Abstract
A general consensus exists that IBD is associated with compositional and metabolic changes in the intestinal microbiota (dysbiosis). However, a direct causal relationship between dysbiosis and IBD has not been definitively established in humans. Findings from animal models have revealed diverse and context-specific roles of the gut microbiota in health and disease, ranging from protective to pro-inflammatory actions. Moreover, evidence from these experimental models suggest that although gut bacteria often drive immune activation, chronic inflammation in turn shapes the gut microbiota and contributes to dysbiosis. The purpose of this Review is to summarize current associations between IBD and dysbiosis, describe the role of the gut microbiota in the context of specific animal models of colitis, and discuss the potential role of microbiota-focused interventions in the treatment of human IBD. Ultimately, more studies will be needed to define host–microbial relationships relevant to human disease and amenable to therapeutic interventions.
IBD, including crohn's disease and ulcerative colitis, affects ∼3.1 million people in the USA and is increasing in incidence worldwide1,2. IBD is characterized by chronic immune-mediated intestinal inflammation that is driven by both genetic predisposition and environmental factors such as diet, antibiotic use and socioeconomic development3.
A key role of the gut microbiota in the pathogenesis of IBD has long been postulated; however, definitive cause–effect mechanistic relationships have been challenging to prove outside of specific animal models. In particular, IBD has been associated with dysbiosis, defined as a decrease in gut microbial diversity owing to a shift in the balance between commensal and potentially pathogenic microorganisms4–7. Indeed, the clinical observation that IBD can respond to antibiotic treatment is consistent with the idea that intestinal bacteria contribute to the inflammatory response8,9. Other observations supporting a role for the gut microbiota in IBD include the predisposition of inflammation for anatomical regions with relative faecal stasis (terminal ileum and rectum), the effectiveness of faecal diversion as a treatment for Crohn's disease10–12, and the rapidly increasing incidence of IBD globally associated with industrialization and accompanying alterations in diet and environmental exposures13,14.
Although these associations are consistent with a role of the gut microbiota in IBD pathogenesis, the precise role of dysbiosis is less clear. Studies attempting to determine whether dysbiosis is truly causative or merely a consequence of inflammation have suffered from a number of limitations, making it difficult to draw definitive conclusions (BOX 1). In this Review, we will describe current associations between IBD and dysbiosis, the role of the gut microbiota in the context of specific animal models, and the potential clinical translation of microbiota-centered therapeutic approaches for human IBD.
Box 1. Limitations of current IBD microbiome research in humans.
Wide clinical spectrum of ulcerative colitis and Crohn's disease cannot be captured in single studies
Many microbial taxa are fastidious and difficult to culture
Microbiome studies have focused on bacteria with relatively little known about other microorganisms, including fungi and viruses, as well as how they interact with each other
Microbiota composition is markedly different between faecal and mucosal samples, yet most analyses of microbiome communities have been based on faecal samples Most studies focus on microbiota composition rather than function
Most studies characterize the gut microbiota using 16S ribosomal RNA tagged sequencing rather than shotgun metagenomics with deep sequencing to provide strain-level taxonomic classifications
Microbiome studies in IBD are confounded by treatment interventions and the effects of inflammation
Most published results are based on cross-sectional and not prospective longitudinal cohort studies
Microbiota composition and IBD
Multiple studies have documented differences in the composition of the gut microbiota between patients with IBD and healthy individuals, particularly with respect to microbial diversity and the relative abundance of specific bacterial taxa. Both expansion of potential pathogens and global changes in composition (that is, increased or decreased abundance of indicator species) have been described. For example, the phylum Firmicutes — specifically Faecailbacterium prausnitzii — is often reduced in proportional abundance in the stool of patients with Crohn's disease7,15–24, although studies focused on mucosal biopsies have questioned this association25,26. Conversely, members of the Proteobacteria phylum, such as Enterobacteriaceae27,28, including Escherichia coli19,29,30, are commonly increased in patients with IBD relative to healthy individuals. Differences in the composition of the gut microbiota have been documented even between members of the same family (including twins) who are discordant for IBD31,32, suggesting that dysbiosis is primarily associated with disease state rather than environmental or genetic factors. Such cross-sectional studies do not provide information about the timing of dysbiosis relative to disease onset and, therefore, should be interpreted with caution particularly with regards to cause–effect relationships. For example, mucosal biopsies from twin pairs discordant for ulcerative colitis have shown a reduction in gut microbiota diversity in both siblings relative to unrelated healthy individuals, but the changes were more pronounced in the affected twin33. However, as the unaffected twins were not followed prospectively, it is not clear whether these findings reflected the gradual development of dysbiosis before the onset of IBD or, on the contrary, a lack of association between dysbiosis and clinical disease.
Studies of paediatric cohorts have attempted to address the temporal relationship between dysbiosis and inflammmation. Although there has been debate over the degree of dysbiosis in paediatric patients, particularly at the mucosal level, most studies have shown substantial differences between diseased and healthy individuals34,35. Thus, dysbiotic changes were described in stool and mucosal biopsies from newly diagnosed, treatment-naive children with Crohn's disease, suggesting that dysbiosis might precede clinical disease and develop independently of long-standing inflammation and/or medical therapy36. A prospective study of paediatric patients with Crohn's disease similarly concluded that dysbiosis reflected the presence and severity of inflammation; however, this study also found an independent association between dysbiosis and other factors such as diet and the use of antibiotics37. Thus, while changes in the gut microbiota might occur early in IBD and perhaps contribute to the onset of disease, over time environmental factors, including inflammation itself, probably further contribute to dysbiosis by altering the metabolic conditions in the gut (discussed later).
Although global changes in the microbiota of patients with IBD have been documented, strong evidence for the existence of specific pathobionts — commensal microorganisms that, under specific environmental or genetic influences, can cause IBD38 — is limited. Enterobacteriaceae, and in particular certain strains of adherent–invasive E. coli (AIEC), have been associated with the ileal mucosa of patients with Crohn's disease39 and have been proposed as potential pathobionts based on their ability replicate in epithelial cells in vitro40. Mycobacterium avium subsp. paratuberculosis has also been investigated as a potential cause of Crohn's disease owing to its ability to cause chronic granulomatous enteritis in sheep and cattle41–44; however, clinical studies have not borne out this hypothesis45,46. Similarly, a specific association between Fusobacterium nucleatum, ulcerative colitis and the development of colorectal cancer has been proposed based on the isolation of a highly invasive strain from patients with ulcerative colitis, but a clear cause-and-effect relationship has not been proven47,48. Pathobionts probably act in concert with the rest of the gut microbiota to cause disease, rather than as individual infectious agents, a possibility supported by observations in animal models (discussed later).
Although it is tempting to postulate an inciting role of the microbiota in IBD pathogenesis, it should be emphasized that the studies discussed so far describe associations and do not prove causation. In fact, evidence suggests that dysbiosis in IBD might, in large part, reflect the response of a complex microbial community to the environmental stress of intestinal inflammation. In the healthy gastrointestinal tract a radial oxygen gradient exists due to the diffusion of oxygen from the host mucosa into the gut lumen49–51. Accordingly, bacteria adherent to the colonic mucosa have higher oxygen tolerance and catalase expression relative to faeces-associated species51. As inflammation is an oxidative state, it might be expected to promote the outgrowth of aerotolerant taxa such as Proteobacteria and Actinobacteria. Indeed, the mouse pathogen Citrobacter rodentium has been shown to gain a fitness advantage by promoting epithelial aerobic respiration and increasing oxygenation of the mucosal surface52. Alternatively, several lines of evidence have shown that intestinal inflammation induces the production of small molecules that serve as terminal electron acceptors for facultative anaerobes such as Enterobacteriaceae37,53. Thus, metabolic alterations associated with inflammation and/or pathobiont colonization might act as microbial stressors and promote the outgrowth of dysbiotic species.
The nonbacterial microbiota and IBD
The virome and IBD
To date, most studies investigating the link between inflammation and the microbiota have focused on bacteria. However, the microbiome also includes fungi and viruses, and the role of these microorganisms in health and disease is being increasingly appreciated. Shotgun metagenomic analyses of viral particles isolated from faecal samples have shown that the gut virome is composed predominantly of bacteriophages54–56. Changes in bacteriophage composition associated with IBD have been described, most notably an increase in Caudovirales bacteriophage sequences in ileal biopsy samples and intestinal washes from paediatric patients with Crohn's disease57,58. Importantly, expansion of Caudovirales bacteriophages was associated with a reduction in bacterial diversity. Through their diverse effects on bacteria — ranging from cell lysis to the transfer of genetic material encoding toxins or antibiotic resistance — phages can confer differential fitness on their hosts and influence the microbial composition of the gut (FIG. 1). Whether bacteriophages have a direct role in IBD pathogenesis, or merely reflect underlying dysbiosis remains to be determined. Similarly, a clear role for eukaryotic viruses in IBD has not been established59.
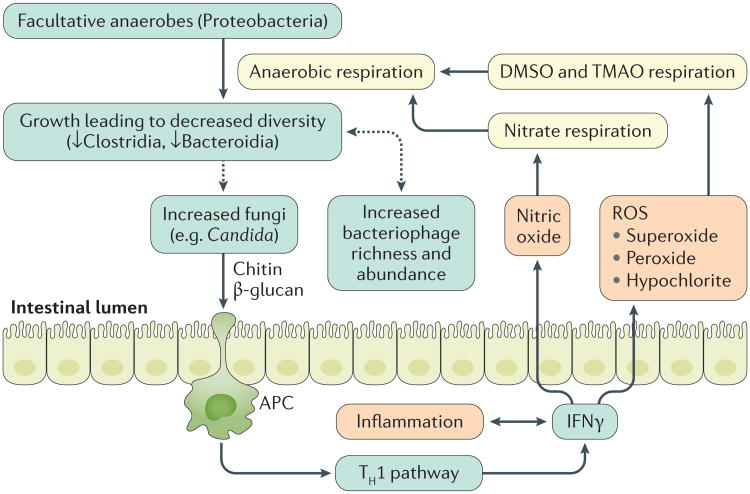
Figure 1. Colonic inflammation in IBD and link to the gut microbiota.
Colonic inflammation stimulates IFNγ production, which generates reactive oxygen species (ROS) by phagocytic innate immune cells. These radicals eventually form products for anaerobic respiration. Facultative anaerobes utilize these products to outgrow, causing decreases in bacterial diversity. The dysbiotic microbiota might further encourage the outgrowth of fungi, especially Candida, which can in turn exacerbate inflammation via chitin and β -glucan antigen-presenting cell (APC) activation of the type 1 T helper (TH1) pathway. Similarly, the dysbiotic microbiota are associated with increased bacteriophage richness and abundance, which can in turn modify the bacterial microbiota via gene transfer. DMSO, dimethyl sulfoxide; TMAO, trimethylamine N-oxide.
The mycobiome and IBD
Antibodies directed against cell wall components of Saccharomyces cerevisiae have been associated with Crohn's disease since 1988 (REF. 60), but the clinical relevance of this finding remains unclear. Owing to technical challenges such as the misattribution of sequences and incorrect annotation of fungi in current genomic databases, relatively few studies have examined the role of fungi in IBD61,62. Fungal sequences can now be identified via culture- independent methods by PCR amplification of the small 18S ribosomal subunit or the internal transcribed spacer region62,63, allowing for more comprehensive assessment of the mycobiome in health and disease.
A study of the stool-associated mycobiome showed an overall increased representation of fungi in paediatric patients with Crohn's disease compared with healthy individuals, however the same five taxa accounted for the majority of fungi in both groups37. Moreover, success ful therapy was associated with partial resolution of bacterial dysbiosis but did not lead to reduction in fungal colonization, putting into question the causative role of fungi in inflammation. Indeed, antibiotic treatment, which is commonly used in Crohn's disease, was independently associated with expansion of the same fungal taxa37. On the other hand, a study of adults with IBD did show differences in the relative abundance of specific fungi between patients with IBD and healthy controls64. Specifically, fungal dysbiosis in IBD was associated with an increased Basidiomycota:Ascomycota ratio, a decreased proportion of Saccharomyces cerevisiae, and an increased abundance of Candida albicans. This study further suggested that the inflammatory environment of Crohn's disease favours the expansion of fungi over bacteria (FIG. 1). Such a conclusion, however, should be interpreted with caution given the multiple confounders inherent in human research (BOX 1).
Studies of the mucosa-associated fungal composition have yielded similarly varied results. One study found that overall fungal diversity was higher in patients with IBD relative to healthy individuals, with several species detected only in Crohn's disease or ulcerative colitis samples, although no consistent pattern could be discerned65. This study also showed substantial differences in the fungal composition of stool and mucosal samples. Another study of paediatric patients failed to show statistically significant changes in the mycobiome of patients with Crohn's disease relative to healthy individuals, except for enrichment of the genus Malassezia which varied by geographical location35. The relative abundance of Basidiomycota and Ascomycota has also been examined in mucosal samples. In a study of paediatric patients, the Basidiomycota:Ascomycota ratio was found to be increased in Crohn's disease compared with healthy individuals, similar to the findings above for stool66. On the other hand, in another study of adults with Crohn's disease, no discernible pattern with regards to these two phyla was seen67.
Mechanistically, the involvement of fungi in IBD pathogenesis is plausible as a number of IBD susceptibility genes in mice and humans are involved in antifungal immune responses (for example, CARD9, CLEC7A and RELA)68. Mice lacking dectin-1 (encoded by Clec7a), the innate immune receptor that recognizes β-glucans in the fungal cell wall, have increased susceptibility to dextran sodium sulfate (DSS) colitis due to the expansion of opportunistic pathogenic fungi69. Similarly, Card9-/- mice have altered bacterial and fungal microbiota that cannot metabolize tryptophan into ligands for the aryl hydrocarbon receptor and, therefore, fail to upregulate IL-22, which is necessary for recovery from colitis70. More recently, S. cerevisiae colonization was shown to promote purine metabolism in mice, leading to elevated levels of uric acid, which has direct pro-inflammatory proper-ties71. Conversely, a protective role for the normal gut mycobiome (including Malassezia spp. and C. albicans) has also been postulated. For example, prolonged treatment of mice with the antifungal agent fluconazole leads to fungal dysbiosis characterized by the expansion of opportunistic species including Aspergillus amstelodami, Epicoccum nigrum, and Wallemia sebi. Mice enriched for these fungal organisms have worse outcomes of both DSS-associated and T-cell-transfer-mediated colitis with increased numbers of IFNγ and IL-17-secreting CD4+ T cells in the intestine72. Collectively, these animal models suggest that fungi might influence intestinal health and disease by suppressing the outgrowth of potential pathobionts, promoting immunoregulatory pathways, and modulating host metabolism. However, as illustrated by the human studies discussed earlier, establishing a direct causal relation ship between specific fungal species and health and disease remains challenging.
Microbial metabolites and IBD
Changes in the the composition of the gut microbiota lead to metabolite alterations that are likely to have a role in IBD pathogenesis. Through metabolomics, correlations between microbial composition and specific bacterial metabolic pathways can be established and the effects of small molecule (<1,500 Da) products on IBD pathogenesis assessed73. When the gut micro biota of healthy individuals and patients with IBD were compared, 12% of metabolic pathways were markedly different, compared with just 2% of genus-level clades74. Specifically, amino acid biosynthesis and carbohydrate metabolism pathways were reduced in the IBD micro-biome in favour of nutrient uptake, virulence and secretion pathways. Moreover, expression of genes related to oxidative stress, such as glutathione and sulfate transport, were increased74. These findings likely reflect the microbial response to the inflammatory intestinal environment of IBD and suggest that functional, rather than compositional, differences might be more informative when studying dysbiosis.
The microbial production of metabolites might affect the host in other ways that are relevant to IBD pathogenesis. For example, bile acid signalling via the nuclear farnesoid-activated X receptor (FXR, also known as bile acid receptor) has been shown to be protective in DSS and 2,4,6-trinitrobenzenesulfonic acid models of colitis by inhibiting NF-κB signalling75. As bile salt hydrolases (BSH) in intestinal bacteria play a key part in bile acid modification, dysbiosis could have a direct effect on FXR signalling. Indeed, in silico analysis showed that the relative abundance of BSH in the gut microbiota was markedly reduced in patients with IBD compared with healthy individuals, and this reduction was most evident among Firmicutes from patients with Crohn's disease76. Consistent with these findings, levels of secondary bile acids are decreased in patients with IBD, particularly during flares77. Collectively, these data are consistent with a model in which impaired microbial enzymatic activity in IBD leads to altered bile salt metabolism and loss of anti-inflammatory signalling through FXR.
Another bacterial metabolic pathway with relevance to IBD is the production of short-chain fatty acids (SCFAs) through the fermentation of undigestable carbohydrates. SCFAs produced by specific clades of Clostridia spp. have been shown to augment regulatory T (Treg)-cell function in the intestinal mucosa through the activation of G protein-coupled receptors as well as via epigenetic effects leading to inhibition of histone deacetylase78. This process promotes restoration of immune tolerance and reduces inflammation in mouse models of colitis and asthma78,79. Efforts to target these pathways, either by altering the gut microbiota or through novel small molecule drugs are currently underway.
Host mucosal immune system: animal models
As discussed earlier, the gastrointestinal tract is populated by trillions of microorganisms that normally have a commensal or mutualistic relationship with their host. This healthy coexistence is maintained by a variety of immune mechanisms including secretion of mucus, immunoglobulin a (IgA), and antimicrobial peptides that shape the gut microbiota and prevent direct contact with the epithelium80. Conversely, the intestinal microbiota influences immune function in both health and disease. For example, germ-free mice have impaired immune development81 and epithelial repair82, while treatment with oral antibiotics can worsen the outcome of viral infections in mice83. In humans, the use of antibiotics in early childhood has been associated with increased risk of Crohn's disease, suggesting the microbiota might help set the threshold for immune activation versus tolerance84. Alternatively, antibiotics can potentiate the expansion of pathobionts such as Clostridium difficile, which is best managed by restoring intestinal microbial diversity using faecal microbiota transplantation85. Thus, the micro biota suppresses pathogens, promotes immune tolerance, initiates epithelial repair, and ensures the development of balanced immune cell subsets.
The healthy relationship between host and gut micro-biota is tested at times of mucosal injury when changes in immune activation and microbiota composition are likely to take place80. Given its vast surface area and constant exposure to the environment, the intestinal epithelial barrier is susceptible to damage by pathogens, ischaemia and environmental toxins. Numerous animal models of colitis have explored the diverse ways in which such insults lead to chronic inflammation in genetically predisposed animals, revealing complex immune– microbial interactions. Notably, although animal models have greatly advanced our understanding of IBD pathogenesis, they have inherent limitations (BOX 2). Nevertheless, these systems have enforced the notion of ‘dysregulated’ immunity as a key driver of IBD and have established diverse and context-dependent roles for the gut micro-biota in health and disease. In the following sections the issue of causation versus correlation of dysbiosis will be considered in the context of specific IBD animal models.
IL-23 signalling, TH17 cells and type 3 innate lymphoid cells
Genome-wide association studies (GWAS) have established a role for IL-23 signalling in the pathogenesis of several immune-mediated diseases including IBD86. Polymorphisms in genes encoding the β subunit of IL-23 (IL-12p40), its receptor, and downstream signalling molecules (STAT3 and JAK2) have been implicated87, while blockade of IL-12p40 is an effective treatment for Crohn's disease according to a phase III clinical trial88. IL-23 is secreted by intestinal dendritic cells (DCs) and promotes the expansion of type 17 T helper (TH17) cells and type 3 innate lymphoid cells (ILC3s). Development of these immune subsets is driven by the transcriptional factor RORγt and their signature cytokines (IL-17, IL-21 and IL-22) have key roles in mucosal defenses and immunopathology89,90.
IL-23 signalling is central in the adoptive T-cell transfer and T-bet−/−RAG−/− ulcerative colitis (TRUC) models of spontaneous colitis91. In the adoptive T-cell transfer model, naive CD4+ donor T cells induce spontaneous colitis when transferred to SCID (severe combined immunodeficiency) or Rag−/− recipients lacking adaptive immunity92. Inflammation in this model is mediated by TH17 cells and T cells of a mixed phenotype capable of secreting both IL-17 and type 1 cytokines (such as IFNγ)93,94. TRUC mice, on the other hand, lack both adaptive immunity and T-bet-dependent components of innate immunity (including ILC1 and NKp46+ ILC3s). In this setting, IL-23 signalling leads to expansion of NKp46− ILC3 that mediate inflammation via IL-17 and IL-22 secretion91.
A key feature of the adoptive transfer and TRUC models is their dependence on the microbiota, as germ-free animals do not develop colitis. In the adoptive T-cell transfer model, several specific micro organisms have been implicated, including Helicobacter muridarum, Helicobacter hepaticus and segmented filamentous bacteria (SFB); however, in most cases, colonization with a defined cocktail of specific pathogen- free (SPF) bacteria is also required for colitis to develop95–97. Conversely, co-colonization with the human symbiont Bacteroides fragilis reverses the inflammatory effects of Helicobacter hepaticus by promoting Treg-cell develop-ment98,99. Similarly, Klebsiella pneumoniae and Proteus mirabilis cause colitis in TRUC mice, but only in the presence of the endogen ous microbiota100. Thus, the inflammatory potential of individual pathobionts is ultimately determined by the overall composition of the gut microbiota. Moreover, TRUC mice can transmit colitis even to wild-type animals, suggesting that the inflammatory environment can condition the gut microbiota to cause disease even in the absence of host risk factors101.
Regulatory T cells and immune tolerance
Treg cells are a distinct subset of helper T cells defined by the transcription factor FOXP3 and anti-inflammatory cytokine IL-10. As with TH17 cells, Treg cells are induced by transforming growth factor (TGF) β, accumulate at mucosal surfaces, and differentiate under the influence of commensal organisms and environmental signals81,102,103. In humans, mutations in FOXP3 lead to chronic enteritis104, whereas blockade of SMAD7, an inhibitor of TGFβ, improves inflammation by augmenting Treg-cell function105,106. Similarly, IL10 polymorphisms have been associated with human IBD in GWAS, and congenital IL-10 deficiency leads to severe childhood colitis107,108. Not surprisingly, spontaneous colitis in Il10−/− mice depends on the gut microbiota and is driven by unopposed TH17 cells109,110. Conversely, specific microbial species — including Bacteroides fragilis98,99 and a consortium of human-derived Clostridia strains111 — can alleviate inflammation in several colitis models by promoting Treg-cell development (FIG. 2). Thus, the relative abundance of TH17 and Treg cells is a key factor in determining intestinal inflammation versus tolerance. By differentially promoting the development of these immune subsets, the gut microbiota have an important role in maintaining or disrupting intestinal homeostasis.
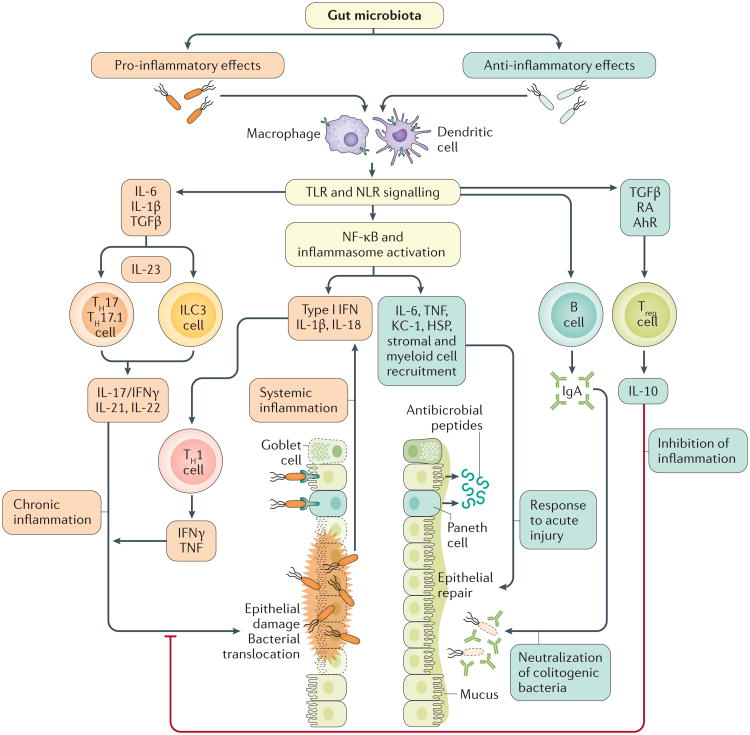
Figure 2. Pro-inflammatory and anti-inflammatory effects of the gut gut microbiota.
Pathogenic microorganisms are sensed via Toll-like receptors (TLR) and NOD-like receptors (NLR) on innate immune cells (dendritic cells (DCs), macrophages), Paneth cells and epithelial cells. This process leads to differentiation of type 17 T helper (TH17) cells and type 3 innate lymphoid cells (ILC3s) under the influence of transforming growth factor (TGF)β, IL-6 and IL-1β, and activation of the IL-23 inflammatory pathway. TLR and NLR signalling also leads to NF-κB and inflammasome activation, secretion of pro-inflammatory cytokines, and type 1 T helper (TH1) cell activation. Ultimately, these inflammatory responses lead to epithelial damage, loss of mucus-secreting goblet cells, and bacterial translocation, which further stimulates the inflammatory response. Anti-inflammatory bacteria are also sensed via TLR and NLR; however, this process leads to regulatory T (Treg)-cell differentiation via TGFβ, retinoic acid (RA) and the aryl hydrocarbon receptor (AhR) signalling. Treg cells exert their immunoregulatory function via IL-10 secretion. Moreover, NF-κB and inflammasome activation lead to secretion of anti-apoptotic factors and antimicrobial peptides, alongside recruitment of stromal and myeloid cells necessary for epithelial repair. Finally, IgA (produced by B cells) prevents colitogenic bacteria from penetrating the mucus layer.
Microbial sensing, the inflammasome and epithelial repair
The complex relationship between the gut microbiota and intestinal inflammation is further illustrated by innate models of colitis. Innate immune cells detect pathogenic and commensal bacteria via a conserved set of pattern-recognition receptors that are linked to NF-κB activation, inflammasome assembly, and epithelial repair pathways (FIG. 2). A number of these receptors — most notably NOD2, NLRP3, and several Toll-like receptors (TLRs) — have been implicated in the pathogenesis of IBD by GWAS112–118. Animal models involving NOD-like receptor (NLR) or TLR signalling defects have revealed a complex and context-dependent role of innate immunity in colitis, ranging from protective to pro-inflammatory.
MYD88 is a central adaptor for signalling through most TLRs, as well as the IL-1 and IL-18 receptors, and disruption of this protein leads to profound innate immune dysfunction82. Administration of DSS to Myd88−/− mice results in severe and lethal colitis which, in contrast to the models discussed earlier, is not driven by the gut microbiota82. Thus, germ-free Myd88−/− mice are not protected from DSS, and in fact antibiotic treatment of wild-type animals exacerbates disease severity82, consistent with a protective role for the gut microbiota. Indeed, bacterial sensing and signalling through TLRs is critical for epithelial repair by promoting secretion of tissue-protective factors, inhibiting apoptosis, and recruiting stromal and myeloid cells to the colonic crypts119-124. Thus, the normal gut microbiota is essential for restoring homeostasis after acute mucosal injury.
A number of other mouse models have linked TLR and NLR signalling to epithelial repair and homeostasis after DSS injury. For example, absence of NF-κB activation in Nemo (also known as Ikbkg)−/− or Ikk1/2 (also known as Chuk–Ikbk)−/− mice results in spontaneous colitis characterized by increased epithelial apoptosis125. Similarly, inflammasome disruption via deletion of NLRP3 (linked to IBD in GWAS), NLRP6, PYCARD, caspase 1, or IL-18 leads to impaired epithelial repair and increased cancer susceptibility126–130. However, in most of these mouse models, defects in innate immunity also lead to the outgrowth and translocation of pathogens that contribute to the overall disease phenotype and respond to antibiotic treatment125,128. For example, DSS colitis in NLRP6-deficient mice is complicated by the outgrowth of Prevotellaceae species, which can exacerbate disease when transferred to wild-type animals130. Similarly, Myd88−/− mice develop severe colitis and bacteraemia when colonized with the pathogen Citrobacter rodentium131,132 (FIG. 2). Thus, sensing of the gut microbiota by innate immune cells maintains intestinal homeostasis by both limiting bacterial growth and promoting epithelial repair.
Paneth cells, autophagy and Crohn's ileitis
Autophagy is a conserved process of targeted degradation of cytoplasmic pathogens and cellular components via the formation of a double-membrane vesicle133. This pathway has been implicated in the pathogenesis of Crohn's disease by GWAS, which have identified susceptibility loci in the ATG16L1 and IRGM autophagy genes134,135. ATG16L1 has been linked to NOD2 activation136,137 and plays a key part in the function of Paneth cells, specialized epithelial cells located in the intestinal crypt base138. Paneth cells shape the gut microbiota via secretion of antimicrobial peptides139; conversely, dysfunction of these cells has been associated with Crohn's disease140. Cadwell et al.141 have shown that mice hypomorphic for ATG16L1 develop Paneth cell defects upon infection with a chronic strain of murine norovirus (MNV). Subsequent administration of DSS to such MNV-infected mice triggers Crohn's disease-like colonic lesions in a microbiota-dependent manner. This model provides an elegant example of the convergence of genetic, environmental and microbial factors to produce the clinical phenotype of Crohn's disease.
Paneth cell dysfunction and dysbiosis have also been implicated in the TNFΔARE model of Crohn's disease ileitis142. TNFΔARE mice have dysregulated TNF expression due to deletions in the AU-rich elements (ARE) of Tnf, leading to chronic ileal inflammation143. Ileitis in conventionally reared TNFΔARE mice is microbiota-dependent and can be attenuated by antibiotic treatment. Remarkably, TNFΔARE mice reared in an SPF environment, develop discrete gradients of disease activity ranging from normal to severe ileitis. These clinical phenotypes correspond to distinct degrees of dysbiosis and Paneth cell dysfunction in individual mice. Moreover, the gut microbiota from inflamed, but not from healthy, TNFΔARE mice can transmit ileitis and Paneth cell defects to germ-free TNFΔARE recipients, pointing to a direct causal relationship between dysbiosis and inflammation.
IgA-coated taxa in the gut microbiota as possible pathobionts in IBD
IgA is the predominant antibody isotype at mucosal surfaces and exerts protective and immuno-regulatory effects by modulating the composition of the gut microbiota144. IgA coating of intestinal bacteria can distinguish colitogenic (IgA+) from commensal (IgA-) species, and IgA+ bacteria identified in this manner include known pathobionts (Prevotellaceae, Helicobacter and SFB) capable of mediating DSS-induced colitis when transferred to SPF mice145. Moreover, IgA+ bacteria isolated from patients with IBD, but not from healthy individuals, led to severe DSS-induced colitis when transferred to SPF mice145. A major feature that dis tinguished colitogenic IgA+ from non-colitogenic IgA− bacteria was the ability of the former to penetrate the mucus layer and trigger T-cell-dependent high-affinity IgA production (FIG. 2). Thus, identifying colitogenic bacteria based on IgA coating could eventually lead to a personalized treatment approach for IBD. These findings further suggest that IgA has an anti-inflammatory role by selectively neutralizing colitogenic bacteria. Consistent with this interpretation, IgA deficiency has been associated with a 3.9 and 5.7 prevalence ratio for ulcerative colitis and Crohn's disease, respectively146.
Human IBD: evidence from clinical studies
As discussed in the preceding sections, dysbiosis in patients with IBD has been well-documented. Moreover, the microbiota have a variety of roles in IBD models of intestinal inflammation, ranging from protective to causative. As many such models have been informed by GWAS data and are based on knockouts of human susceptibility genes, one might speculate that bacteria, fungi and viruses have similar roles in human IBD. However, as discussed in BOXES 1, 2, results from animal studies have a number of limitations and must be interpre ted with caution, especially with regards to their relevance to human disease. The question of whether dysbiosis precedes the development of IBD and sets the inflammatory process in motion, or merely reflects the altered immune and metabolic environment of the inflamed mucosa, remains to be answered. Although a number of human participant studies support a causal role of dys biosis in IBD pathogenesis, conclusions are limited by the lack of prospective data.
Box 2. Limitations of current IBD mouse models.
Most mouse models rely on gene knockouts, whereashuman risk alleles seldom lead to complete loss of function
Mouse models usually explore the effect of a single gene, whereas in humans there are often multiple alleles involved
Immune responses differ between mice and humans
Mice do not capture the genetic and environmental diversity of human populations
Mouse experiments fail to account for variables such as medication exposure, smoking and diet that are inherent in human research
IBD preferentially affects intestinal regions with the highest abundance of bacteria, and both faecal diversion and antibiotics can be effective in the management of Crohn's disease10–12. Individuals with active Crohn's disease who undergo ileocolonic resection and placement of a diverting ileostomy often have a normal neoterminal ileum 3–6 months after surgery11,147. Moreover, infusion of the proximal ileum effluent into the excluded distal ileum can lead to disease recurrence, although the precise factor(s) mediating inflammation have not been determined147. Of note, faecal stream diversion is not universally effective and can itself be colitogenic by depriving colonocytes of SCFAs normally produced by resident bacteria148.
The possibility that inflammation in IBD might be driven by the gut microbiota has informed a number of clinical approaches aimed at correcting dysbiosis by dietary or microbial interventions. Examples include the use of probiotics, antibiotics, defined enteral nutritional therapy (ENT), and faecal microbiota transplantation (FMT). Probiotics are mixtures of bacteria or yeasts with perceived beneficial health effects, utilized to restore gut microbial balance149. To date, evidence for the efficacy of probiotics in the treatment of IBD is equivocal149. The most compelling findings come from post-surgical patients with ulcerative colitis who have undergone ileal pouch-anal anastomosis. Such patients are predisposed to inflammation of the ileal pouch (pouchitis) and probiotics have been shown to be effective in preventing this complication following successful antibiotic treatment149,150. However, basic questions regarding the optimal composition of probiotics, timing of administration and durability of the response remain unanswered.
Antibiotics can alleviate inflammation in various animal models of colitis; however, their effectiveness in human IBD has been limited. Two meta-analyses of randomized controlled trials did show a statistically significant benefit of antibiotics relative to placebo in the treatment of both Crohn's disease and ulcerative colitis (relative risk of remission or relapse between 0.62 and 0.85 (REF. 8); odds ratio 1.35 to 2.17 in favour of antibiotics9. Moreover, several studies have shown that broad-spectrum antibiotics improved rates of steroid-free remission in ulcerative colitis151–153. Given the phenotypic heterogeneity of IBD and the diversity of human populations, it is not surprising that inconsistent outcomes of antibiotic treatment have been reported. Indiscriminately targeting the gut microbiota with broad-spectrum antibiotics probably depletes beneficial, as well as pathogenic, microorganisms with potentially unpredictable consequences. As with probiotics, future antibiotic regimens might have to be tailored to individual patients based on their specific gut microbiota composition and genetic makeup.
Diet has been shown to have a major effect on the composition of the gut microbiota by altering its functionality and metabolism at the genomic level154. Epidemiological evidence points to a role of diet in the pathogenesis of IBD, and dietary modification has long been used as therapy for Crohn's disease155. ENT with elemental, semi-elemental, or polymeric formulas can be used as first-line therapy for the induction of remission in Crohn's disease and has been associated with both clinical improvement and mucosal healing156–161. Although the mechanism of action of ENT has not been well defined, hypotheses include reduction in luminal antigens secondary to food exclusion and/or modulation of the gut microbiota and its metabolome. Several studies have shown a change in faecal microbiota composition following ENT therapy37,162,163, as well as functional changes leading to increased levels of anti-inflammatory SCFAs in children with Crohn's disease164. ENT was effective in inducing remission in paediatric patients with Crohn's disease, and was associated with rapid changes in gut microbiota composition37. Another study further showed that ENT was associated with reduced numbers of Treg cells in the lamina propria, reflecting the resolution of intestinal inflammation165. Notably, favourable therapeutic outcomes of ENT that we and others have reported was associated with initial shifts in microbial composition towards even greater dysbiosis relative to healthy individuals37,166. These observations highlight the need to further explore the effects of diet on microbial composition and disease activity, and further point to the complex relationship between dysbiosis and IBD.
FMT has been explored as a strategy to correct dysbiosis. The success of FMT for the treatment of refractory Clostridium difficile infection167 has generated strong interest in using this approach in IBD. However, clinical results so far have been varied, possibly reflecting the higher complexity of IBD compared to C. difficile colitis168–171. Two randomized, placebo-controlled trials for the treatment of ulcerative colitis showed that FMT was markedly more effective than placebo in inducing clinical and endoscopic remission in ulcerative colitis in adults172,173. However, one other recent randomized, placebo-controlled trail failed to show a similar benefit of FMT174. Even fewer studies have explored FMT for the treatment of Crohn's disease and no randomized-controlled clinical trials have yet been completed although several are ongoing175,176. In children, FMT has shown clinical benefit in a small cohort of patients with Crohn's disease177, whereas a pilot study of FMT in adult refractory Crohn's disease resulted in high rates of clinical remission and clinical improvement178. Overall, data regarding FMT are scarce and questions remain regarding the safety and durability of this approach (short-term and long-term), particularly in immunosuppressed patients; the most effective mode of administration; and how to select appropriate donors and recipients. Larger randomized controlled trials are necessary to better define the role of FMT in the treatment of IBD.
Conclusions
In this Review, we have summarized human microbiome and metabolome associations with IBD, provided an overview of animal models with a focus on host–microbial interactions and their effect on mucosal homeostasis, and discussed the potential therapeutic role of the gut micro-biota in IBD management. Our analysis seems to suggest a greater effect of the gut microbiota on disease phenotype and activity in mice than in humans receiving microbial-based therapies. What is the basis for this difference? One interpretation is that the micro biota plays, at most, a limited part in the pathogenesis of human IBD and dysbiosis is simply a marker of disease (BOX 3). Such a conclusion is supported by a longitudinal analysis published in 2017 of faecal microbiota from patients with IBD, which failed to show a correlation between the degree of dysbiosis and Crohn's disease severity24. It should be noted, however, that the authors used faecal calprotectin levels as a marker of disease activity and, therefore, might have underestimated the severity of ileal inflammation. In fact, the same study showed that recent steroid use (which probably reflected disease exacerbation) did correlate with increased microbiome instability. Similarly, a study of paediatric patients with IBD showed a significant correlation between microbota composition and disease severity, with resolution of dysbiosis in patients responding to anit-TNF therapy179. Nevertheless, a clear cause-effect relationship between dysbiosis and human IBD reamins to be definitively established.
Box 3. Outstanding questions for the role of the gut microbiota in IBD.
What are the very early events in IBD pathogenesis? Specifically, does dysbiosis precede the development of inflammation, or does inflammation arise independently of the microbiota and lead to dysbiosis?
Can microbiota testing be used as a reliable marker of disease onset and progression?
What are the antigens in the intestinal lumen that drive T-cell activation in IBD?
What is the best way to administer faecal microbiota transplantation (FMT) with regards to patient selection, donor selection, and mode of administration?
What will be the long-term outcomes of FMT for IBD? Specifically, how durable will the effects be?
How can we refine microbial therapies beyond FMT? Can we re-engineer the microbiota of individual patients based on their specific disease phenotype, genetic makeup, and microbiome?
Can microbial-based therapies be used to prevent, rather than treat, IBD?
What is the role of the virome in IBD?
We believe in a nuanced interpretation of the differences between mouse and human studies. First, responses to the gut microbiota are likely to differ between mammalian species. Second, mice are genetically homogenous, consume a monotonous diet, and inhabit a well-defined environment, in which the micro-biota is shared between co-housed cagemates through coprophagia. The latter observation probably accounts for the substantial ‘cage effects’ observed in microbiome studies180. Third, similarities between mice are amplified further in germ-free or gnotobiotic studies in which even fewer differences are present between individual animals. This uniformity contributes to a high signal-to-noise ratio and enables investigators to observe reproducible outcomes with relatively low numbers of experimental animals. Such tightly controlled experimental conditions have been critical in establishing cause–effect relationships and uncovering novel biological responses and pathways.
By contrast, humans live in a highly variable environment, exhibit genetic diversity, and consume a variable diet. This inherent variability is even more pronounced in IBD in which disease activity, medication use, and numerous environmental factors such as smoking can modulate the individual response to the gut microbiota and their metabolites. Indeed, intersubject variability is often one of the largest sources of variance in human gut microbiome studies, leading to a low signal-to-noise ratio that can obscure meaningful biological outcomes. Thus, one of the lessons learned from microbiota-based interventional studies in humans might be that such approaches show a hint of promise but will require further adjustment to amplify the favourable clinical response above the threshold of normal human variation. To address these issues, there is a need for large prospective longitudinal studies such as the Genetic Environmental Microbial (GEM) Project to define interactions between human genetics, environmental factors, and microbial composition that contribute to the development of IBD181.
It is important to recognize that the relationship between dysbiosis and IBD is probably complex and dynamic, rather than one of simple cause–effect. Thus, the view of dysbiosis as the response of a complex micro-bial community to the environmental stress of inflammation or medication use is not incompatible with the notion that dysbiosis plays a direct role in IBD pathogenesis. For example, dysbiosis might not be the inciting event but might develop later in the course of IBD and contribute to disease progression and chronicity. Studies of next-generation probiotics and larger controlled FMT trials could provide proof-of-concept in support of this notion.
Alternatively, it is possible that the gut microbiota have a critical role in the initiation of disease but that the window for such an effect occurs early in life. Concurrent with industrialization, the incidence of Crohn's disease and ulcerative colitis has risen globally, revealing a number of important associations between early life exposures and IBD13. Thus, birth by caesarean section, childhood exposure to antibiotics, use of infant formula, and residence in a sanitized environment have emerged as risk factors for the development of IBD and other immune-mediated diseases13. Conversely, childhood exposure to house dust in a rural setting is protective against the development of asthma, providing a cogent example of this principal182. Moreover, mouse models in which the gut microbiota are altered in early life phenocopy the associations with diseases observed in humans183,184. Although it might be difficult to demonstrate efficacy of a microbiota-based strategy for disease prevention, such an approach could be enourmously impactful, making it worth the investment in time, energy and resources.
In conclusion, we believe that association studies, animal models and early therapeutic trials collectively point to an important role of the gut microbiota and their metabolites in IBD pathogenesis. Translating these insights into viable therapeutic approaches might be challenging and require an investment in human subject research to definitively demonstrate cause–effect relation ships. Nevertheless, given the growing incidence of IBD worldwide and its association with environmental triggers, research into the gut microbiota with an eye towards therapeutics will be critically important.
Key points.
Alterations in intestinal microbial composition have long been associated with chronic inflammation; however, a definitive cause–effect relationship between dysbiosis and IBD has been difficult to prove, especially in humans
Dysbiosis alters not only the composition of the intestinal microbiota, but also its metabolome, thereby exerting a wide range of effects on the host
While the microbiota plays a key pathogenic role in IBD, chronic inflammation, in turn, promotes dysbiosis by altering the oxidative and metabolic environment of the gut
Animal studies have elucidated key immunological pathways in the pathogenesis of IBD, established both pro-inflammatory and anti-inflammatory roles of the gut microbiota, and shown that the gut microbiota is indispensable for pathogenesis in most colitis models
Microbial-based treatments will likely have a role in the future management of IBD; however, many questions remain regarding the bacterial composition, timing of administration, and patient selection for such therapies
Acknowledgments
J.N. acknowledges support from the National Institute of Diabetes and Digestive and Kidney Diseases (T32DK00706640) and holds the AGA-Takeda Pharmaceuticals Research Scholar Award in Inflammatory Bowel Disease. G.D.W. acknowledges funding from PennCHOP Microbiome Program, NIH grants R01 DK107565, R24 AI 118629, and the Crohn's and Colitis Foundation Microbiome Initiative. L.A. acknowledges support from grant K23DK109136-01. V.T.T. acknowledges support from NIH grant K08-DK097301.
Glossary
- Crohn's disease
A chronic inflammatory bowel disease that can involve the entire gastrointestinal tract and is characterized by areas of transmural inflammation surrounded by normal mucosa
- Ulcerative colitis
A chronic inflammatory bowel disease characterized by diffuse mucosal inflammation limited to the colon
- Dysbiosis
An alteration of the microbiota that is associated with disease
- Faecal diversion
Surgical diversion of the faecal stream by means of a loop ileostomy or colostomy
- Virome
The collection of prokaryotic and eukaryotic viruses that are part of the human microbiota
- Bacteriophages
Viruses that infect and replicate within bacteria
- Bile acids
Steroid acids found in bile that aid in fat emulsification and nutrient digestion
- Short-chain fatty acids
Fatty acids with less than 6 carbons produced by bacterial fermentation of dietary carbohydrates
- Immunoglobulin A
(IgA). The most abundant antibody type, mostly associated with mucosal surfaces
- Ileocolonic resection
Resection of the terminal ileum, caecum and ascending colon, followed by an ileocolonic anastomosis
- Ileostomy
A surgical operation in which a piece of the ileum is diverted to an artificial opening in the abdominal wall
- Probiotics
Living microorganisms which, when administered in adequate amounts, might confer health benefits on the host
- Enteral nutritional therapy
(ENT). Nutritional supplementation via a nasoenteric feeding tube
- Faecal microbiota transplantation
(FMT). The administration of microorganisms derived from the stool of a healthy donor to the gastrointestinal tract of a patient
- Coprophagia
The consumption of faeces
Footnotes
Competing interests statement: Seres Therapeutics has an option agreement with the University of Pennsylvania for some intellectual property, listing G.D.W. as an inventor.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dahlhamer JM, Zammitti EP, Ward BW, Wheaton AG, Croft JB. Prevalence of inflammatory bowel disease among adults aged ≥18 Years — United States, 2015. MMWR Morb Mortal Wkly Rep. 2016;65:1166–1169. doi: 10.15585/mmwr.mm6542a3. [DOI] [PubMed] [Google Scholar]
- 2.Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 4.Sartor RB, Mazmanian SK. Intestinal microbes in inflammatory bowel diseases. Am J Gastroenterol Suppl. 2012;1:15–21. [Google Scholar]
- 5.Mosca A, Leclerc M, Hugot JP. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol. 2016;7:455. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott SJ, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manichanh C, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan KJ, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106:661–673. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 9.Wang SL, Wang ZR, Yang CQ. Meta-analysis of broad-spectrum antibiotic therapy in patients with active inflammatory bowel disease. Exp Ther Med. 2012;4:1051–1056. doi: 10.3892/etm.2012.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper PH, Lee EC, Kettlewell MG, Bennett MK, Jewell DP. Role of the faecal stream in the maintenance of Crohn's colitis. Gut. 1985;26:279–284. doi: 10.1136/gut.26.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutgeerts P, et al. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet. 1991;338:771–774. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 12.Janowitz HD, Croen EC, Sachar DB. The role of the fecal stream in Crohn's disease: an historical and analytic review. Inflamm Bowel Dis. 1998;4:29–39. doi: 10.1097/00054725-199802000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan GG, Ng SC. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology. 2017;152:313–321.e2. doi: 10.1053/j.gastro.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 14.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van de Merwe JP, Schroder AM, Wensinck F, Hazenberg MP. The obligate anaerobic faecal flora of patients with Crohn's disease and their first-degree relatives. Scand J Gastroenterol. 1988;23:1125–1131. doi: 10.3109/00365528809090179. [DOI] [PubMed] [Google Scholar]
- 17.Walker AW, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. Culture-independent phylogenetic analysis to characterize the gut microbiotas of patients with IBD and non-IBD controls showed that a subset of Crohn's disease and ulcerative colitis samples contained abnormal gut microbiota, characterized by depletion of commensal bacteria, notably members of the phyla Firmicutes and Bacteroidetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 21.Prescott NJ, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 22.Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 23.Rehman A, et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut. 2016;65:238–248. doi: 10.1136/gutjnl-2014-308341. [DOI] [PubMed] [Google Scholar]
- 24.Halfvarson J, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen R, et al. Microbiota of de-novo pediatric IBD: increased Faecalibacterium prausnitzii and reduced bacterial diversity in Crohn's but not in ulcerative colitis. Am J Gastroenterol. 2012;107:1913–1922. doi: 10.1038/ajg.2012.335. [DOI] [PubMed] [Google Scholar]
- 26.Assa A, et al. Mucosa-associated ileal microbiota in new-onset pediatric crohn's disease. Inflamm Bowel Dis. 2016;22:1533–1539. doi: 10.1097/MIB.0000000000000776. [DOI] [PubMed] [Google Scholar]
- 27.Seksik P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumgart M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 29.Sartor RB. Therapeutic correction of bacterial dysbiosis discovered by molecular techniques. Proc Natl Acad Sci USA. 2008;105:16413–16414. doi: 10.1073/pnas.0809363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangin I, et al. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS Microbiol Ecol. 2004;50:25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Joossens M, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 32.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854.e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 33.Lepage P, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Shah R, et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microbes. 2016;7:384–396. doi: 10.1080/19490976.2016.1190073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellermayer R, et al. Microbiota separation and C-reactive protein elevation in treatment-naive pediatric granulomatous Crohn disease. J Pediatr Gastroenterol Nutr. 2012;55:243–250. doi: 10.1097/MPG.0b013e3182617c16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. This paper analysed microbiomes from multiple gastrointestinal locations in new-onset Crohn's disease cases, identifying co-occurring and co-excluded Crohn's-disease-associated microorganism and finding that antibiotics amplify the dysbiosis associated with Crohn's disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis JD, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darfeuille-Michaud A, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 40.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun. 1999;67:4499–4509. doi: 10.1128/iai.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiodini RJ, Van Kruiningen HJ, Thayer WR, Merkal RS, Coutu JA. Possible role of mycobacteria in inflammatory bowel disease. I. An unclassified Mycobacterium species isolated from patients with Crohn's disease. Dig Dis Sci. 1984;29:1073–1079. doi: 10.1007/BF01317078. [DOI] [PubMed] [Google Scholar]
- 42.Feller M, et al. Mycobacterium avium subspecies paratuberculosis and Crohn's disease: a systematic review and meta-analysis. Lancet Infect Dis. 2007;7:607–613. doi: 10.1016/S1473-3099(07)70211-6. [DOI] [PubMed] [Google Scholar]
- 43.Wagner J, et al. Mycobacterium avium subspecies paratuberculosis in children with early-onset Crohn's disease: a longitudinal follow-up study. Inflamm Bowel Dis. 2011;17:1825–1826. doi: 10.1002/ibd.21603. [DOI] [PubMed] [Google Scholar]
- 44.Timms VJ, Daskalopoulos G, Mitchell HM, Neilan BA. The association of Mycobacterium avium subsp paratuberculosis with inflammatory bowel disease. PLoS ONE. 2016;11:e0148731. doi: 10.1371/journal.pone.0148731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selby W, et al. Two-year combination antibiotic therapy with clarithromycin, rifabutin, and clofazimine for Crohn's disease. Gastroenterology. 2007;132:2313–2319. doi: 10.1053/j.gastro.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 46.Liverani E, Scaioli E, Cardamone C, Dal Monte P, Belluzzi A. Mycobacterium avium subspecies paratuberculosis in the etiology of Crohn's disease, cause or epiphenomenon? World J Gastroenterol. 2014;20:13060–13070. doi: 10.3748/wjg.v20.i36.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011;79:2597–2607. doi: 10.1128/IAI.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeller G, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glover LE, Lee JS, Colgan SP. Oxygen metabolism and barrier regulation in the intestinal mucosa. J Clin Invest. 2016;126:3680–3688. doi: 10.1172/JCI84429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marteyn B, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albenberg L, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota in humans and mice. Gastroenterology. 2014;147:1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez CA, et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016;353:1249–1253. doi: 10.1126/science.aag3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winter SE, Lopez CA, Baumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breitbart M, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minot S, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner J, et al. Bacteriophages in gut samples from pediatric Crohn's disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm Bowel Dis. 2013;19:1598–1608. doi: 10.1097/MIB.0b013e318292477c. [DOI] [PubMed] [Google Scholar]
- 58.Norman JM, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. This study analysed the enteric virome, which was abnormal in patients with Crohn's disease and ulcerative colitis and had a substantial expansion of Caudovirales bacteriophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magin WS, Van Kruiningen HJ, Colombel JF. Immunohistochemical search for viral and bacterial antigens in Crohn's disease. J Crohns Colitis. 2013;7:161–166. doi: 10.1016/j.crohns.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 60.Main J, et al. Antibody to Saccharomyces cerevisiae (bakers' yeast) in Crohn's disease. BMJ. 1988;297:1105–1106. doi: 10.1136/bmj.297.6656.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nilsson RH, et al. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE. 2006;1:e59. doi: 10.1371/journal.pone.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Findley K, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sokol H, et al. Fungal microbiota dysbiosis in IBD. Gut. 2016;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ott SJ, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 66.Mukhopadhya I, et al. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect. 2015;17:304–310. doi: 10.1016/j.micinf.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liguori G, et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn's disease patients. J Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:656–665. doi: 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 69.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. This study shows that mice lacking dectin-1 exhibited increased susceptibility to chemically induced colitis, which was the result of altered responses to indigenous fungi; in humans, a polymorphism in the gene for dectin-1 (CLEC7A) was strongly linked to a severe form of ulcerative colitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiaro TR, et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017;9:eaaf9044. doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wheeler ML, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. 2016;19:865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wishart DS, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gadaleta RM, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. Nuclear farnesoid X receptor is a bile salt that is implicated in intestinal antibacterial defence and barrier function; this study shows that its activation prevents chemically induced intestinal inflammation in mice and inhibits pro-inflammatory cytokine production. [DOI] [PubMed] [Google Scholar]
- 76.Ogilvie LA, Jones BV. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: a mechanism and marker of disease? Gut. 2012;61:1642–1643. doi: 10.1136/gutjnl-2012-302137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duboc H, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 78.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grigg JB, Sonnenberg GF. Host–microbiota interactions shape local and systemic inflammatory diseases. J Immunol. 2017;198:564–571. doi: 10.4049/jimmunol.1601621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol. 2015;6:639. doi: 10.3389/fimmu.2015.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 83.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol. 2010;105:2687–2692. doi: 10.1038/ajg.2010.398. [DOI] [PubMed] [Google Scholar]
- 85.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 86.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sandborn W, et al. O-001 A multicenter, double-blind, placebo-controlled phase3 study of ustekinumab, a human IL-12/23P40 mAB, in moderate-service Crohn's disease refractory to anti-TNFalpha: UNITI-1 [abstract] Inflamm Bowel Dis. 2016;22(Suppl. 1):S1. [Google Scholar]
- 89.Almeida FF, Belz GT. Innate lymphoid cells:models of plasticity for immune homeostasis and rapid responsiveness in protection. Mucosal Immunol. 2016;9:1103–1112. doi: 10.1038/mi.2016.64. [DOI] [PubMed] [Google Scholar]
- 90.Ignacio A, Morales CI, Camara NO, Almeida RR. Innate sensing of the gut microbiota: modulation of inflammatory and autoimmune diseases. Front Immunol. 2016;7:54. doi: 10.3389/fimmu.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mohamed R, Lord GM. T-Bet as a key regulator of mucosal immunity. Immunology. 2016;147:367–376. doi: 10.1111/imm.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kiesler P, Fuss IJ, Strober W. Experimental models of inflammatory bowel diseases. Cell Mol Gastroenterol Hepatol. 2015;1:154–170. doi: 10.1016/j.jcmgh.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Globig AM, et al. Comprehensive intestinal T helper cell profiling reveals specific accumulation of IFN-gamma+ IL- 17+ coproducing CD4+ T cells in active inflammatory bowel disease. Inflamm Bowel Dis. 2014;20:2321–2329. doi: 10.1097/MIB.0000000000000210. [DOI] [PubMed] [Google Scholar]
- 94.Ramesh R, et al. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J Exp Med. 2014;211:89–104. doi: 10.1084/jem.20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stepankova R, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 96.Jiang HQ, Kushnir N, Thurnheer MC, Bos NA, Cebra JJ. Monoassociation of SCID mice with Helicobacter muridarum, but not four other enterics, provokes IBD upon receipt of T cells. Gastroenterology. 2002;122:1346–1354. doi: 10.1053/gast.2002.32959. [DOI] [PubMed] [Google Scholar]
- 97.Cahill RJ, et al. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 99.Round JL, Mazmanian SK. Inducible Foxp 3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. This study showed that Bacteroides fragilis capsular polysaccharide A (PSA) can alleviate inflammation in several colitis mouse models by promoting development of regulatory T cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. This study shows that T-bet deficiency in the innate immune system results in spontaneous and communicable ulcerative colitis, and T-bet−/−RAG−/− ulcerative colitis (TRUC) mice can transmit colitis even to wild-type animals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 103.Quintana FJ, et al. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 104.Alroqi FJ, Chatila TA. T regulatory cell biology in health and disease. Curr Allergy Asthma Rep. 2016;16:27. doi: 10.1007/s11882-016-0606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fantini MC, et al. Smad7 controls resistance of colitogenic T cells to regulatory T cell-mediated suppression. Gastroenterology. 2009;136:1308–1316.e1–e3. doi: 10.1053/j.gastro.2008.12.053. [DOI] [PubMed] [Google Scholar]
- 106.Monteleone G, et al. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn's disease. N Engl J Med. 2015;372:1104–1113. doi: 10.1056/NEJMoa1407250. [DOI] [PubMed] [Google Scholar]
- 107.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Franke A, et al. Sequence variants in IL10. ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 109.Hoshi N, et al. MyD88 signalling in colonic mononuclear phagocytes drives colitis in IL-10-deficient mice. Nat Commun. 2012;3:1120. doi: 10.1038/ncomms2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rakoff-Nahoum S, Hao L, Medzhitov R. Role of Toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25:319–329. doi: 10.1016/j.immuni.2006.06.010. This study demonstrated a differential role of innate immune recognition by Toll-like receptors (TLRs) in the development of commensal-dependent colitis and a key pathogenic role of TLR signalling for chronic intestinal inflammation. [DOI] [PubMed] [Google Scholar]
- 111.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 112.Asquith M, Powrie F. An innately dangerous balancing act: intestinal homeostasis, inflammation, and colitis-associated cancer. J Exp Med. 2010;207:1573–1577. doi: 10.1084/jem.20101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Jager PL, et al. The role of the Toll receptor pathway in susceptibility to inflammatory bowel disease. Genes Immun. 2007;8:387–397. doi: 10.1038/sj.gene.6364398. [DOI] [PubMed] [Google Scholar]
- 114.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 115.Ogura Y, et al. A frameshit mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 116.Pierek M, et al. Toll-like receptor-1, -2. and -6 polymorphisms influence disease extension in inflammatory bowel diseases. Inflamm Bowel Dis. 2006;12:1–8. doi: 10.1097/01.mib.0000195389.11645.ab. [DOI] [PubMed] [Google Scholar]
- 117.Torok HP, et al. Crohn's disease is associated with Toll-like receptor-9 polymorphism. Gastroenterology. 2004;127:365–366. doi: 10.1053/j.gastro.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 118.Villani AC, et al. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brown SL, et al. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Invest. 2007;117:258–269. doi: 10.1172/JCI29159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim JY, Kajino-Sakamoto R, Omori E, Jobin C, Ninomiya-Tsuji J. Intestinal epithelial-derived TAK1 signaling is essential for cytoprotection against chemical-induced colitis. PLoS ONE. 2009;4:e4561. doi: 10.1371/journal.pone.0004561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Malvin NP, Seno H, Stappenbeck TS. Colonic epithelial response to injury requires Myd88 signaling in myeloid cells. Mucosal Immunol. 2012;5:194–206. doi: 10.1038/mi.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Podolsky DK, Gerken G, Eyking A, Cario E. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology. 2009;137:209–220. doi: 10.1053/j.gastro.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci USA. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zheng L, Riehl TE, Stenson W. Regulation of colonic epithelial repair in mice by Toll-like receptors and hyaluronic acid. Gastroenterology. 2009;137:2041–2051. doi: 10.1053/j.gastro.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nenci A, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 126.Allen IC, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dupaul-Chicoine J, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–378. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 128.Zaki MH, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takagi H, et al. Contrasting action of IL-12 and IL-18 in the development of dextran sodium sulphate colitis in mice. Scand J Gastroenterol. 2003;38:837–844. doi: 10.1080/00365520310004047. [DOI] [PubMed] [Google Scholar]
- 130.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gibson DL, et al. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cell Microbiol. 2008;10:618–631. doi: 10.1111/j.1462-5822.2007.01071.x. [DOI] [PubMed] [Google Scholar]
- 132.Lebeis SL, Bommarius B, Parkos CA, Sherman MA, Kalman D. TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. J Immunol. 2007;179:566–577. doi: 10.4049/jimmunol.179.1.566. [DOI] [PubMed] [Google Scholar]
- 133.Cadwell K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat Rev Immunol. 2016;16:661–675. doi: 10.1038/nri.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 135.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 137.Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 138.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wehkamp J, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. This study demonstrates that an interaction between a specific virus infection (murine norovirus) and a gene mutation induces Crohn's-disease-like intestinal pathologies in mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schaubeck M, et al. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut. 2016;65:225–237. doi: 10.1136/gutjnl-2015-309333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 144.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 145.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. This paper shows that IgA coating of intestinal bacteria can distinguish colitogenic (IgA+) from commensal (IgA−) species, and IgA+ bacteria identified in this manner include known pathobionts (Prevotellaceae, Helicobacter and segmented filamentous bacteria) capable of mediating chemically induced colitis when transferred to specific pathogen-free mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ludvigsson JF, Neovius M, Hammarstrom L. Association between IgA deficiency and other autoimmune conditions: a population-based matched cohort study. J Clin Immunol. 2014;34:444–451. doi: 10.1007/s10875-014-0009-4. [DOI] [PubMed] [Google Scholar]
- 147.D'Haens GR, et al. Early lesions of recurrent Crohn's disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology. 1998;114:262–267. doi: 10.1016/s0016-5085(98)70476-7. [DOI] [PubMed] [Google Scholar]
- 148.Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med. 1989;320:23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- 149.Sanders ME, et al. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Singh S, Stroud AM, Holubar SD, Sandborn WJ, Pardi DS. Treatment and prevention of pouchitis after ileal pouch-anal anastomosis for chronic ulcerative colitis. Cochrane Database Syst Rev. 2015;6:CD001176. doi: 10.1002/14651858.CD001176.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Terao S, et al. Antibiotic combination therapy for steroid withdrawal in steroid-dependent ulcerative colitis. Digestion. 2011;83:198–203. doi: 10.1159/000321811. [DOI] [PubMed] [Google Scholar]
- 152.Ohkusa T, et al. Newly developed antibioticcombination therapy for ulcerative colitis: a double-blind placebo-controlled multicenter trial. Am J Gastroenterol. 2010;105:1820–1829. doi: 10.1038/ajg.2010.84. [DOI] [PubMed] [Google Scholar]
- 153.Uehara T, et al. Efficacy of antibiotic combinationtherapy in patients with active ulcerative colitis, including refractory or steroid-dependent cases. J Gastroenterol Hepatol. 2010;25(Suppl. 1):S62–S66. doi: 10.1111/j.1440-1746.2010.06231.x. [DOI] [PubMed] [Google Scholar]
- 154.Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. 2014;146:1564–1572. doi: 10.1053/j.gastro.2014.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lewis JD, Abreu MT. Diet as a trigger or therapy for inflammatory bowel diseases. Gastroenterology. 2017;152:398–414.e6. doi: 10.1053/j.gastro.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 156.Sandhu BK, et al. Guidelines for the management of inflammatory bowel disease in children in the United Kingdom. J Pediatr Gastroenterol Nutr. 2010;50(Suppl. 1):S1–S13. doi: 10.1097/MPG.0b013e3181c92c53. [DOI] [PubMed] [Google Scholar]
- 157.Caprilli R, et al. European evidence based consensus on the diagnosis and management of Crohn's disease: special situations. Gut. 2006;55(Suppl. 1):i36–i58. doi: 10.1136/gut.2005.081950c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fell JM, et al. Mucosal healing and a fall in mucosal pro-inflammatory cytokine mRNA induced by a specific oral polymeric diet in paediatric Crohn's disease. Aliment Pharmacol Ther. 2000;14:281–289. doi: 10.1046/j.1365-2036.2000.00707.x. [DOI] [PubMed] [Google Scholar]
- 159.Borrelli O, et al. Polymeric diet alone versus corticosteroids in the treatment of active pediatric Crohn's disease: a randomized controlled open-label trial. Clin Gastroenterol Hepatol. 2006;4:744–753. doi: 10.1016/j.cgh.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 160.Afzal NA, et al. Improvement in quality of life of children with acute Crohn's disease does not parallel mucosal healing after treatment with exclusive enteral nutrition. Aliment Pharmacol Ther. 2004;20:167–172. doi: 10.1111/j.1365-2036.2004.02002.x. [DOI] [PubMed] [Google Scholar]
- 161.Froslie KF, Jahnsen J, Moum BA, Vatn MH, Group I. Mucosal healing in inflammatory bowel disease: results from a Norwegian population-based cohort. Gastroenterology. 2007;133:412–422. doi: 10.1053/j.gastro.2007.05.051. [DOI] [PubMed] [Google Scholar]
- 162.Lionetti P, et al. Enteral nutrition and microflora in pediatric Crohn's disease. JPEN J Parenter Enteral Nutr. 2005;29(4 Suppl.):S173–S175. doi: 10.1177/01486071050290S4S173. [DOI] [PubMed] [Google Scholar]
- 163.Leach ST, Mitchell HM, Eng WR, Zhang L, Day AS. Sustained modulation of intestinal bacteria by exclusive enteral nutrition used to treat children with Crohn's disease. Aliment Pharmacol Ther. 2008;28:724–733. doi: 10.1111/j.1365-2036.2008.03796.x. [DOI] [PubMed] [Google Scholar]
- 164.Tjellstrom B, et al. Effect of exclusive enteral nutrition on gut microflora function in children with Crohn's disease. Scand J Gastroenterol. 2012;47:1454–1459. doi: 10.3109/00365521.2012.703234. [DOI] [PubMed] [Google Scholar]
- 165.Schwerd T, et al. Exclusive enteral nutrition in active pediatric Crohn disease: effects on intestinal microbiota and immune regulation. J Allergy Clin Immunol. 2016;138:592–596. doi: 10.1016/j.jaci.2015.12.1331. [DOI] [PubMed] [Google Scholar]
- 166.Gerasimidis K, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn's disease during enteral nutrition. Inflamm Bowel Dis. 2014;20:861–871. doi: 10.1097/MIB.0000000000000023. [DOI] [PubMed] [Google Scholar]
- 167.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 168.Angelberger S, et al. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol. 2013;108:1620–1630. doi: 10.1038/ajg.2013.257. [DOI] [PubMed] [Google Scholar]
- 169.Cui B, et al. Step-up fecal microbiota transplantation strategy: a pilot study for steroid-dependent ulcerative colitis. J Transl Med. 2015;13:298. doi: 10.1186/s12967-015-0646-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kunde S, et al. Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J Pediatr Gastroenterol Nutr. 2013;56:597–601. doi: 10.1097/MPG.0b013e318292fa0d. [DOI] [PubMed] [Google Scholar]
- 171.Suskind DL, Singh N, Nielson H, Wahbeh G. Fecal microbial transplant via nasogastric tube for active pediatric ulcerative colitis. J Pediatr Gastroenterol Nutr. 2015;60:27–29. doi: 10.1097/MPG.0000000000000544. [DOI] [PubMed] [Google Scholar]
- 172.Paramsothy S, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. This randomized, placebo-controlled clinical trial of faecal microbiota transplantation (FMT) for treatment of ulcerative colitis showed that intensive-dosing, multidonor FMT induces clinical remission and endoscopic improvement in active ulcerative colitis and is associated with distinct microbial changes that relate to outcome. [DOI] [PubMed] [Google Scholar]
- 173.Moayyedi P, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149:102–109.e6. doi: 10.1053/j.gastro.2015.04.001. First randomized, placebo-controlled trial of faecal microbiota transplantation in ulcerative colitis showing potential benefit. [DOI] [PubMed] [Google Scholar]
- 174.Rossen NG, et al. Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology. 2015;149:110–118.e4. doi: 10.1053/j.gastro.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 175.US National Library of Medicine. 2017 doi: 10.1080/15360280801989377. ClinicalTrials.gov [online], https://clinicaltrials.gov/ct2/show/NCT03078803. [DOI] [PubMed]
- 176.US National Library of Medicine. 2016 doi: 10.1080/15360280801989377. ClinicalTrials.gov [online], https://clinicaltrials.gov/ct2/show/NCT02097797. [DOI] [PubMed]
- 177.Suskind DL, et al. Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn's disease. Inflamm Bowel Dis. 2015;21:556–563. doi: 10.1097/MIB.0000000000000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cui B, et al. Fecal microbiota transplantation through mid-gut for refractory Crohn's disease: safety, feasibility, and efficacy trial results. J Gastroenterol Hepatol. 2015;30:51–58. doi: 10.1111/jgh.12727. [DOI] [PubMed] [Google Scholar]
- 179.Kolho KL, et al. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol. 2015;110:921–930. doi: 10.1038/ajg.2015.149. [DOI] [PubMed] [Google Scholar]
- 180.Hildebrand F, et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Turpin W, et al. Assoication of host genome with intestinal microbiota composition in a large healthy cohort. Nat Genet. 2016;48:1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 182.Stein MM, et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med. 2016;3751:411–42. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488(62):1–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]