Abstract
Recently, several innovative approaches have been developed that allow one to directly screen or select for improved protein folding in the cellular context. These methods have the potential of not just leading to a better understanding of the in vivo folding process, they may also allow for improved production of proteins of biotechnological interest.
Introduction
Most proteins are only marginally stable, exhibiting net free energies of folding in the range of only a few kcal/mol [1,2••]. This marginal stability is not only a source of experimental frustration to scientists working with proteins but also greatly inhibits the use of proteins for biotechnological purposes. Optimizing protein folding would thus be of great practical value. Moreover, in vivo optimization of folding would give us a better understanding of how proteins normally fold in the cell, possibly granting us insight into how protein misfolding can lead to disease. Improving protein stability either in vivo or in vitro, however, is rather challenging as most amino acid substitutions are destabilizing [1], and those rare stabilizing variants that can be found often interfere with protein function [1,3,4]. Attempts to circumvent this function-stability tradeoff by manipulating cellular chaperones in beneficial ways is also challenging; this is not surprising as millions of years of evolution have been at work optimizing these in vivo folding machines. Most chaperones are designed to work on many proteins [5]. This fact may help explain why chaperone variants specifically selected for improved function with one protein often show impaired function in their folding of other proteins [6]. These issues may help account for the mixed results that have been obtained with protein stabilization efforts.
Although our general understanding of protein folding has been significantly advanced by in vitro experiments, applying the lessons learned in vitro to improve folding in vivo has been difficult. In vivo folding differs substantially from in vitro folding due to the effects of macromolecular crowding, hindered diffusion, co-translational folding, and chaperone-facilitated folding that are at play in the cellular environment. These areas have been the subject of recent excellent reviews and so will not be further discussed here [7–10]. Innovative in-cell reporting systems that allow for the fluorescent detection of in vivo protein denaturation after in-cell urea titration suggest that at least protein thermodynamic stability is not radically different in the in vivo and in vitro environments [11,12••,13]. However, in vivo stability is not just thermodynamic stability; in vivo stability also entails the protein’s persistence in a functional, non-aggregated form in the cell. While in vivo protease susceptibility is roughly correlated with in vitro thermodynamic stability [2••,14], aggregation susceptibility is poorly assessable as it is strongly affected by the in vivo environment. Aggregation is a crucial factor for rationally designed proteins, as proteins often fold into insoluble oligomeric states in vivo due to the effect of unanticipated intermolecular interactions that occur within the cell [15].
Optimizing protein folding in the cell could at least in principle fix many of these problems. The simplest and most generic methods commonly used to improve protein folding in vivo are by optimizing growth and expression conditions, including growth temperature, time of induction, promoter strength, inducer concentration, codon usage, and the use of solubility-enhancing fusion tags. These approaches are well documented and will thus not be discussed further here [16–19]. A more targeted way to improve protein folding is by using directed evolution. These methods typically involve the generation of a pool of mutant variants followed by a selection process to find those with improved folding properties. Through multiple iterative rounds of Darwinian selection, folding-optimized variants with multiple mutations can be obtained, including many that are unlikely to have been generated using rational design or phylogenetic comparison approaches. In addition to evolving the proteins themselves for improved stability, directed evolution can also be used to customize host organisms to provide an optimized folding environment for specific proteins [20].
In this review, we outline recent advances in harnessing the power of directed evolution to optimize protein folding in the cell. Approaches include novel selection and screening methods for protein variants and host strains as well as the evolution of chaperones.
Harnessing the power of molecular biology for genetic diversification
Genetic variation can be generated using several different methods. Classic random mutagenesis techniques such as chemical and physical mutagenesis, and error prone PCR, transposon insertion mutagenesis, gene shuffling as well as more recently developed technologies for targeted mutagenesis including Multiplex Automated Genome Engineering (MAGE) facilitate the introduction of genetic changes in vitro and in vivo. The diverse methods are described in several excellent reviews and articles and will not be discussed further here [21–24].
Systems to assay for improved protein folding
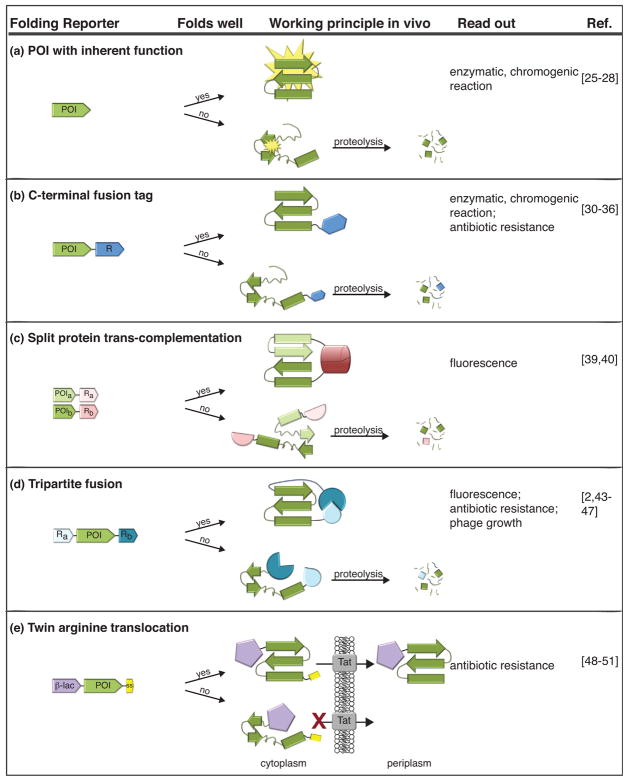
The accuracy, power, throughput, and stringency of selection or screening approaches are crucial to the success of a Darwinian optimization process. Various selection and screening systems have recently been developed to allow for the identification of stabilized protein variants (Figure 1).
Figure 1.
Selection and screening systems for improved protein folding in vivo. (a) An inherent, measurable function (e.g. enzymatic, chromogenic reaction) of a protein of interest (POI) is exploited to screen for stabilized protein variants. (b) A screen or selection for stabilized POI variants is enabled by fusion to a reporter protein with a measurable function. (c) Tight interaction of two portions of POI variants leads to proper complementation of fused fluorescent reporter protein portions and thereby allows screening for stabilized protein variants. Unstable protein variants will be depleted from the screen by degradation. (d) Split halves of a reporter protein will only interact and confer its inherent selectable/screenable function if the inserted protein variant folds well and doesn’t get proteolyzed or aggregated. (e) The twin arginine translocation (Tat) system relies on translocation of exclusively well-folded protein variants into the periplasm where proper folding is additionally selected for by a fused β-lactamase tag. POI, protein of interest; POIa/POIb, POIa half or POIb half, respectively, of split POI; Ra/Rb, Ra half or Rb half, respectively, of split reporter protein R; P, proteolysis; Tat, twin arginine translocase; ss, Tat signal sequence; β-lac, β-lactamase resistance marker.
Selection for improved folding based on an endogenous property
One very straightforward way to select for increased stability is to take advantage of an inherent property of the protein of interest, such as its enzymatic activity or any other property that can be easily screened for (Figure 1a). Unfortunately, the limited availability of simple assays for these properties makes this approach very protein-specific and generally relatively low-throughput [25–28]. However, one recent and very promising approach [29••] allows one to directly assay protein solubility in vivo, bypassing the need for protein-specific assays. In this assay, cells expressing the protein are incubated at elevated temperatures, lysed on a Durapore membrane filter which blocks variants that tend to aggregate but allows variants that remain soluble to pass through. These soluble variants are then retained on a nitrocellulose membrane and detected either with antibodies against the protein of interest itself or against an affinity tag attached to the protein. This method is not protein-specific, thus potentially broadly applicable and can be conducted in high-throughput.
Improving protein folding using folding reporter tags
Another way around the lack of an easily assayed property is to fuse the protein of interest to a reporter protein with the hope that the activity of the reporter will reflect the folding of its fusion partner. Improved folding of a tagged protein variant, for instance, could result in a parallel increase in the amount of the reporter protein. Quantification of the reporter then provides an indirect measure of the effective abundance of the protein of interest (Figure 1b). Green fluorescent protein (GFP) is one of the first and most commonly used fusion tags employed in screens for improved stability in vivo [30,31]. By combining GFP fusions with fluorescence activated cell sorting (FACS), folding variants can be screened and isolated in high throughput [32]. Other C-terminal reporter fusion tags that rely on chromogenic, enzymatic, or antibiotic resistance-conferring properties have also been developed and applied to screen or select for improved folding [33–36]. For example, dihydrofolate reductase has been used as a solubility reporter to select for soluble expression constructs [37]. These reporter fusion tag systems have disadvantages, however. They entail a propensity for false positives due to truncations or cleavage artifacts and fusions can alter the solubility of the protein variant [38].
To help eliminate these problems, split GFP systems have been developed in which only a short, non-fluorescent portion of GFP is fused to the C terminus of the protein of interest (Figure 1c). Expression of the remainder of GFP from a second plasmid will only complement the short portion well (and thereby emit fluorescence) if the protein of interest is soluble and remains intact (i.e. it is not degraded). The short nature of the GFP portion of these fusion constructs links it more closely to the folding and solubility properties of the protein of interest [39•].
In an alternative approach, Lindman et al. successfully used trans-complementation of fragments of GFP to screen for improved thermodynamic stabilization of the B1 domain of protein G (PGB1) [40,41•]. The rationale here is that mutations that stabilize a protein chain, in this case of PGB1, will tend to increase the affinity between two fragments of that chain. If these two fragments are fused to different portions of GFP, fragment stabilization will tend to drive the GFP portions together, resulting in better complementation and increased fluorescence [41•].
Tripartite protein folding sensors to optimize protein stability
More recently, advanced tripartite folding reporters have been developed in which a protein of interest is inserted at a permissive site within a reporter protein (Figure 1d). In the Proside (Protein stability increased by directed evolution) approach, the protein is inserted between two domains of an essential bacteriophage capsid protein. Stable variants will be more resistant to in vitro proteolysis and can be selected on that basis [42,43]. Several in vivo folding biosensors have also been developed based on a similar rationale; that is, the two parts of the reporter protein will only be able to fold together and confer the reporter’s intrinsic function if the inserted protein folds well. If the inserted protein is poorly folded, it will be cleaved by the plethora of proteases in the cell. This will separate the two halves of the reporter, resulting in lower levels of reporter function. This cis-complementation tripartite fusion approach is advantageous in that it discriminates against artifacts arising from internal ribosome initiation sites and other events that can untether the reporter from the target protein. Several tripartite protein folding reporter systems have been developed based on GFP [44]. Tripartite protein-based systems that rely on antibiotic resistance markers enable an efficient ‘fold or die’ selection for improved stability [2••,45]. The protein of interest is fused between the split marker halves of an antibiotic resistance gene. Improved folding of the inserted protein will result in complementation of the split marker halves and in turn, increased antibiotic resistance. Foit et al. optimized folding of Immunity protein 7 (Im7) in the Escherichia coli periplasm by applying a β-lactamase-based tripartite system. Interestingly, mutants that enhanced the thermodynamic stability of Im7 almost entirely mapped to surface residues involved in binding to its natural binding partner E7, suggesting that a stability-function tradeoff exists for this protein. This β-lactamase-based tripartite system has been applied to evaluate and further evolve the folding of rationally designed proteins [46,47] and to identify small molecule inhibitors of aggregation in vivo [48•].
Another tripartite selection also couples proper folding of a test protein to antibiotic resistance but is based on an entirely different principle: only folded proteins will be efficiently exported to the periplasm by the twin arginine transport (Tat) quality control system (Figure 1e). In this approach [49,50,51••], the Tat signal sequence is fused to the N-terminus of the protein of interest followed by fusion to β-lactamase, which will only encode antibiotic resistance if it is exported into the periplasm. The test protein must be properly folded to be recognized and exported by the Tat apparatus. This selection has been used to improve the in vivo solubility of several proteins [49,50,52,53]. A two-hybrid type version of this approach, based on the ability of the Tat translocase to carry with it non-covalently interacting proteins, has enabled the selection of protein variants with stronger protein-protein interactions [54–56] and enhanced intracellular stability [57].
Evolving an improved folding environment
Protein folding is not exclusively dependent on the protein sequence — the cellular folding environment is also important. There has been significant interest in engineering bacterial strains for the improved folding and expression of recombinant proteins [58]. Folding can be optimized by evolving the redox capacity to facilitate disulfide bond isomerization [59,60]. A specially designed strain called SHuffle has been generated that contains several cleverly targeted alterations and shows substantially improved folding of proteins with multiple disulfide bonds [61•].
General chaperone overexpression can promote in vivo protein folding [56]. It fosters protein evolution through buffering the destabilizing effect of thermodynamically unfavorable mutations of folding intermediates [58,62,63]. However, this approach is not aimed at improving the folding of specific proteins and is not broadly applicable given that even the generalist chaperone GroEL is estimated to interact with only about 10% of proteins in E. coli [64,65]. Chaperone expression has already been efficiently balanced in the cell by evolution, perhaps explaining why chaperone co-expression is only occasionally successful in improving the in vivo expression of specific proteins [66,67].
A more effective approach to improve in vivo folding may be to specifically evolve the cellular environment for the folding of a single protein of interest. Our group tested this strategy by using the tripartite β-lactamase approach in E. coli to select for host variants that improved the folding of Im7 [68]. The selected variants overproduced the periplasmic chaperone Spy. This protein, when purified, was shown to inhibit the aggregation and facilitate the refolding of a variety of proteins including Im7.
As protein folding can be improved by overexpressing client-specific chaperones, expression of a specifically optimized chaperone should have a similarly beneficial effect. It appears that the foldability of proteins and the sequence of highly specific chaperones has co-evolved [69]. Thus, it seems unlikely that these types of chaperone–client interactions can be easily further optimized. In contrast, promiscuous chaperones that normally interact with many different binding partners can be evolved to enhance their interaction with one specific client protein. A detailed review about chaperone enhancement has been published recently by Mack et al. [70].
For example, Wang et al. evolved variants of the Hsp60 chaperone GroEL and co-chaperone GroES that showed an enhanced ability to stabilize GFP in E. coli [6]. Unfortunately, these GroEL/S mutants were defective in their ability to fold other proteins, reflecting the specificity-promiscuity tradeoff that one is faced with in the evolution of most chaperones.
Aponte et al. focused on improving another chaperone, Hsp70 DnaK. The evolved chaperone showed several fold improved refolding ability for soluble, denatured luciferase as compared to wild-type DnaK [71]. Their selection system was based on a destabilized antibiotic resistance marker that only confers antibiotic resistance in vivo if properly folded (a variation on the schema illustrated in Figure 1a). Overexpression of effective DnaK variants allowed the phenotype to be rescued.
Our group has isolated Spy variants that not only improve the stability of unstable Im7 mutants, but other proteins as well, implying that they may be generally more effective [72•,73,74]. One interesting class appears to work by enhancing the flexibility of a segment in Spy whose flexibility is known to be important for Spy’s action. Interestingly, the residue change found in one of these ‘super Spy’ mutants is actually quite common in evolution. It seems likely that the affected residue may act as an evolutionary rheostat, tuning the flexibility of this segment to fit the organism’s need for Spy’s substrate diversity, balanced with its need to maintain some minimal stability for Spy.
A comparable concept has been observed with potentiated Hsp104 variants selected for increased disaggregation activity. In this case as well, some mutants appear to act by increasing the flexibility of the chaperone [75], highlighting the importance of chaperone flexibility in their action [76].
Conclusions and outlook
Insights from directed evolution studies highlight the various and sometimes unexpected ways that protein folding, the product of many years of evolution, can actually be improved upon in vivo. Newly developed fusion approaches that allow for the direct selection of proteins with improved folding in vivo and direct ways of screening for improved solubility present a diverse platform for future creative endeavors.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM102829). J.C.A.B. is a Howard Hughes Medical Institute Investigator.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 2••.Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, Warriner SL, Radford SE, Bardwell JCA. Optimizing protein stability in vivo. Mol Cell. 2009;36:861–871. doi: 10.1016/j.molcel.2009.11.022. Foit et al. developed a direct selection system for stable protein variants in vivo based on a tripartite fusion with a split antibiotic marker protein. Based on their isolated stabilized protein variants, it was concluded that thermodynamic and kinetic stability seem to be the decisive factors for expression and protein stability in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taverna DM, Goldstein RA. Why are proteins marginally stable? Proteins Struct Funct Genet. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- 4.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLoS Comput Biol. 2008;4:35–37. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koldewey P, Horowitz S, Bardwell JCA. Chaperone–client interactions: non-specificity engenders multifunctionality. J Biol Chem. 2017;292:12010–12017. doi: 10.1074/jbc.R117.796862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell. 2002;111:1027–1039. doi: 10.1016/s0092-8674(02)01198-4. [DOI] [PubMed] [Google Scholar]
- 7.Gershenson A, Gierasch LM. Protein folding in the cell: challenges and progress. Curr Opin Struct Biol. 2011;21:32–41. doi: 10.1016/j.sbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hingorani KS, Gierasch LM. Comparing protein folding in vitro and in vivo: foldability meets the fitness challenge. Curr Opin Struct Biol. 2014;24:81–90. doi: 10.1016/j.sbi.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science (80-) 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 10.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Ulrich Hartl F. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 11.Ghaemmaghami S, Oas TG. Quantitative protein stability measurement in vivo. Nat Struct Biol. 2001;8:879–882. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 12••.Ignatova Z, Gierasch LM. Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling. Proc Natl Acad Sci U S A. 2004;101:523–528. doi: 10.1073/pnas.0304533101. The authors genetically engineered a tetracysteine motif in an unconserved loop of a protein of interest and by adding the membrane-permeable bis-arsenical fluorescein-based dye FlAsH were able to monitor folding and aggregation in vivo in real time by urea titration. In vivo and in vitro aggregation displayed similar kinetic properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ignatova Z, Krishnan B, Bombardier JP, Marcelino AMC, Hong J, Gierasch LM. From the test tube to the cell: exploring the folding and aggregation of a beta-clam protein. Biopolymers. 2007:157–163. doi: 10.1002/bip.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J Biol Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 15.Huang P-S, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 16.Donovan RS, Robinson CW, Click BR. Review: Optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter. J Ind Microbiol. 1996;16:145–154. doi: 10.1007/BF01569997. [DOI] [PubMed] [Google Scholar]
- 17.Francis DM, Page R. Strategies to optimize protein expression in E. coli. Curr Protoc Protein Sci. 2010 doi: 10.1002/0471140864.ps0524s61. http://dx.doi.org/10.1002/0471140864.ps0524s61. [DOI] [PMC free article] [PubMed]
- 18.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014:5. doi: 10.3389/fmicb.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia B, Jeon CO. High-throughput recombinant protein expression in Escherichia coli: current status and future perspectives. Open Biol. 2016;6:1–17. doi: 10.1098/rsob.160196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlegel S, Genevaux P, de Gier J-W. Isolating Escherichia coli strains for recombinant protein production. Cell Mol Life Sci. 2017;74:891–908. doi: 10.1007/s00018-016-2371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 22.Zaugg J, Gumulya Y, Gillam EMJ, Bodén M. Directed evolution library creation: methods and protocols. Directed Evolution Library Creation. 2014:315–333. doi: 10.1007/978-1-4939-1053-3_21. [DOI] [PubMed] [Google Scholar]
- 23.Bassalo MC, Liu R, Gill RT. Directed evolution and synthetic biology applications to microbial systems. Curr Opin Biotechnol. 2016;39:126–133. doi: 10.1016/j.copbio.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Zheng X, Xing X-H, Zhang C. Targeted mutagenesis: a sniper-like diversity generator in microbial engineering. Synth Syst Biotechnol. 2017;2:75–86. doi: 10.1016/j.synbio.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Speck J, Hecky J, Tam HK, Arndt KM, Einsle O, Müller KM. Exploring the molecular linkage of protein stability traits for enzyme optimization by iterative truncation and evolution. Biochemistry. 2012;51:4850–4867. doi: 10.1021/bi2018738. [DOI] [PubMed] [Google Scholar]
- 26.Khersonsky O, Rosenblat M, Toker L, Yacobson S, Hugenmatter A, Silman I, Sussman JL, Aviram M, Tawfik DS. Directed evolution of serum paraoxonase PON3 by family shuffling and ancestor/consensus mutagenesis, and its biochemical characterization. Biochemistry. 2009;48:6644–6654. doi: 10.1021/bi900583y. [DOI] [PubMed] [Google Scholar]
- 27.Xie X, Pashkov I, Gao X, Guerrero JL, Yeates TO, Tang Y. Rational improvement of simvastatin synthase solubility in Escherichia coli leads to higher whole-cell biocatalytic activity. Biotechnol Bioeng. 2009;102:20–28. doi: 10.1002/bit.22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khersonsky O, Kiss G, Rothlisberger D, Dym O, Albeck S, Houk KN, Baker D, Tawfik DS. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc Natl Acad Sci. 2012;109:10358–10363. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Asial I, Cheng YX, Engman H, Dollhopf M, Wu B, Nordlund P, Cornvik T. Engineering protein thermostability using a generic activity-independent biophysical screen inside the cell. Nat Commun. 2013;4:2901. doi: 10.1038/ncomms3901. The authors developed a high-throughput colony-based stability screen that provides a direct biophysical read-out of intrinsic protein stability. The screen allowed for the isolation of thermostabilized protein variants from a randomly mutagenized sequence pool and was successfully demonstrated on a diverse range of proteins. [DOI] [PubMed] [Google Scholar]
- 30.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 31.Van Den Berg S, Löfdahl PÅ, Härd T, Berglund H. Improved solubility of TEV protease by directed evolution. J Biotechnol. 2006;121:291–298. doi: 10.1016/j.jbiotec.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Seitz T, Thoma R, Schoch GA, Stihle M, Benz J, D’Arcy B, Wiget A, Ruf A, Hennig M, Sterner R. Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening. J Mol Biol. 2010;403:562–577. doi: 10.1016/j.jmb.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 33.Wigley WC, Stidham RD, Smith NM, Hunt JF, Thomas PJ. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat Biotechnol. 2001;19:131–136. doi: 10.1038/84389. [DOI] [PubMed] [Google Scholar]
- 34.Maxwell KL, Mittermaier AK, Forman-Kay JD, Davidson AR. A simple in vivo assay for increased protein solubility. Protein Sci. 1999;8:1908–1911. doi: 10.1110/ps.8.9.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sieber V, Martinez CA, Arnold FH. Libraries of hybrid proteins from distantly related sequences. Nat Biotechnol. 2001;19:456–460. doi: 10.1038/88129. [DOI] [PubMed] [Google Scholar]
- 36.Gul N, Linares DM, Ho FY, Poolman B. Evolved Escherichia coli strains for amplified, functional expression of membrane proteins. J Mol Biol. 2014;426:136–149. doi: 10.1016/j.jmb.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Dyson MR, Perera RL, Shadbolt SP, Biderman L, Bromek K, Murzina NV, Mccafferty J. Identification of soluble protein fragments by gene fragmentation and genetic selection. Nucleic Acids Res. 2008:36. doi: 10.1093/nar/gkn151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawasaki M, Inagaki F. Random PCR-based screening for soluble domains using green fluorescent protein. Biochem Biophys Res Commun. 2001;280:842–844. doi: 10.1006/bbrc.2000.4229. [DOI] [PubMed] [Google Scholar]
- 39•.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. This study describes a simple protein tagging system involving soluble, self-associating fragments of GFP. The trans-complementation method developed here enables one to distinguish soluble and insoluble proteins in living cells and in cell lysates. [DOI] [PubMed] [Google Scholar]
- 40.Lindman S, Johansson I, Thulin E, Linse S. Green fluorescence induced by EF-hand assembly in a split GFP system. Protein Sci. 2009;18:1221–1229. doi: 10.1002/pro.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Lindman S, Hernandez-Garcia A, Szczepankiewicz O, Frohm B, Linse S. In vivo protein stabilization based on fragment complementation and a split GFP system. Proc Natl Acad Sci U S A. 2010;107:19826–19831. doi: 10.1073/pnas.1005689107. Lindman et al. describe the stabilization of protein PGB1 using a novel complementation system. The method is based on the affinity between two similar PGB1 mutant chain variants, which is thermodynamically linked to complementation of fused GFP fragments. Fluorescence-based screening allows one to identify stabilized PGB1 protein variants in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kristensen P, Winter G. Proteolytic selection for protein folding using filamentous bacteriophages. Fold Des. 1998;3:321–328. doi: 10.1016/S1359-0278(98)00044-3. [DOI] [PubMed] [Google Scholar]
- 43.Sieber V, Plückthun A, Schmid FX. Selecting proteins with improved stability by a phage-based method. Nat Biotechnol. 1998;16:955–960. doi: 10.1038/nbt1098-955. [DOI] [PubMed] [Google Scholar]; Current Opinion in Structural Biology. 2018;48:117–123. doi: 10.1016/j.sbi.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabantous S, Rogers Y, Terwilliger TC, Waldo GS. New molecular reporters for rapid protein folding assays. PLoS ONE. 2008:3. doi: 10.1371/journal.pone.0002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik A, Mueller-Schickert A, Bardwell JCA. Cytosolic selection systems to study protein stability. J Bacteriol. 2014;196:4333–4343. doi: 10.1128/JB.02215-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong P, Wang M, Zhou X, Zhang T, Zhang J, Chen Q, Liu H. Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability. Nat Commun. 2014;5:5330. doi: 10.1038/ncomms6330. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Zhang T, Liu R, Song M, Wang J, Hong J, Chen Q, Liu H. Recurring sequence-structure motifs in (βα)8-barrel proteins and experimental optimization of a chimeric protein designed based on such motifs. Biochim Biophys Acta – Proteins Proteomics. 2017;1865:165–175. doi: 10.1016/j.bbapap.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 48•.Saunders JC, Young LM, Mahood RA, Jackson MP, Revill CH, Foster RJ, Smith DA, Ashcroft AE, Brockwell DJ, Radford SE. An in vivo platform for identifying inhibitors of protein aggregation. Nat Chem Biol. 2015;12:94–101. doi: 10.1038/nchembio.1988. This study describes an E. coli periplasmic sensor for detecting the aggregation of proteins into both amyloid and amorphous aggregates and its inhibition by small molecules. By fusing the protein variants between a split antibiotic marker in cis, the protein variants’ aggregation propensity in vivo was directly linked to antibiotic resistance. Using this clever system, the authors were able to identify compounds binding to test proteins and characterize their interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher AC, Kim W, DeLisa MP. Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci. 2006;15:449–458. doi: 10.1110/ps.051902606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher AC, DeLisa MP. Efficient isolation of soluble intracellular single-chain antibodies using the twin-arginine translocation machinery. J Mol Biol. 2009;385:299–311. doi: 10.1016/j.jmb.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Fisher AC, Rocco MA, Delisa MP. Genetic selection of solubility-enhanced proteins using the twin-arginine translocation system. Methods Mol Biol. 2011;705:53–67. doi: 10.1007/978-1-61737-967-3_4. The authors developed a genetic selection system that directly linked the Tat folding quality control mechanism with antibiotic resistance. A Tat mutant strain exports folded as well as misfolded protein variants into the periplasmic space. The efficient selection of stable and soluble protein variants is enabled by fusing a periplasmic selectable marker to the protein variants. [DOI] [PubMed] [Google Scholar]
- 52.Kim D-S, Song H-N, Nam HJ, Kim S-G, Park Y-S, Park J-C, Woo E-J, Lim H-K. Directed evolution of human heavy chain variable domain (VH) using in vivo protein fitness filter. PLOS ONE. 2014;9:e98178. doi: 10.1371/journal.pone.0098178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boock JT, King BC, Taw MN, Conrado RJ, Siu KH, Stark JC, Walker LP, Gibson DM, Delisa MP. Repurposing a bacterial quality control mechanism to enhance enzyme production in living cells. J Mol Biol. 2015;427:1451–1463. doi: 10.1016/j.jmb.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waraho D, DeLisa MP. Versatile selection technology for intracellular protein–protein interactions mediated by a unique bacterial hitchhiker transport mechanism. Proc Natl Acad Sci U S A. 2009;106:3692–3697. doi: 10.1073/pnas.0704048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waraho D, Delisa MP. Identifying and optimizing intracellular protein–protein interactions using bacterial genetic selection. Methods Mol Biol. 2012;813:125–143. doi: 10.1007/978-1-61779-412-4_7. [DOI] [PubMed] [Google Scholar]
- 56.Waraho-Zhmayev D, Gkogka L, Yu T-Y, DeLisa MP. A microbial sensor for discovering structural probes of protein misfolding and aggregation 1. Prion. 2013;7:151–156. doi: 10.4161/pri.23328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waraho-Zhmayev D, Meksiriporn B, Portnoff AD, DeLisa MP, Bradbury A. Optimizing recombinant antibodies for intracellular function using hitchhiker-mediated survival selection. Protein Engineering, Design and Selection. 2014:351–358. doi: 10.1093/protein/gzu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact. 2011;10:32. doi: 10.1186/1475-2859-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Marco A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb Cell Fact. 2009;8:26. doi: 10.1186/1475-2859-8-26. www.sciencedirect.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Marco A. Recent contributions in the field of the recombinant expression of disulfide bonded protein in bacteria. Microb Cell Fact. 2012;11:129. doi: 10.1186/1475-2859-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61•.Lobstein J, Emrich CA, Jeans C, Faulkner M, Riggs P, Berkmen M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb Cell Fact. 2012;11:753. doi: 10.1186/1475-2859-11-56. This study describes a novel strain for cytoplasmic protein production. The engineered SHuffle strain permits oxidative folding in the cytoplasm and additional over-expression of the DsbC isomerase improves oxidative folding. Optimal expression conditions and the effect of co-expressing helper proteins on the folding state were characterized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 63.Wyganowski KT, Kaltenbach M, Tokuriki N. GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J Mol Biol. 2013;425:3403–3414. doi: 10.1016/j.jmb.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 64.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 65.Fujiwara K, Ishihama Y, Nakahigashi K, Soga T, Taguchi H. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 2010;29:1552–1564. doi: 10.1038/emboj.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santra M, Farrell DW, Dill KA. Bacterial proteostasis balances energy and chaperone utilization efficiently. Proc Natl Acad Sci. 2017;114:E2654–E2661. doi: 10.1073/pnas.1620646114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kolaj O, Spada S, Robin S, Wall JG. Use of folding modulators to improve heterologous protein production in Escherichia coli. Microb Cell Fact. 2009;8:9. doi: 10.1186/1475-2859-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, et al. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat Struct Mol Biol. 2011;18:262–269. doi: 10.1038/nsmb.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mapa K, Tiwari S, Kumar V, Jayaraj GG, Maiti S. Information encoded in Non-native states drives substrate-chaperone pairing. Structure. 2012;20:1562–1573. doi: 10.1016/j.str.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 70.Mack KL, Shorter J. Engineering and evolution of molecular chaperones and protein disaggregases with enhanced activity. Front Mol Biosci. 2016:3. doi: 10.3389/fmolb.2016.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aponte RA, Zimmermann S, Reinstein J. Directed evolution of the DnaK chaperone: mutations in the lid domain result in enhanced chaperone activity. J Mol Biol. 2010;399:154–167. doi: 10.1016/j.jmb.2010.03.060. [DOI] [PubMed] [Google Scholar]
- 72•.Quan S, Wang L, Petrotchenko EV, Makepeace KAT, Horowitz S, Yang J, Zhang Y, Borchers CH, Bardwell JCA. Super Spy variants implicate flexibility in chaperone action. Elife. 2014;2014:e01584. doi: 10.7554/eLife.01584. This study describes the selection of several more efficiently folding Spy chaperone variants compared to wild-type Spy by using an antibiotic resistance-based folding assay to select for improved folding in vivo. Evolved Spy variants were shown to have a more hydrophobic binding surface and exhibited greater flexibility compared to wild-type Spy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stull F, Koldewey P, Humes JR, Radford SE, Bardwell JCA. Substrate protein folds while it is bound to the ATP-independent chaperone Spy. Nat Struct Mol Biol. 2015;23:53–58. doi: 10.1038/nsmb.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salmon L, Ahlstrom LS, Horowitz S, Dickson A, Brooks CL, Bardwell JCA. Capturing a dynamic chaperone–substrate interaction using NMR-informed molecular modeling. J Am Chem Soc. 2016;138:9826–9839. doi: 10.1021/jacs.6b02382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jackrel ME, Desantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, Shorter J. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bardwell JCA, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37:517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]