585–589. Chinese [with English abstract] [PubMed] [Google Scholar]
- 32.Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 33.McCrea PD, Gottardi CJ. Beyond β-catenin: prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016;17:55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(334):re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bao Z, Xu X, Liu Y, et al. CBX7 negatively regulates migration and invasion in glioma via Wnt/beta-catenin pathway inactivation. Oncotarget. 2017;8(24):39048–39063. doi: 10.18632/oncotarget.16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu CC, Cai DL, Sun F, et al. FERMT1 mediates epithelial-mesenchymal transition to promote colon cancer metastasis via modulation of beta-catenin transcriptional activity. Oncogene. 2017;36(13):1779–1792. doi: 10.1038/onc.2016.339. [DOI] [PubMed] [Google Scholar]
- 37.Fan G, Ye D, Zhu S, et al. RTL1 promotes melanoma proliferation by regulating Wnt/β-catenin signalling. Oncotarget. 2017;8(62):106026–106037. doi: 10.18632/oncotarget.22523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ju H, Li Y, Xing X, et al. Manganese-12 acetate suppresses the migration, invasion, and epithelial-mesenchymal transition by inhibitingWnt/β-catenin and PI3K/AKT signaling pathways in breast cancer cells. Thoracic Cancer. 2018 Jan;:8. doi: 10.1111/1759-7714.12584. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Yang L, Pei YY, et al. Overexpressed ACBD3 has prognostic value in human breast cancer and promotes the self-renewal potential of breast cancer cells by activating the Wnt/beta-catenin signaling pathway. Exp Cell Res. 2018;363(1):39–47. doi: 10.1016/j.yexcr.2018.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overexpression of KIF18B promotes Caski cell proliferation, invasion, and migration.
Notes: (A) The wound-healing assay showed that the migration of oe-KIF18B Caski cells was increased. (B) CCK-8 assays revealed that overexpression of KIF18B promoted the proliferation of Caski cells. (C) oe-KIF18B-transfected Caski cells had more colonies than did those transfected with control pcDNA3.1. (D) The migration and invasion assay showed that oe-KIF18B treatment impaired the migration and invasion capacities of Caski cells. Scale bar 100μm. **P<0.01, ***P<0.001. Error bars indicate the SEM.
Abbreviation: CCK-8, Cell Counting kit-8.