Abstract
Background
Enlargement of the vestibular aqueduct (EVA) is the most common radiological abnormality in children with sensorineural hearing loss. Mutations in coding regions and splice sites of the SLC26A4 gene are often detected in Caucasians with EVA. Approximately one-fourth of patients with EVA have two mutant alleles (M2), one-fourth have one mutant allele (M1) and one-half have no mutant alleles (M0). The M2 genotype is correlated with a more severe phenotype.
Methods
We performed genotype–haplotype analysis and massively parallel sequencing of the SLC26A4 region in patients with M1 EVA and their families.
Results
We identified a shared novel haplotype, termed CEVA (Caucasian EVA), composed of 12 uncommon variants upstream of SLC26A4. The presence of the CEVA haplotype on seven of ten ’mutation-negative’ chromosomes in a National Institutes of Health M1 EVA discovery cohort and six of six mutation-negative chromosomes in a Danish M1 EVA replication cohort is higher than the observed prevalence of 28 of 1006 Caucasian control chromosomes (p<0.0001 for each EVA cohort). The corresponding heterozygous carrier rate is 28/503 (5.6%). The prevalence of CEVA (11 of 126) is also increased among M0 EVA chromosomes (p=0.0042).
Conclusions
T he CEVA haplotype causally contributes to most cases of Caucasian M1 EVA and, possibly, some cases of M0 EVA. The CEVA haplotype of SLC26A4 defines the most common allele associated with hereditary hearing loss in Caucasians. The diagnostic yield and prognostic utility of sequence analysis of SLC26A4 exons and splice sites will be markedly increased by addition of testing for the CEVA haplotype.
Introduction
Enlargement of the vestibular aqueduct (EVA (MIM#6 00 791)) is the most common radiological malformation associated with childhood sensorineural hearing loss.1 Hearing loss and EVA can be non-syndromic (NSEVA) or associated with an iodine organification defect that can lead to goitre as part of Pendred syndrome (PS (MIM#2 74 600)).2,3 Both phenotypes can be associated with recessive mutant alleles of the SLC26A4 gene (MIM#605646) on chromosome 7. Approximately one-fourth of North American or European Caucasians with NSEVA or PS have homozygous or compound heterozygous mutations in the coding regions or splice sites of SLC26A4, as expected for recessive inheritance.2,3 We refer to this genotype as ‘M2’, in contrast to the one-half of patients with EVA with no detectable mutations (M0) or the one-fourth of patients with only one allele with a detectable mutation of SLC26A4 (M1).4
The number of mutant alleles of SLC26A4 is strongly correlated with the auditory and thyroid phenotypes. The severity of hearing loss in EVA ears is greater in M2 patients than in M0 or M1 patients.5,6 With rare exceptions, M2 patients exhibit bilateral EVA, while patients with unilateral EVA are almost always M1 or M0.2,3,7,8 Moreover, the iodine organification defect underlying the development of thyroid goitre is directly correlated with the M2 genotype.3,7,9
The probability (approximately one in four) of EVA among siblings of M1 probands is not significantly different from the probability of EVA in siblings of M2 probands, suggesting the inheritance of an undetected recessive mutation of SLC26A4 in M1 EVA subjects.4 Two previous studies proposed that EVA could be caused by digenic inheritance of a pathogenic variant of FOXI1 (MIM#601093) or KCNJ10 (MIM#602208) in combination with a mutation of SLC26A4.10–12 However, the methodologies did not permit definitive conclusions, and subsequent studies have been unable to replicate the findings.13–15 Alternatively, we have consistently observed co-segregation of SLC26A4-linked short tandem repeat markers with EVA in M1 sibships, suggesting the existence of an undetected pathogenic variant(s) on the allele that has no mutation in the exons or splice sites of SLC26A4.4 The purpose of our current study was to explore this hypothesis in a National Institutes of Health (NIH) discovery cohort and a Danish replication cohort of patients with M1 EVA.
Methods
Study and subjects
The current study was approved by the Combined Neuroscience Institutional Review Board, NIH, Bethesda, Maryland, USA. Danish subjects were studied in accordance with approval from the Danish Research Ethical Committee (KF 01-108/03, 11-112/04, KF120/03). Written informed consent was obtained for all subjects. NIH subjects were evaluated at the NIH Clinical Center as described.3,5–9,16 Some subjects have been previously reported.3,6–8,16,17 We originally defined a vestibular aqueduct as enlarged if the midpoint diameter was >1.5 mm or if it had a grossly malformed overall morphology,3 but we later modified the midpoint diameter criterion to >1.0 mm for the NIH cohort.16 The midpoint diameter criterion was >1.5 mm for the Danish cohort. To distinguish between PS and NSEVA, the thyroid gland was evaluated by serological tests of thyroid function, ultrasonographic examination of thyroid volume and texture, and perchlorate discharge testing.9 Results were interpreted according to published objective criteria.9 We classified the pathogenicity of SLC26A4 variants based on the American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) variant interpretation guidelines.18,19 Based on these criteria, we classified EVA subjects as M2, M1 or M0 if they carried two, one or zero mutations in the coding regions or splice sites of SLC26A4.4 We classified a variant as indeterminate if it is benign in trans with an allele with no mutations but pathogenic in trans with a mutant allele of SLC26A4.7 There were 34 M2 subjects, 15 M1 subjects and 83 M0 subjects in the NIH cohort.
Massively parallel sequencing
We used Agilent SureDesign to design biotinylated RNA probes targeting the smallest region of overlap (SRO) and subsequently the shared chromosomal segments defined by homozygosity haplotype mapping. The designs achieved 95% coverage of targeted intervals. Library preparation and capture were prepared using SureSelectXT Target Enrichment System (see additional details in online supplementary material). Sequences were aligned to the hg19 version of the human genome using Burrows-Wheeler Aligner (version 0.7.13).20 Variants were called using the best-practices pipeline for GATK variant analysis package (version 3.5).21,22 After deduplication, the average read coverage over the targeted region was 234×. ANNOVAR23 software was used to annotate predicted pathogenicity, allele frequency in reference populations and dbSNP number (see online supplementary methods).
Statistical analyses
We used GraphPad Prism 7 (http://www.graphpad.com/) to perform a two-tailed Fisher’s exact test to compare the prevalence of the CEVA (Caucasian EVA) haplotype, as defined by genotypes of all 12 variants, among chromosomes from different cohorts (see online supplementary methods) and from the 1000 Genomes database (http://phase3browser.1000genomes.org/index.html). For EVA cohorts, identical-by-descent chromosomes within a single family were counted as only one chromosome for the comparisons (see online supplementary methods).
Homozygosity haplotype mapping
For each sibling pair in the NIH cohort, we identified homozygous SNPs throughout the region until we reached a SNP at each end of the region that was discordant within that pair of affected siblings.24 We then performed pairwise comparisons of homozygous SNP genotypes between all possible combinations of sibling pairs.
In silico prediction of pathogenic potential
We used Combined Annotation Dependent Depletion (CADD; http://cadd.gs.washington.edu/) to predict the pathogenic potential of sequence variants. CADD integrates a range of diverse annotations into a single measure (C-score) of the likelihood of deleteriousness for each variant.25 The scaled C-scores are based on the rank of the C-score of any variant relative to those of 8.6 billion possible single nucleotide variants from the human genome.25
Results
SLC26A4-linked haplotype shared among M1 EVA chromosomes
Our strategy was based on the hypothesis that many M1 EVA families co-segregate the same undetected pathogenic variant affecting the expression of an allele of SLC26A4 that has no detectable mutations in the coding regions or splice sites of this gene. We henceforth refer to these chromosomes as ‘mutation-negative.’
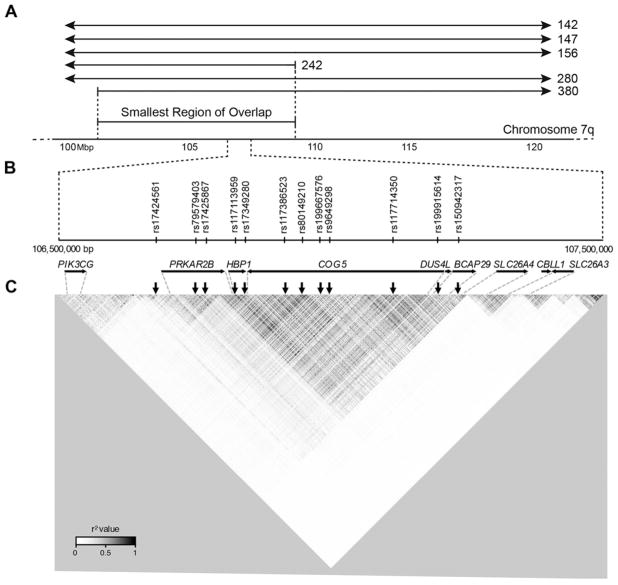
We performed a haplotype analysis of short tandem repeat markers (see online supplementary methods) spanning the SLC26A4 region of chromosome 7 in six NIH sibling pairs affected with EVA. Five were M1 families (142, 156, 242, 280 and 380), and one family (147) was initially included with the M1 cohort but was later moved to the M0 cohort based on reclassification of p.Met775Thr as a variant of indeterminate pathogenic potential.7 We identified a centromeric meiotic breakpoint on the mutation-negative chromosome in family 380 and a telomeric breakpoint on the mutation-negative chromosome in family 242 (figure 1A). These breakpoints define an 8.218-Mb SRO of the mutation-negative chromosomes co-segregating with EVA in the six sibling pairs.
Figure 1.
Region of the ‘mutation-negative’ allele of SLC26A4 on chromosome 7q that co-segregates with enlargement of the vestibular aqueduct (EVA). (A) Region on chromosome 7q that co-segregates with EVA among families 142, 147, 156, 242, 280 and 380. The dotted vertical lines delineate the centromeric recombination breakpoint in family 380 and the telomeric breakpoint in family 242 that define a smallest region of overlap among the families. (B) Twelve uncommon variants that define the Caucasian EVA haplotype that co-segregates with EVA in families 142, 147, 156, 280 and 380. Two variants (rs199667576 and rs199915614) are single-nucleotide deletions and ten are SNPs. (C) Linkage disequilibrium (LD) map shows the 12 variants in a single region of LD spanning from upstream of PRKAR2B to intron 3 of SLC26A4. The colour scale represents pairwise r2 values with darker shades reflecting higher co-inheritance. The 12 variants are all in modest LD with each other (r2>0.6). The nine telomeric variants are in a large region of higher LD, and the three centromeric variants are in an adjacent region of weaker LD. Genes and their orientations are shown as horizontal arrows. Vertical arrows indicate the location of the variants.
We identified 23 159 variants by massively parallel sequencing (MPS) of the entire SRO in the six sibling pairs. There are 16 820 variants, located in non-repetitive genomic regions, which co-segregate among any one or more affected sibling pairs. We searched for variants with a minor allele frequency (MAF) ≤0.05 that are shared among the six mutation-negative chromosomes. We used MAFs calculated for the Caucasian-European superpopulation that includes CEU (Utah residents with Northern and Western European ancestry), TSI (Toscani in Italy), FIN (Finnish in Finland), GBR (British in England and Scotland) and IBS (Iberian populations in Spain) populations. The Caucasian-European population has 1785 variants with MAF ≤0.05 in the SRO. None of the 1785 variants are shared among all six families, but 12 variants (table 1) with MAFs between 0.019 and 0.049 are shared by five of six families (142, 147, 156, 280 and 380, but not 242). This indicates that the pathogenic variant(s) in family 242 may be different than that (those) in the other five families. Since the telomeric breakpoint of the original SRO was identified in family 242, the actual SRO for the other five families extends further toward the telomere than shown in figure 1A. The 12 variants include ten single-nucleotide substitutions and two single-nucleotide deletions spanning a 613-kb region upstream of SLC26A4. Some are located in intergenic regions, and the others are found within introns of other genes (figure 1B). We confirmed the genotypes of all 12 variants by Sanger sequencing. Sequencing of parental DNA samples demonstrated that the 12 variants are in cis with each other and in trans with the known mutation(s) of SLC26A4. The 12 variants thus comprise a previously unrecognised haplotype that we refer to as the CEVA haplotype.
Table 1.
SLC26A4-linked variants comprising the CEVA haplotype
| Variant | Chromosome 7q position (bp)* | Reference genotype | CEVA genotype | Minor allele frequency† | CADD score‡ |
|---|---|---|---|---|---|
| rs17424561 | 106669858 | G | A | 0.049 | 3.9 |
| rs79579403 | 106741374 | T | C | 0.048 | 2.9 |
| rs17425867 | 106764419 | T | A | 0.048 | 11.7 |
| rs117113959 | 106815154 | T | C | 0.047 | 0.1 |
| rs17349280 | 106837681 | G | A | 0.047 | 2.2 |
| rs117386523 | 106930234 | C | T | 0.047 | 13.5 |
| rs80149210 | 106967931 | A | G | 0.047 | 1.6 |
| rs199667576 | 106993159 | T | – | 0.046 | 5.9 |
| rs9649298 | 107014419 | A | G | 0.046 | 13.7 |
| rs117714350 | 107147622 | T | C | 0.035 | 7.8 |
| rs199915614 | 107242636 | T | – | 0.035 | 21.6 |
| rs150942317 | 107282469 | A | C | 0.035 | 10.4 |
Based on hg19.
From European populations in 1000 Genomes phase 3.
CADD scaled C-scores for predicted deleteriousness. Larger scores are more strongly predictive of deleteriousness. CADD, Combined Annotation Dependent Depletion; CEVA, Caucasian enlargement of the vestibular aqueduct.
To understand the haplotype structure of the region containing the CEVA haplotype and SLC26A4, we used Haploview to generate a linkage disequilibrium (LD) map (see online supplementary methods) of all SNPs with MAF >0.01 among 503 unrelated Caucasian-European individuals (figure 1C). The 12 CEVA variants are located within a single region of LD that spans from upstream of the PRKAR2B gene to intron 3 of SLC26A4. Pairwise comparison of the 12 variants showed that they are all in modest LD (r2 >0.6) with each other (figure 1C, see online supplementary table 1).
Association of SLC26A4-linked CEVA haplotype with M1 EVA
We queried the 1000 Genomes database for the prevalence of the 12-variant CEVA haplotype in the Caucasian-European and other superpopulations. Of a total of 1006 Caucasian-European alleles, 947 had the reference haplotype, and 28 had the CEVA haplotype (table 2). Twenty-two other alleles were composed of different combinations of CEVA and reference genotypes (table 2). The prevalence of the CEVA haplotype is lower in other superpopulations: 1 (0.0756%) of 1322 African chromosomes, 11 (1.59%) of 694 Admixed American Chromosomes, zero of 1008 East Asian chromosomes and 1 (0.102%) of 978 South Asian chromosomes (table 2).
Table 2.
Numbers of SLC26A4-linked haplotypes in 1000 Genomes
| Haplotype | EUR | AFR | AMR | EAS | SAS | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CEU | GBR | TSI | IBS | FIN | ALL | |||||
| GTTTGCATATTA (reference) | 188 | 177 | 197 | 200 | 185 | 947 | 1314 | 676 | 1008 | 971 |
| ACACATG-GC-C (CEVA) | 3 | 2 | 8 | 7 | 8 | 28 | 1 | 11 | 0 | 1 |
| ACACATG-GTTA | 2 | 0 | 1 | 3 | 3 | 9 | 5 | 1 | 0 | 0 |
| ATTTGCATATTA | 1 | 1 | 1 | 1 | 0 | 4 | 0 | 3 | 0 | 0 |
| ACATGCATATTA | 1 | 1 | 0 | 1 | 0 | 3 | 1 | 0 | 0 | 0 |
| GCACATG-GTTA | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| GCACATG-GC-C | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| GTTCATG-GC-C | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 |
| GTTCATG-GTTA | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ACTCATG-GC-C | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ACACACGTAC-C | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GTATGCATATTA | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GTTTGTATATTA | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 0 |
| ACACATA-GC-C | 1* | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GTTTGCGTATTA | 1* | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ACACATG-GCTC | 0 | 0 | 1† | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GTTTGCATAT-A | 0 | 0 | 1† | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ACACATG-GCTA | 0 | 0 | 1‡ | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GTTTGCATAT-C | 0 | 0 | 1‡ | 0 | 0 | 1 | 0 | 0 | 0 | 1§ |
| GTTCATG-GCTA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1§ |
| GCATGCATATTA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| GTTCACATATTA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| GTTCGCATATTA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| GTTTGCA-ATTA | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Total number of haplotypes | 198 | 182 | 214 | 214 | 198 | 1006 | 1322 | 694 | 1008 | 978 |
Superpopulations are shown in italic: European (EUR), African (AFR), Admixed American (AMR), East Asian (EAS) and South Asian (SAS). European subpopulations are Utah residents with Northern and Western European ancestry (CEU), Toscani in Italy (TSI), Finnish in Finland (FIN), British in England and Scotland (GBR), and Iberian populations in Spain (IBS). Genotypes for CEVA variants are underlined. Footnotes indicate individuals with two rare haplotypes due to allele flip error.
NA12400.
NA20761.
NA20768.
NA20858.
In order to determine the prevalence of the CEVA haplotype among all M1 mutation-negative chromosomes in the NIH cohort, we analysed five additional M1 EVA subjects from simplex families (table 3). Three of the five subjects have the CEVA haplotype in trans with their SLC26A4 mutation. One subject (1580) had the reference haplotype, and one subject (1805) had the reference-CEVA combination haplotype GTTCATG-GC-C (CEVA genotypes are underlined; table 2). Taken together with the familial M1 EVA data, seven of ten M1 mutation-negative chromosomes have the CEVA haplotype (table 3). This is significantly different from the observed prevalence (28/1006) in Caucasian-European controls (p<0.0001).
Table 3.
SLC26A4 genotypes, haplotypes and phenotypes of M1 subjects with enlargement of the vestibular aqueduct and one mutated allele
| Family | Subject | Ancestry | Allele 1 genotype | Allele 2 haplotype | HL/EVA* | Thyroid |
|---|---|---|---|---|---|---|
| 142 | 1303 | Caucasian | p.Leu445Trp | CEVA | B | NS |
| 142 | 1304 | Caucasian | p.Leu445Trp | CEVA | R | NS |
| 156 | 1425 | Caucasian | c.1001+1G>A | CEVA | B | NS |
| 156 | 1426 | Caucasian | c.1001+1G>A | CEVA | B | I |
| 217 | 1580 | Caucasian | p.Glu384Gly | Reference | R | NS |
| 239 | 1645 | Caucasian | p.Thr416Pro | CEVA | B | NS |
| 242 | 1630 | Caucasian | p.Met1Thr | Reference | B | I |
| 242 | 1631 | Caucasian | p.Met1Thr | Reference | B | I |
| 264 | 1762 | Caucasian | p.Leu445Trp | CEVA | B | I |
| 273 | 1805 | Caucasian | p.Met1Thr | Other† | B | NS |
| 280 | 1802 | Caucasian | p.Thr416Pro | CEVA‡ | B | I |
| 280 | 1803 | Caucasian | p.Thr416Pro | CEVA‡ | B | I |
| 293 | 1849 | Caucasian | p.V138F | CEVA | B | I |
| 380 | 2083 | Caucasian | c.365insT | CEVA | L | NS |
| 380 | 2085 | Caucasian | c.365insT | CEVA | B | NS |
| DK-1 | 12 624–11 | Caucasian | p.Thr416Pro | CEVA | B | NT |
| DK-2 | 11 934–10 | Caucasian | p.Thr416Pro | CEVA | B | NT |
| DK-3 | 11 890–10 | Caucasian | c.1614+1G>A | CEVA | B | NT |
| DK-4 | kc102065 | Caucasian | p.Leu236Pro | CEVA | B§ | NT |
| DK-5 | 2531–02 | Caucasian | p.Thr416Pro | CEVA | B | NS |
| DK-6 | 13 317–12 | Caucasian | p.Val138Phe | CEVA | B | NS |
Hearing loss with enlargement of the vestibular aqueduct affecting right (R), left (L) or both ears (B).
Neither reference nor CEVA.
Phase configuration not definitive but likely to be in trans to p.Thr416Pro.
Bilateral EVA but left ear has normal hearing.
CEVA, Caucasian enlargement of the vestibular aqueduct; EVA, enlargement of the vestibular aqueduct; I, indeterminate; HL, hearing loss; NS, non-syndromic; NT, not tested.
Interestingly, the mixed reference-CEVA haplotype in subject 1805 was present on one chromosome, of Finnish ancestry, among the 1006 Caucasian-European alleles in the 1000 Genomes database (table 2). The different haplotype in subject 1805 may indicate the existence of a different pathogenic variant than is present in cis with the 12-variant CEVA haplotype in other M1 subjects. Alternatively, it could refine the pathogenic CEVA haplotype to the nine telomeric variants (figure 1C, table 2). The nine telomeric variants are located in their own region of higher LD adjacent to and including the 5′ end of SLC26A4 (figure 1C; see online supplementary table 1).
We then determined the prevalence of the 12-variant CEVA haplotype in a Danish replication cohort of six unrelated M1 EVA subjects. Six of the six Danish M1 mutation-negative chromosomes carry the CEVA haplotype in trans with a pathogenic variant of SLC26A4 (table 3). This is significantly different from the observed prevalence of the CEVA haplotype in Caucasian-European controls (p<0.0001). We conclude that there is a specific association of the CEVA haplotype with EVA in M1 subjects with a mutation of the trans allele of SLC26A4.
To explore the penetrance of the CEVA haplotype in trans with an SLC26A4 mutation, we analysed all unaffected members of M1 families in the NIH and Danish cohorts. We identified two unaffected individuals, one from the NIH cohort (2086, family 380) and one from the Danish cohort (15 283–14), who have the CEVA haplotype in trans with a mutation of SLC26A4. NIH subject 2086 had normal audiometric test results at the age of 10 years and no evidence of EVA by MRI. Danish subject 15 283–14 had normal audiometric test results at the age of 22 years and did not undergo any temporal bone imaging studies. Seventeen (85%) of 19 subjects in the NIH and Danish cohorts who have the CEVA haplotype in trans with a mutation of SLC26A4 thus have hearing loss and EVA (table 3). Therefore, the penetrance of the CEVA haplotype in trans with an SLC26A4 mutation is incomplete. This result contrasts with the complete penetrance of EVA in homozygous or compound heterozygous carriers of mutations affecting splice sites or exons of SLC26A4.3–6,8
We also determined the prevalence of the 12-variant CEVA haplotype among M2 EVA chromosomes from the NIH cohort. Since all of the M2 chromosomes already have an SLC26A4 mutation, we hypothesised that the prevalence of the CEVA haplotype would not be increased in comparison to normal control chromosomes. Four of 54 NIH M2 chromosomes have the CEVA haplotype, which is not different (p=0.0749) from the prevalence among Caucasian-European controls. We obtained a similar result (p=0.0871) using a non-conservative estimate (see ‘Statistical analyses’ in the Methods section) of the total number (57) of M2 chromosomes. When we excluded chromosomes from subjects whose ethnicity was either unknown, multiracial or not Caucasian, we obtained the same results (p=0.06) for both conservative and non-conservative estimates of numbers of chromosomes, respectively. When we compared the prevalence of the CEVA haplotype among M1 chromosomes versus M2 chromosomes in the NIH cohort, the difference was statistically significant irrespective of whether we employed conservative or non-conservative estimates of the number of M2 chromosomes (p<0.0001). These results further support the specific association of the CEVA haplotype with EVA in M1 subjects.
Association of SLC26A4-linked CEVA haplotype with M0 EVA
Since Mendelian inheritance is a rare pathogenic mechanism for M0 EVA,4 we hypothesised that the prevalence of the CEVA haplotype would be the same or only slightly increased among M0 EVA chromosomes in comparison to Caucasian-European controls. However, we observed that 11 of 126 chromosomes from the NIH M0 cohort carry the 12-variant CEVA haplotype, which is significantly higher (p=0.0042) than the observed prevalence among Caucasian-European controls. When we excluded chromosomes from subjects whose ethnicity was either unknown or not Caucasian, we obtained similar results (p=0.0129 and p=0.0117) for conservative and non-conservative estimates of numbers of chromosomes, respectively. One possible reason for this observation is that homozygosity for the CEVA haplotype can cause EVA. Indeed, four of 84 M0 EVA subjects in the NIH cohort are homozygous for the CEVA haplotype. One CEVA haplotype homozygote (subject 1693, family 258) is a singleton with an indeterminate thyroid phenotype.7 Two CEVA haplotype homozygotes are dizygotic twins (subjects 1702 and 1703, family 147) who each carry one of their CEVA haplotypes in cis with an SLC26A4 variant (p.Met775Thr) with residual pendrin activity (50%–60% of wild type).7 p.Met775Thr is a hypomorphic ‘indeterminate’ variant previously proposed to be pathogenic in trans with a mutation of SLC26A4.7 It seems likely that the combination of p.Met775Thr with homozygosity for the CEVA haplotype is aetiologic in these subjects. The fourth CEVA haplotype homozygote (subject 2246, family 443) is the oldest of three affected siblings.16 His father (subject 2244, family 443) and siblings (subjects 2247 and 2248, family 443) are heterozygous for the CEVA haplotype. The discordant segregation of EVA with homozygosity for the CEVA haplotype suggests the existence of other factors, genetic or non-genetic, which contribute to the aetiology of EVA in family 443 (online supplementary figure 1).
There are seven heterozygotes for the CEVA haplotype among five NIH M0 families, but the CEVA haplotype does not co-segregate with EVA in four of the five families (online supplementary figure 1). The EVA subject in the fifth family has no siblings, so co-segregation cannot be assessed. Therefore, a Mendelian contribution of heterozygosity for the CEVA haplotype to EVA seems unlikely in M0 subjects.
In addition to subjects 1702 and 1703 of family 147, there are 21 additional M0 EVA subjects from 17 families in the NIH cohort that carry an SLC26A4 coding or splice site variant whose pathogenic potential is indeterminate or is thought to be pathogenic only in trans with a coding or splice site mutation of SLC26A4 (table 4). Two of the 23 subjects are black, and two are multiracial. None of the 21 subjects carry the CEVA haplotype, extending our previous conclusion that their SLC26A4 variants are either benign or pathogenic only in trans with a mutation of exons or splice sites of SLC26A4.7 Moreover, this result further demonstrates that the association of the CEVA haplotype with EVA in M1 subjects is specific.
Table 4.
SLC26A4 genotypes, haplotypes and phenotypes of NIH subjects with enlargement of the vestibular aqueduct and a benign or indeterminate variant on one allele
| Family | Subject | Ancestry | Allele 1 genotype* | Allele 2 haplotype | HL/EVA† | Thyroid |
|---|---|---|---|---|---|---|
| 118 | 1166 | Caucasian | p.Leu597Ser | Reference | B | NS |
| 118 | 1167 | Caucasian | p.Leu597Ser | Reference | R | NS |
| 133 | 1281 | Caucasian | p.Arg776Cys | Reference | B | NS |
| 133 | 1282 | Caucasian | p.Arg776Cys | Reference | R | NS |
| 145 | 1422 | Caucasian | p.Leu597Ser | Reference | B | I |
| 147 | 1702 | Caucasian | p.Met775Thr‡ | CEVA | R | I |
| 147 | 1703 | Caucasian | p.Met775Thr‡ | CEVA | B | I |
| 176 | 1473 | Caucasian | p.Leu597Ser | Reference | B | NS |
| 182 | 1495 | Caucasian | p.Phe335Leu | Reference | B | NS |
| 218 | 1590 | Black | p.Val609Gly | Reference | B | I |
| 219 | 1598 | Caucasian | c.-5A>G | Reference | B | NS |
| 223 | 1643 | Caucasian | c.-66C>G | Reference | B | I |
| 229 | 1619 | Caucasian | p.Leu597Ser | Reference | B | I |
| 231 | 1639 | Caucasian | p.Arg776Cys | Reference | L | I |
| 240 | 1649 | Multiracial | p.Leu597Ser | Reference | B | I |
| 255 | 1691 | Multiracial | p.Leu597Ser | Reference | B | NS |
| 259 | 1726 | Black | c.-60A>G | Reference§ | L | I |
| 278 | 1791 | Caucasian | p.Leu597Ser | Reference | B | NS |
| 341 | 1986 | Caucasian | p.Gly6Val;p.Leu597Ser | Reference | B | NS |
| 341 | 1981 | Caucasian | p.Leu597Ser | Reference | B | I |
| 388 | 2106 | Caucasian | p.Asp324Tyr | Reference | B | NS |
| 388 | 2107 | Caucasian | p.Asp324Tyr | Reference | B | NS |
| 388 | 2108 | Caucasian | p.Asp324Tyr | Reference | B | NS |
Alleles are considered benign, indeterminate or pathogenic only in trans with a mutation affecting coding regions or splice sites of SLC26A4.
Hearing loss with enlargement of the vestibular aqueduct affecting right (R), left (L) or both ears (B).
p.Met775Thr is in cis and trans with CEVA; the subject is homozygous for CEVA.
p.Val609Gly carried on allele 2.
I, indeterminate; NS, non-syndromic.
Other SLC26A4-linked haplotypes in M0 EVA subjects
Four M0 subjects from three families have haplotypes with a combination of reference and CEVA genotypes. Two affected monozygotic twins in family 379 have the reference haplotype GTTTGCATATTA and a CEVA-reference combination haplotype ACACATG-GTTA (CEVA genotypes are underlined) which is present in nine European chromosomes in 1000 Genomes (table 2). A third subject (1422) has the reference haplotype and the CEVA-reference combination haplotype GTTTGTG-GT-A. This haplotype was not observed in 1000 Genomes (table 2). The reference-CEVA combination haplotype in these three subjects may be coincidental and unrelated to EVA, since they were not present in trans with a mutation or another CEVA haplotype. In contrast, another subject (1470) has two reference-CEVA combination haplotypes, GTTCATG-GC-C and GCACATG-GC-C. The former haplotype is composed of the nine telomeric variants of the CEVA haplotype and is identical to that observed in M1 subject 1805 and a single Finnish chromosome in 1000 Genomes (table 2). The latter haplotype carries the 11 telomeric variants of the CEVA haplotype and is identical to that observed in a single Iberian-Spanish chromosome in 1000 Genomes. Taken together, these results suggest that the CEVA haplotype can be refined to its nine telomeric variants (figure 1C).
Candidate pathogenic sequence variants
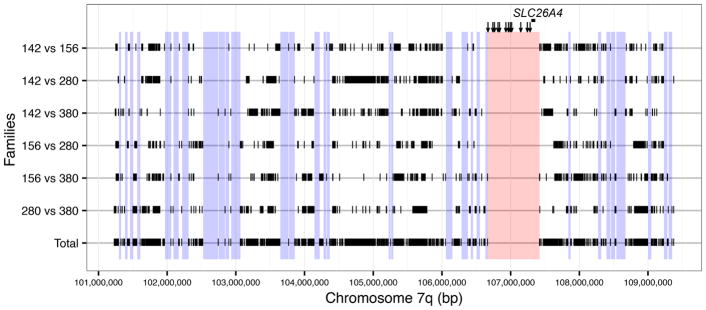
One or more of the variants comprising the CEVA haplotype could be pathogenic alone or in combination. It is also possible that they are in LD with a true pathogenic variant(s) that we did not detect because it was not annotated in 1000 Genomes or had a MAF >0.05. Therefore, we performed MPS to search for additional variants linked to the CEVA haplotype region in three additional simplex trios. We also performed MPS of DNA from parents from four initial M1 multiplex families (142, 156, 280 and 380) to define the meiotic phase configuration of variants with the CEVA haplotype. To define the regions to be sequenced by MPS, we used homozygosity haplotype mapping (figure 2) to delineate chromosomal segments that were not shared among the M1 families and could thus be excluded from MPS analysis.24 The longest contiguous segment (756 kb) for MPS included the 12 CEVA variants and the entire SLC26A4 gene. We designed a second set of Agilent XT SureSelect probes for MPS to cover this segment as well as the other top 32 longest (>35 kb) shared segments. We also included exons and splice sites for FOXI1 and KCNJ10, since Yang et al10,11 reported potential pathogenic roles for these genes in EVA. We did not find any exonic or splice site variants in FOXI1 or KCNJ10. We could not identify any variants, with MAF <0.1 or not annotated in the 1000 Genomes database, in cis configuration with the 12 CEVA variants.
Figure 2.
Homozygosity haplotype analysis of alleles of SLC26A4 with no detectable mutations in the coding regions or splice sites in M1 families. Homozygous SNPs in the 8.218 Mb smallest region of overlap (figure 1A) were compared between pairs of chromosomes from four M1 families. Black vertical tick marks (|) show unmatched homozygous SNPs between each pair of chromosomes (ie, GG vs CC). Segments bounded by two tick marks indicate regions potentially shared by the two families. All unmatched pairwise homozygous SNPs from the six comparisons are shown at the bottom (‘Total’), delineating intervening regions that may be shared among the four chromosomes from the four families. The longest segment (shown in red) includes the Caucasian enlarged vestibular aqueduct haplotype markers (vertical arrows) and SLC26A4. Other segments longer than 35 kb (shown in blue) were analysed by massively parallel sequencing.
We used the CADD analysis to estimate the pathogenic potential of each of the 12 CEVA sequence variants.25 Any variant with C≥20 is considered to be within the top 1% of variants likely to have a deleterious effect.25 The scores for our variants range from 0.071 to 21.6 (table 1). The variant (rs199915614) with the highest score (C=21.6) is located within a sequence with partial similarity to the consensus binding sequence for the FOX family of transcription factors (RegulomeDB accessed on 11/4/16). However, we have thus far not detected binding of FOXI1 protein to the reference sequence in an electrophoretic mobility shift assay.
Discussion
We have identified a haplotype, which we have designated the CEVA haplotype, composed of 12 variants upstream of wildtype exonic and splice site sequence of SLC26A4, associated with EVA in patients with a mutation of the coding regions or splice sites of the trans allele. We did not detect any other potential pathogenic variants in cis with the CEVA haplotype by MPS. It is possible that we missed the true pathogenic variant due to the technical caveats of assembling the short reads produced by this methodology. However, we were previously unable to detect copy number variants by comparative genomic hybridisation microarray analyses of SLC26A4 and its flanking regions in M0 and M1 EVA subjects.4 We conclude that one or more of the 12 variants of the CEVA haplotype, either alone or in combination, exert a pathogenic effect on the expression or function of SLC26A4. The mixed reference-CEVA haplotype in subjects 1470 and 1805 suggests that it is one or more of the nine telomeric variants of the CEVA haplotype that are pathogenic (figure 1C). It is possible that these variants act in combination with other variants that have a minor allele frequency greater than 0.1 and thus escaped our analysis.
The phenotype in M1 patients is generally less severe than in M2 patients. Since our current study shows that nearly all of these M1 patients carry the CEVA haplotype, this indicates that a modest effect of the CEVA haplotype on SLC26A4 expression underlies this genotype–phenotype correlation. This difference in expression would be difficult to detect with conventional measures of RNA levels. Furthermore, alternative transcription initiation sites and alternative splicing resulting in a family of shorter transcripts may contribute to complex regulation and function of the ‘full-length’ transcript.26 Finally, inner ear tissue from human patients is not accessible for research purposes, and peripheral leucocytes express SLC26A4 at low levels that cannot be reliably measured or compared (unpublished observations). Other potential surrogate tissues for measurement of SLC26A4 expression levels include the thyroid gland, but the mature thyroid may not be an appropriate surrogate for embryonic endolymphatic sac.27,28 In our view, an invasive procedure to obtain thyroid or other tissue is not justified.
We presently lack data supporting the hypothesis that the CEVA haplotype affects expression of SLC26A4. However, rs199915614 had the highest CADD score of any of the variants associated with the CEVA haplotype, rendering it a top candidate for exploration of a pathogenic role in EVA. The reference sequence, TGTTCGA, matches five (underlined) of seven nucleotide positions of the consensus binding sequence, TRTTKRY (R=A/G; K=G/T; Y=C/T), reported for FOXI1 proteins.11 rs199915614 is a deletion of the first T in the reference sequence, resulting in CGTTCGA in chromosomes with the CEVA haplotype and a reduced match to the consensus binding sequence. Although we have not detected binding of FOXI1 protein to the reference sequence, this may be a false-negative result due to experimental procedures and conditions or the affected sequence may have some other functional role. Alternatively, rs199915614 may not be pathogenic.
This study’s elucidation of the contribution of the CEVA haplotype to NSEVA will have an important impact on routine genetic testing for EVA. The detection of the CEVA haplotype can provide a conclusive diagnostic result for most Caucasian patients with EVA previously classified as having only one (M1) detectable mutation of SLC26A4. Moreover, our study advances the interpretation of SLC26A4 test results in which a single hypomorphic variant is detected.7 We observed that the only indeterminate SLC26A4 coding variant (p.Met775Thr) in trans with the CEVA haplotype is also in cis with the CEVA haplotype (table 4). Similarly, there are two unrelated Danish EVA subjects who are heterozygous for the indeterminate variant p.Glu29Gln and homozygous for the CEVA haplotype (not shown). These observations suggest that hypomorphic variants such as p.Met775Thr and p.Glu29Gln can be pathogenic when they are in cis with the CEVA haplotype. Conversely, none of the other indeterminate or benign variants were observed in trans with the CEVA haplotype, indicating that they are coincidental non-pathogenic variants in those subjects. Finally, detection of homozygosity for the CEVA haplotype may even provide a diagnostic result for patients with M0 EVA. We expect the diagnostic yield and prognostic utility of sequence analysis of SLC26A4 exons and splice sites to be markedly increased by the addition of testing for the CEVA haplotype.
We previously reported two unilateral EVA subjects in the NIH cohort with two mutant alleles of SLC26A4, leading us to propose an expansion of the indications for SLC26A4 testing to include unilateral EVA.8 In our current study, two (18%) of 11 NIH M1 subjects carrying the CEVA haplotype had unilateral hearing loss and EVA (table 3). These results further support the diagnostic utility of SLC26A4 testing for unilateral EVA.
The heterozygous carrier frequency (5.567%) of the CEVA haplotype is high among Caucasian controls. The CEVA haplotype defines, to our knowledge, the most common pathogenic allele of any gene implicated in hereditary hearing loss in these populations. In contrast, we did not observe the CEVA haplotype in East Asian populations, such as Koreans and Japanese, in 1000 Genomes (table 2). This may explain why M1 genotypes are rare in Japanese and Korean patients with EVA.3,5–9,29–32 If there are EVA-associated haplotypes in East Asians, they likely have a lower prevalence or lower penetrance.
In summary, this study elucidates the genetic architecture of non-syndromic hearing loss associated with EVA, the most common radiological abnormality observed in ears of children with sensorineural hearing loss. Elucidating the mechanistic link between the CEVA haplotype and its phenotypic associations may reveal therapeutic opportunities to restore SLC26A4 expression and preserve hearing in ears with EVA.
Supplementary Material
Acknowledgments
We thank the study subjects for their participation, Lone Sandbjerg Hindbæk at the Kennedy Centre, Glostrup, for haplotyping the Danish samples, NIH Clinical Center staff and NIDCD clinic staff for support of the study and subjects, NIDCD colleagues for advice and discussion, and Steven Boyden and Dennis Drayna for critical review of this manuscript. This work used the computational resources of the NIH HPC Biowulf cluster.
Funding Work in the NIH authors’ laboratories was supported by NIH Intramural Research Program funds Z01-DC000060, Z01-DC-000086, Z01-DC-000046, Z01-DC000039 and Z01-LM000097.
Footnotes
Contributors PC, TM, KH, NDR, IR and RJM contributed to molecular and functional analyses. PC, TM, KH, DSR and RJM contributed to bioinformatic analyses. JSR, JAM, LT and AJG generated and reviewed clinical data. PC, TM, KH, DSR, EMG, AAS, TBF, RJM and AJG contributed to design of experiments and interpretation of molecular and bioinformatic data. PC, TM, KH, RJM and AJG contributed to the first draft of the manuscript. All authors critically reviewed the manuscript. AJG conceptualised the study.
Competing interests None declared.
Ethics approval Combined Neuroscience Institutional Review Board, National Institutes of Health (NIH), Bethesda, Maryland; Danish Research Ethical Committee (KF 01-108/03, 11-112/04, KF120/03).
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Valvassori GE, Clemis JD. The large vestibular aqueduct syndrome. Laryngoscope. 1978;88:723–8. doi: 10.1002/lary.1978.88.5.723. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Choi BY, King KA, Zalewski CK, Muskett J, Chattaraj P, Shawker T, Reynolds JC, Butman JA, Brewer CC, Wangemann P, Alper SL, Griffith AJ. SLC26A4 genotypes and phenotypes associated with enlargement of the vestibular aqueduct. Cell Physiol Biochem. 2011;28:545–52. doi: 10.1159/000335119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pryor SP, Madeo AC, Reynolds JC, Sarlis NJ, Arnos KS, Nance WE, Yang Y, Zalewski CK, Brewer CC, Butman JA, Griffith AJ. SLC26A4/PDS genotype-phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J Med Genet. 2005;42:159–65. doi: 10.1136/jmg.2004.024208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi BY, Madeo AC, King KA, Zalewski CK, Pryor SP, Muskett JA, Nance WE, Butman JA, Brewer CC, Griffith AJ. Segregation of enlarged vestibular aqueducts in families with non-diagnostic SLC26A4 genotypes. J Med Genet. 2009;46:856–61. doi: 10.1136/jmg.2009.067892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King KA, Choi BY, Zalewski C, Madeo AC, Manichaikul A, Pryor SP, Ferruggiaro A, Eisenman D, Kim HJ, Niparko J, Thomsen J, Butman JA, Griffith AJ, Brewer CC. SLC26A4 genotype, but not cochlear radiologic structure, is correlated with hearing loss in ears with an enlarged vestibular aqueduct. Laryngoscope. 2010;120:384–9. doi: 10.1002/lary.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rose J, Muskett JA, King KA, Zalewski CK, Chattaraj P, Butman JA, Kenna MA, Chien WW, Brewer CC, Griffith AJ. Hearing loss associated with enlarged vestibular aqueduct and zero or one mutant allele of SLC26A4. Laryngoscope. 2017;127:E238–E243. doi: 10.1002/lary.26418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi BY, Stewart AK, Madeo AC, Pryor SP, Lenhard S, Kittles R, Eisenman D, Kim HJ, Niparko J, Thomsen J, Arnos KS, Nance WE, King KA, Zalewski CK, Brewer CC, Shawker T, Reynolds JC, Butman JA, Karniski LP, Alper SL, Griffith AJ. Hypo-functional SLC26A4 variants associated with nonsyndromic hearing loss and enlargement of the vestibular aqueduct: genotype-phenotype correlation or coincidental polymorphisms? Hum Mutat. 2009;30:599–608. doi: 10.1002/humu.20884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattaraj P, Reimold FR, Muskett JA, Shmukler BE, Chien WW, Madeo AC, Pryor SP, Zalewski CK, Butman JA, Brewer CC, Kenna MA, Alper SL, Griffith AJ. Use of SLC26A4 mutation testing for unilateral enlargement of the vestibular aqueduct. JAMA Otolaryngol Head Neck Surg. 2013;139:907–13. doi: 10.1001/jamaoto.2013.4185. [DOI] [PubMed] [Google Scholar]
- 9.Madeo AC, Manichaikul A, Reynolds JC, Sarlis NJ, Pryor SP, Shawker TH, Griffith AJ. Evaluation of the thyroid in patients with hearing loss and enlarged vestibular aqueducts. Arch Otolaryngol Head Neck Surg. 2009;135:670–6. doi: 10.1001/archoto.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang T, Gurrola JG, Wu H, Chiu SM, Wangemann P, Snyder PM, Smith RJ. Mutations of KCNJ10 together with mutations of SLC26A4 cause digenic nonsyndromic hearing loss associated with enlarged vestibular aqueduct syndrome. Am J Hum Genet. 2009;84:651–7. doi: 10.1016/j.ajhg.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang T, Vidarsson H, Rodrigo-Blomqvist S, Rosengren SS, Enerback S, Smith RJ. Transcriptional control of SLC26A4 is involved in Pendred syndrome and nonsyndromic enlargement of vestibular aqueduct (DFNB4) Am J Hum Genet. 2007;80:1055–63. doi: 10.1086/518314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landa P, Differ AM, Rajput K, Jenkins L, Bitner-Glindzicz M. Lack of significant association between mutations of KCNJ10 or FOXI1 and SLC26A4 mutations in Pendred syndrome/enlarged vestibular aqueducts. BMC Med Genet. 2013;14:85. doi: 10.1186/1471-2350-14-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi BY, Alper SL, Griffith AJ. The c.-103T>C variant in the 5′-UTR of SLC26A4 gene: a pathogenic mutation or coincidental polymorphism? Hum Mutat. 2009;30:1471–70. doi: 10.1002/humu.21097. [DOI] [PubMed] [Google Scholar]
- 14.Jonard L, Niasme-Grare M, Bonnet C, Feldmann D, Rouillon I, Loundon N, Calais C, Catros H, David A, Dollfus H, Drouin-Garraud V, Duriez F, Eliot MM, Fellmann F, Francannet C, Gilbert-Dussardier B, Gohler C, Goizet C, Journel H, Mom T, Thuillier-Obstoy MF, Couderc R, Garabédian EN, Denoyelle F, Marlin S. Screening of SLC26A4, FOXI1 and KCNJ10 genes in unilateral hearing impairment with ipsilateral enlarged vestibular aqueduct. Int J Pediatr Otorhinolaryngol. 2010;74:1049–53. doi: 10.1016/j.ijporl.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Wu CC, Lu YC, Chen PJ, Yeh PL, Su YN, Hwu WL, Hsu CJ. Phenotypic analyses and mutation screening of the SLC26A4 and FOXI1 genes in 101 Taiwanese families with bilateral nonsyndromic enlarged vestibular aqueduct (DFNB4) or Pendred syndrome. Audiol Neurootol. 2010;15:57–66. doi: 10.1159/000231567. [DOI] [PubMed] [Google Scholar]
- 16.Muskett JA, Chattaraj P, Heneghan JF, Reimold FR, Shmukler BE, Brewer CC, King KA, Zalewski CK, Shawker TH, Butman JA, Kenna MA, Chien WW, Alper SL, Griffith AJ. Atypical patterns of segregation of familial enlargement of the vestibular aqueduct. Laryngoscope. 2016;126:E240–E247. doi: 10.1002/lary.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rendtorff ND, Schrijver I, Lodahl M, Rodriguez-Paris J, Johnsen T, Hansén EC, Nickelsen LA, Tümer Z, Fagerheim T, Wetke R, Tranebjaerg L. SLC26A4 mutation frequency and spectrum in 109 Danish Pendred syndrome/DFNB4 probands and a report of nine novel mutations. Clin Genet. 2013;84:388–91. doi: 10.1111/cge.12074. [DOI] [PubMed] [Google Scholar]
- 18.Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, Berg JS, Biswas S, Bowling KM, Conlin LK, Cooper GM, Dorschner MO, Dulik MC, Ghazani AA, Ghosh R, Green RC, Hart R, Horton C, Johnston JJ, Lebo MS, Milosavljevic A, Ou J, Pak CM, Patel RY, Punj S, Richards CS, Salama J, Strande NT, Yang Y, Plon SE, Biesecker LG, Rehm HL. Performance of AC MG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98:1067–76. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL AC MG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Li M, Hakonarson H. ANOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazawa H, Kato M, Awata T, Kohda M, Iwasa H, Koyama N, Tanaka T, Huqun KS, Okazaki Y, Hagiwara K. Homozygosity haplotype allows a genomewide search for the autosomal segments shared among patients. Am J Hum Genet. 2007;80:1090–102. doi: 10.1086/518176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishio A, Ito T, Cheng H, Fitzgerald TS, Wangemann P, Griffith AJ. Slc26a4 expression prevents fluctuation of hearing in a mouse model of large vestibular aqueduct syndrome. Neuroscience. 2016;329:74–82. doi: 10.1016/j.neuroscience.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi BY, Kim HM, Ito T, Lee KY, Li X, Monahan K, Wen Y, Wilson E, Kurima K, Saunders TL, Petralia RS, Wangemann P, Friedman TB, Griffith AJ. Mouse model of enlarged vestibular aqueducts defines temporal requirement of Slc26a4 expression for hearing acquisition. J Clin Invest. 2011;121:4516–25. doi: 10.1172/JCI59353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Sanneman JD, Harbidge DG, Zhou F, Ito T, Nelson R, Picard N, Chambrey R, Eladari D, Miesner T, Griffith AJ, Marcus DC, Wangemann P. SLC26A4 targeted to the endolymphatic sac rescues hearing and balance in Slc26a4 mutant mice. PLoS Genet. 2013;9:e1003641. doi: 10.1371/journal.pgen.1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park HJ, Lee SJ, Jin HS, Lee JO, Go SH, Jang HS, Moon SK, Lee SC, Chun YM, Lee HK, Choi JY, Jung SC, Griffith AJ, Koo SK. Genetic basis of hearing loss associated with enlarged vestibular aqueducts in Koreans. Clin Genet. 2005;67:160–5. doi: 10.1111/j.1399-0004.2004.00386.x. [DOI] [PubMed] [Google Scholar]
- 30.Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, Kim HN, Moon SK, Abe S, Tukamoto K, Riazuddin S, Kabra M, Erdenetungalag R, Radnaabazar J, Khan S, Pandya A, Usami SI, Nance WE, Wilcox ER, Riazuddin S, Griffith AJ. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003;40:242–8. doi: 10.1136/jmg.40.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukamoto K, Suzuki H, Harada D, Namba A, Abe S, Usami S. Distribution and frequencies of PDS (SLC26A4) mutations in Pendred syndrome and nonsyndromic hearing loss associated with enlarged vestibular aqueduct: a unique spectrum of mutations in Japanese. Eur J Hum Genet. 2003;11:916–22. doi: 10.1038/sj.ejhg.5201073. [DOI] [PubMed] [Google Scholar]
- 32.Pryor SP, Demmler GJ, Madeo AC, Yang Y, Zalewski CK, Brewer CC, Butman JA, Fowler KB, Griffith AJ. Investigation of the role of congenital Cytomegalovirus infection in the etiology of enlarged vestibular aqueducts. Arch Otolaryngol Head Neck Surg. 2005;131:388–92. doi: 10.1001/archotol.131.5.388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.