Abstract
The three-dimensional structures of avian H5 and swine H9 influenza hemagglutinins (HAs) from viruses closely related to those that caused outbreaks of human disease in Hong Kong in 1997 and 1999 were determined bound to avian and human cell receptor analogs. Emerging influenza pandemics have been accompanied by the evolution of receptor-binding specificity from the preference of avian viruses for sialic acid receptors in α2,3 linkage to the preference of human viruses for α2,6 linkages. The four new structures show that HA binding sites specific for human receptors appear to be wider than those preferring avian receptors and how avian and human receptors are distinguished by atomic contacts at the glycosidic linkage. α2,3-Linked sialosides bind the avian HA in a trans conformation to form an α2,3 linkage-specific motif, made by the glycosidic oxygen and 4-OH of the penultimate galactose, that is complementary to the hydrogen-bonding capacity of Gln-226, an avian-specific residue. α2,6-Linked sialosides bind in a cis conformation, exposing the glycosidic oxygen to solution and nonpolar atoms of the receptor to Leu-226, a human-specific residue. The new structures are compared with previously reported crystal structures of HA/sialoside complexes of the H3 subtype that caused the 1968 Hong Kong Influenza virus pandemic and analyzed in relation to HA sequences of all 15 subtypes and to receptor affinity data to make clearer how receptor-binding sites of HAs from avian viruses evolve as the virus adapts to humans.
During the past century, influenza viruses with hemagglutinin (HA) glycoproteins from 3 of the 15 influenza A virus subtypes (H1–H15) emerged from avian or animal hosts to cause worldwide epidemics: in 1918, H1; in 1957, H2; and in 1968, H3 (1). Extensive surveys of H1, H2, and H3 viruses isolated from avian, swine, equine, and human sources show a correlation of receptor-binding preference with species of origin: human isolates prefer terminal sialic acids of glycoprotein and glycolipid receptors in α2,6 linkage to galactose, avian and equine prefer α2,3 linkages, and viruses from swine bind to both (2–4). The source of this specificity may derive from the selective pressure of an abundance of α2,6 linkages on lung and airway epithelial cells in humans in which the infection is respiratory and of α2,3 linkages on intestinal cells of birds in which the infection is enteric (3, 5, 6). The presence of potentially inhibitory mucins rich in α2,3 linkages in human lungs and glycoproteins with sialic acid in α2,6 linkages in horse sera also may be a factor in the selection of linkage specificity (7–9).
In 1997, an avian virus containing an H5 subtype HA with α2,3 linkage preference caused 18 cases of human influenza in Hong Kong, 6 of them fatal (10–13), and in 1999, a very similar virus containing the HA of the H9 subtype with α2,6 specificity (14) caused milder disease in 7 people in Hong Kong (15, 16) and Southern China (17). Neither of these incidents of avian viruses infecting humans resulted in widespread disease (18).
We report here the three-dimensional structures of HAs from avian and swine viruses closely related to those causing the H5 and H9 human outbreaks, each complexed with two sialylated pentasaccharide analogues of avian and human cell surface receptors. We compare these avian and swine HA–receptor complexes with complexes formed by a human H3 HA from the virus that caused the 1968 human pandemic (19). The sialic acid receptor is bound in the same orientation in sites that are conserved largely in all three HAs but that differ in two residues of a loop that forms one edge of the sites; Leu or Gln at residue 226 and Ser or Gly at residue 228. The new structures suggest how avian and human receptors are distinguished: avian H5 HA hydrogen bonds through Gln-226 to the glycosidic oxygen that is exposed in a trans conformation in the α2,3 sialic acid-to-galactose link; Leu-226 of swine H9 HA makes nonpolar interactions with the C6 atom of the α2,6 linkage, in which the glycosidic oxygen is exposed to solvent by a cis conformation of the linkage. HA receptor-binding sites favoring α2,6 linkages (H9 swine and H3 human) are also “wider” than the site in the avian HA (H5 avian) that favors α2,3 linkages.
Materials and Methods
Hemagglutinin Purification, Crystal Growth, and Ligand Soaking.
Bromelain-released hemagglutinins (BHA) from A/Duck/Singapore/3/97 (H5N1) virus and A/Swine/Hong Kong/9/98 (H9N2) virus were prepared as described previously (Y.H., D.J.S., J.J.S., and D.C.W., unpublished data). Crystals were grown and cryoprotected as reported previously (Y.H., et al., unpublished data). Crystals to be soaked in 16 mM LSTa or LSTc (BioCarb, Lund, Sweden) were washed first in 200 μl of well solution; subsequent soaking steps all contained ligand.
Data Collection, Structure Determination, and Refinement.
Diffraction data were collected at 14BMC at APS, 100 K. All data sets were processed with denzo and scalepack (20). LSTa and LSTc were modeled with 2Fo-Fc and Fo-Fc Fourier maps based on phases calculated from the H5 and H9 HA coordinates (Y.H. et al., unpublished data). Sialic acid (Sia) binds similarly in the four structures. Modeling of galactose was based on three density features: a planar pyranose ring, linkages to Sia-1 and GlcNAc, a protrusion normal to the pyranose ring corresponding to the axial 4-OH. The structures were refined with cns (21); statistics are shown in Table 2.
Table 2.
Crystallographic data statistics
| Data collection | H5/LSTa | H5/LSTc | H9/LSTa | H9/LSTc |
|---|---|---|---|---|
| Resolution (Å) | 40.0–2.4 | 40.0–2.4 | 40.0–2.4 | 40.0–2.4 |
| Unique reflections | 30,623 | 29,860 | 29,860 | 38,054 |
| Completeness (%), redundancy, I/〈σ〉 | 92.7 (94.0), 7, 8.8 | 90.6 (87.5), 9, 17.8 | 90.6 (92.9), 9, 17.8 | 97.7 (91.2), 7, 13.9 |
| Rmerge (%) | 10.6 (70.9) | 5.7 (25.9) | 5.7 (36.3) | 6.7 (29.1) |
| Refinement | ||||
| Rcryst (%), Rfree (%) | 24.0 (58.8), 29.6 (63.4) | 22.1 (26.7), 25.1 (32.0) | 20.0 (31.2), 22.7 (27.1) | 20.1 (42.5), 23.5 (45.1) |
| rmsd bond lengths (Å), angles (°) | 0.009, 1.5 | 0.007, 1.3 | 0.006, 1.3 | 0.006, 1.3 |
| Number of protein, water, and carbohydrate atoms | 3,738, 216, 144 | 3,738, 290, 119 | 3,725, 158, 172 | 3,725, 142, 208 |
H5 crystallized in space group R32 (a = 131.6 Å, c = 254.1 Å) and H9 in space group P63 (a = 108.5 Å, c = 149.3 Å). The statistics for the last shell is in parentheses. The higher R and Rfree values for the H5/LSTa complex are due to poorer quality of the diffraction data.
Results and Discussion
Common Features of the HA/Receptor Complexes.
By using x-ray crystallography, the structures of the H5 HA from the A/Duck/Singapore/3/97 virus and the H9 HA from A/Swine/Hong Kong/9/98 that have 26- and 39-aa sequence differences relative to the HAs from the 1997 and 1999 human isolates from Hong Kong [aligned sequences and substitution locations are shown in Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org (refs. 10–13 and 16); K. Cameron and A. J. Hay (H5) and D. Markwell and K. F. Shortridge (H9), personal communications] have been determined to 1.9 Å and 1.8 Å resolution, respectively (Y.H. et al., unpublished data). We report here the structures to 2.4 Å resolution of both HAs complexed with the two sialo-pentasaccharides LSTa and LSTc that are natural sialosides from human milk, which have sequences characteristic of the polylactosamine-like repeats (Gal-GlcNAc) containing the three terminal saccharides (Sia-Gal-GlcNAc) common to complex oligosaccharides on glycoproteins and glycolipids (19).
The terminal sialic acid moiety (Sia-1) of the pentasaccharides is positioned almost identically in all four structures (Fig. 1 a and b) and essentially as observed earlier for the human H3 HA in complexes with trisaccharides, designed inhibitors, and sialo-pentasaccharides (Fig. 1c) (19, 22–24). Four hydrogen bonds (dashed red in Fig. 1) and contacts with four residues conserved in most influenza A HAs, Tyr-98, Ser-136, Trp-153, and Leu-194, are found in all of the complexes. In the H5 and H9 complexes, a water molecule bridges a hydrogen-bonding contact between the 9-OH of Sia-1 and the main-chain amide of Gly-228 that, in the H3 complex, is a direct contact between Sia-1 9-OH and OH-group of Ser-228 (Fig. 1). Small differences in the position of the sialic acids apparently are caused by contacts with six other residues (133a, 155, 183, 186, 190, and 226) that vary in these HAs (Fig. 1; Table 1).
Figure 1.
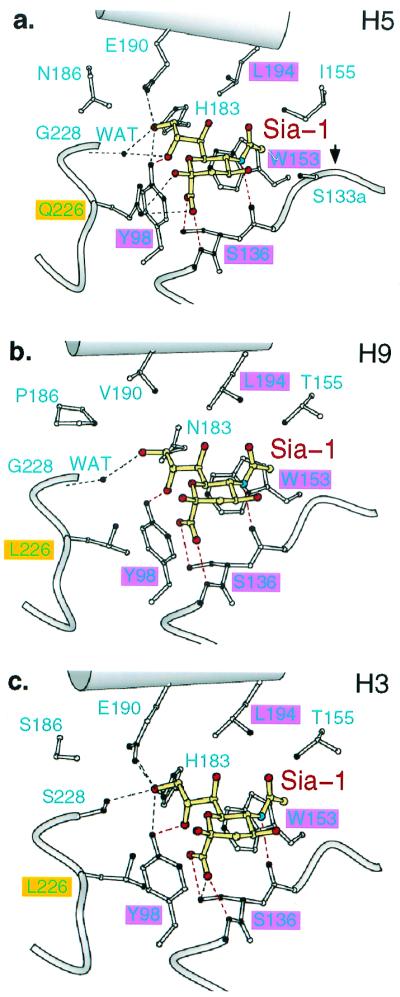
Conserved and variable interactions of H5 avian, H9 swine, and H3 human influenza virus subtype HAs with the sialic acid of pentasialosides. (a) 1997 H5 avian HA. (b) 1999 H9 swine HA. (c) 1968 H3 human HA. Red highlighted areas, conserved residues; yellow, sialic acid (Sia-1); red, oxygen; blue, nitrogen; filled HA atoms contact Sia-1; dashed lines, hydrogen bonds; red dashes, conserved hydrogen bonds.
Table 1.
Atomic contacts between sialosides (LSTa, α2,3, and LSTc α2,6) and H5 (avian), H9 (swine), and H3 (human) subtype influenza virus HAs
| H3
human
|
H5 avian
|
H9 swine
|
||||
|---|---|---|---|---|---|---|
| 2,3 | 2,6 | 2,3 | 2,6 | 2,3 | 2,6 | |
| 98Y | Y-Sia1 | Y-Sia1 | Y-Sia1 | Y-Sia1 | Y-Sia1 | Y-Sia1 |
| 131 | S-Glc5 | |||||
| 133 | S-Glc5 | |||||
| 133a | S-Sia1 | S-Sia1 | ||||
| 134 | G-Sia1 | G-Sia1 | G-Sia1 | G-Sia1 | G-Sia1, G-Glc5 | |
| 135mc | G-Sia1 | G-Sia1 | V-Sia1 | V-Sia1 | T-Sia1 | T-Sia1, T-Glc5 |
| 136S | S-Sia1 | S-Sia1 | S-Sia1 | S-Sia1 | S-Sia1 | S-Sia1 |
| 137mc | N-Sia1 | N-Sia1 | S-Sia1 | S-Sia1 | K-Sia1, K-Gal2 | K-Sia1 |
| 153W | W-Sia1 | W-Sia1 | W-Sia1 | W-Sia1 | W-Sia1 | W-Sia1 |
| 155 | T-Sia1 | T-Sia1 | I-Sia1 | I-Sia1 | T-Sia1 | T-Sia1, T-Gal4 |
| 156 | K-Glc5 | Q-Gal4 | ||||
| 158 | G-Glc5 | |||||
| 165 | CHO-NAG3 | |||||
| 183 | H-Sia1 | H-Sia1 | H-Sia1 | H-Sia1 | ||
| 186 | N-Sia1 | P-Sia1 | P-Sia1 | |||
| 189 | Q-Gal4 | |||||
| 190 | E-Sia1 | E-Sia1 | E-Sia1, E-Gal2 | E-Sia1 | V-Sia1 | V-Sia1, V-NAG3 |
| 193 | S-Glc5 | N-NAG3, N-Gal4 | ||||
| 194L | L-Sia1 | L-Sia1 | L-Sia1 | L-Sia1 | L-Sia1 | L-Sia1, L-Gal4 |
| 222 | W-Gal2, W-NAG3 | |||||
| 225 | G-Gal2 | G-Gal2 | ||||
| 226 | L-Sia1, L-Gal2 | L-Sia1, L-Gal2 | Q-Sia1, Q-Gal2 | Q-Sia1 | L-Sia1, L-Gal2 | L-Sia1, L-Gal2 |
| 228 | S-Sia1 | S-Sia1 | H2O-Sia1 | H2O-Sia1 | H2O-Sia1 | H2O-Sia1 |
| Receptor | ||||||
| Sia1 | NAG3 | NAG3, Gal4, Glc5 | ||||
| Gal2 | NAG3 | |||||
Conserved interactions are underlined. Contacts with hydrogen bonds are in bold. The first column is the sequence according to H3 (X31) numbering. The first elements in each entry represent the amino acid at the corresponding position. Intramolecular interactions within the receptor are listed at the bottom of the table. At position 165 in H3/LSTa, the interaction is between NAG3 and the N linked CHO at Asn-165 across the trimeric interface. At position 228 in H5 and H9, a water molecule mediates the hydrogen bond from Sia1 and Gly-228.
Avian Receptor Binding by H5 Avian Virus HA.
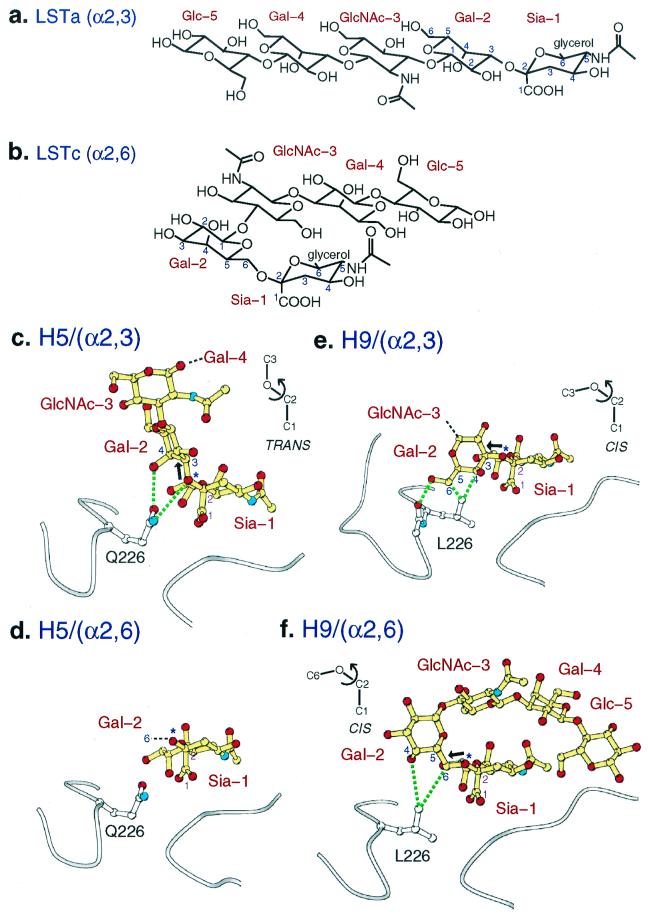
The H5 avian HA, which contains Gln-226/Gly-228 and prefers α2,3 linkages (25), binds to α2,3 LSTa (Fig. 2 a and b) through hydrogen bonds to the glycosidic oxygen of the linkage and the axial, 4-OH of Gal-2 to the amine and carbonyl groups of Gln-226 (dotted lines in Fig. 2c). The glycosidic oxygen is exposed to Gln-226 because LSTa binds in a trans conformation (Fig. 2c Inset) around the bond between the C2 and glycosidic oxygen (star in Fig. 2c). The trans conformation projects the Gal-2 ring up and out of the site (bold arrow in Fig. 2c), away from the protein surface. The orientation of the Gal-2 ring (edge-on in Fig. 2c) around the bond from the glycosidic oxygen (star in Fig. 2c) to the C3 atom of galactose is determined by steric factors within the ligand, which avoid interactions between the 3- and 4-OH groups of Gal-2 and the ring of sialic acid. GlcNAc-3 is positioned outside and in front of the binding pocket and makes no contacts with the protein (Fig. 2c). Gal-4 and Glc-5 are not visible in the electron density map.
Figure 2.
The interaction of H5 and H9 influenza virus subtype HAs with avian α2,3 and human α2,6 sialosides. (a and b) Chemical formula of the pentasaccharides LSTa and LSTc. (c) Avian H5 HA with LSTa (α2,3) pentasaccharide bound. The α2,3 linkage is in trans conformation (bold arrow points up, 180° from the bond to the carboxylate of Sia-1). Hydrogen bonds (dashed lines) from Gln-226 to the α2,3-specific motif [4-OH of Gal-1 and glycosidic-O (at *) of trans linkage]. (d) Avian H5 HA with LSTc (α2,6) pentasaccharide bound. Only sialic acid of the ligand is ordered. (e) Swine H9 HA with LSTa (α2,3) pentasaccharide bound. Only two saccharides are ordered. The bound α2,3 linkage conformation is in cis conformation [bold arrow points nearly horizontal, 60° from the bond to the carboxylate of Sia-1; compare with bold arrow (trans) in c]. Contacts and hydrogen bonds (dotted lines) to Leu-226 and the carbonyl-O of residue 225 are shown. (f) Swine H9 HA with LSTc (α2,6) pentasaccharide bound. All five saccharides are ordered. The α2,6 linkage is in a cis conformation (bold arrow points nearly horizontal).
Thus, H5 avian HA contacts only the sialic acid, the α2,3-linkage glycosidic oxygen, and the axial 4-OH of Gal-2. The trans conformation of the α2,3 glycosidic linkage forms a motif of the glycosidic oxygen and the axial 4-OH of Gal-2 that is complementary to the hydrogen-bonding capacity of Gln-226 (Fig. 2c). [Both cis and trans are low-energy conformations of α2,3 linkages, and both have been observed in studies of sialosides in solution and bound to other proteins (26–31).]
Human Receptor Binding by H5 Avian Virus HA.
Electron density for α2,6 LSTc bound to the H5 avian HA (the nonpreferred combination) shows a poorly defined sialic acid moiety: three segments of density are visible, representing C2 and the attached carboxylate group, C5 and its N-acetyl substituent, and C6 with the glycerol group (Fig. 2d). The remaining four residues of the pentasaccharide are not visible. The quality of the x-ray data (better Rsym than the α2,3 data: 0.057 vs. 0.106) and the statistics from the crystallographic refinement (better Rfree than the α2,3 model: 0.22 vs. 0.240) suggest that the poor quality of the image of the α2,6 ligand indicates low affinity binding and/or the binding of multiple conformations.
In solution, α2,6 linkages are predominantly in a cis conformation, with a cis-to-trans ratio of 9:1 (27) (for cis definition, see Fig. 2e). If LSTc bound in a cis conformation to the H5 HA, then the Gal-2 C6 atom would cover Gln-226 (number 6 in Fig. 2d), shielding it from making hydrogen bonds, and the glycosidic oxygen (* in Fig. 2d) would be exposed to solvent (upward in Fig. 2 c– f). If LSTc bound in a trans conformation, the glycosidic oxygen may hydrogen-bond to Gln-226, but the 4-OH of Gal-2 would be too far away to mimic the α2,3-specific motif, and there would be an energetic penalty for choosing the less stable conformer.
Avian and Human Receptor Binding by H9 Swine Virus HA.
The H9 swine virus HA, which contains Leu-226 and Gly-228, interacts with both α2,3 LSTa and α2,6 LSTc by van der Wall contacts with a methyl group of Leu-226 (Fig. 2 e and f). Both ligands bind to H9 HA in a cis conformation (Fig. 2e Inset) around the bond between Sia-1 C2 and the glycosidic oxygen (star in Fig. 2 e and f), which leaves the glycosidic oxygen exposed away from the nonpolar Leu-226 and up toward solution (up in Fig. 2 c– f), where it might hydrogen-bond with water. This cis conformation around the glycosidic bond also projects the linkage carbon atoms of Gal-2, C3 in LSTa (bold arrows in Fig. 2e), C6 in LSTc (bold arrows in Fig. 2f), and the Gal-2 ring of LSTa toward nonpolar contacts with Leu-226. Steric factors within the two ligands when bound in a cis conformation orient the Gal-2 ring perpendicular (face-on in Fig. 2 e and f) to the sialic acid ring, permitting a hydrogen bond from the 6-OH of LSTa and the main-chain carbonyl oxygen of residue 225.
Only the first (distal) two saccharides are observed in the H9/α2,3 complex, suggesting that the remainder are disordered (Fig. 2e).
HA Contacts with the Asialo Segment of the Pentasaccharides.
In the preferred pairing, H9 with LSTc (Fig. 2f), the three proximal saccharides (GlcNAc-3, Gal-4, Glc-5) fold back over the top of the sialic acid and exit from the site at the front-right. Approximately 59 Å2 of nonpolar surface area is buried between Sia-1 and GlcNAc-3. Gal-4 and Glc-5 also contact Sia-1 (Table 2) and contact the binding site at the loop formed by residues 125–137, at the 150 loop, and at Asn-193, burying ≈201 Å2 of protein surface (Fig. 3a). Up to 11 hydrogen bonds could form between these proximal saccharides and the HA and two with the 4-OH group of the sialic acid (Fig. 3a).
Figure 3.
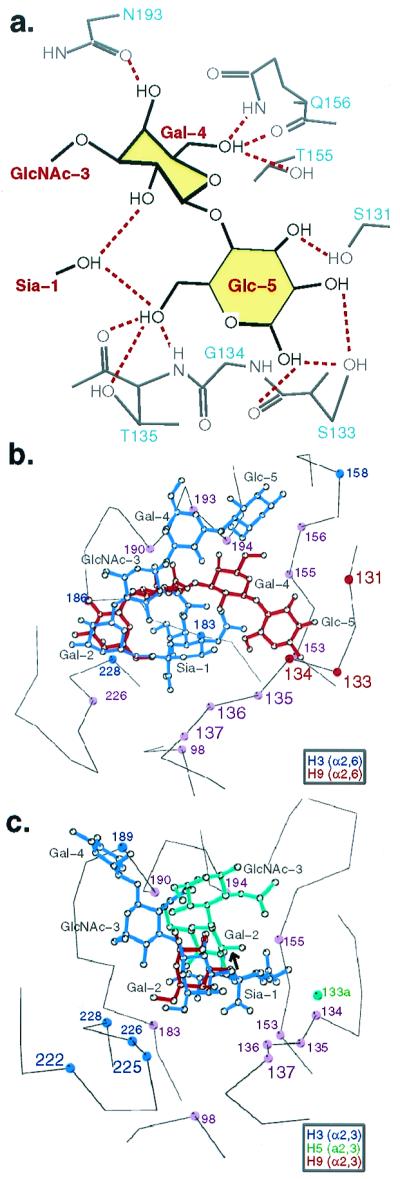
Atomic interactions between H5, H9, and H3 HAs and sialosides. (a) Potential hydrogen bonds (dashed red) between LSTc and the swine H9 HA. (b) Comparison of the binding of LSTc (α2,6) with the 1999 swine H9 (red) and 1968 human H3 (blue) subtype HA. Purple circles, common contacts; blue, H3 contacts; red, H9 contacts. (c) Comparison of the binding of LSTa (α2,3) with the 1997 avian H5 (green), 1999 swine H9 (red), and 1968 human H3 (blue) subtype HAs. Purple circles, common contacts; blue, H3 contacts; green, H5 contacts; red, H9 contacts.
The bound pentasaccharides in the four complexes reported here (Fig. 2 c– f) are not near crystal lattice contacts (>10 Å), suggesting that the oligosaccharide conformations observed result from inherent conformational preferences within the oligosaccharides and from the contacts observed with the HA. A folded conformation for LSTc was observed earlier in the H3 HA/LSTc complex, but the ligand conformation differed in positioning the proximal three saccharides (GlcNAc-3, Gal-4, and Glc-5) toward the back and top of the binding site (blue in Fig. 3b) (19) rather than toward the front and right as seen here (red in Fig. 3b). The different bound conformations of LSTc, mainly a difference of about a 60o rotation about one bond between Gal-2 and GlcNAc-3, may be due to the slightly different position of the HA main chain at residues 133–135 which, in H9 HA, position the carbonyl oxygens of 133 and 135 and the amide of 133 closer into the binding site, where they may hydrogen-bond to Glc-5 (Fig. 3a). Sequence differences between the H3 and H9 HAs at 135 (Thr-H9; Gly-H3) and 133 (Ser-133; Asn-H3) and the fact that Ser-133 in H9 points into the binding site whereas the H3 Asn-133 points outward contribute two more potential hydrogen bonds in the H9/LSTc case (Fig. 3a). Mutagenesis studies could access the contribution to the saccharide conformation of these interactions and others with residues 193, 156, and 155 (Fig. 3a). Another possible source of the difference in the LSTc conformations may result from the difference in the position of Gal-2 and GlcNAc-3, resulting from the sequence differences at 228 and 190 (Gly/Val-H9; Ser/Glu-H3).
The first two saccharides of LSTa (α2,3) bind almost identically to the Leu-226-containing H9 swine and H3 human virus HAs (red and blue in Fig. 3c), although two more of the proximal saccharides were observed in the H3 crystal structure (19). In the preferred combination, LSTa with the Gln-226-containing avian H5 HA, the trans conformation at the α(2,3) linkage (green and bold arrow in Fig. 3c) projects the proximal saccharides up and away from the 220s loop, and a contact from Gal-2 to Glu-190 is observed.
By contrast, only sialic acid and fragments of the penultimate Gal-2 are visible in electron density of the nonpreferred pairings, H5 with α2,6 LSTc (Fig. 2d) and H9 with α2,3 LSTa (Fig. 2e).
Human and Swine HA Receptor-Binding Sites Are Wider than Those of the Avian HA.
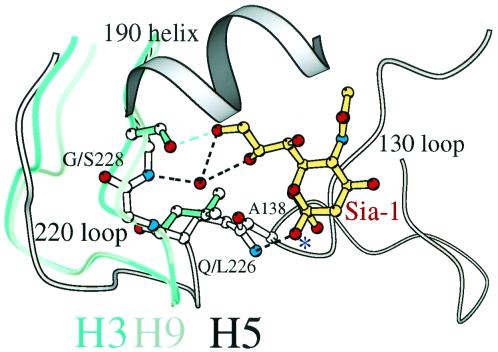
The distance between the 130 loop and the loop formed by residues 223–228, the 220 loop, which form two sides of the receptor-binding sites, is greater by ≈0.5Å (rms backbone atom average) in the human H3 (Leu-226/Ser-228) and swine H9 (Leu-226/Gly-228) HAs than in the Avian H5 HA (Gln-226/Gly-228) (Fig. 4). The narrower avian HA-binding site has Gln-226 close under the sialic acid in position to hydrogen-bond to the carboxylate group, and the amide group of Gly-228 of the avian HA hydrogen-bonds via a water molecule to the 8- and 9-OH of sialic acid (white in Fig. 4). In the swine H9 site, Gly-228 is positioned approximately the same as in the H5 avian site, with a water molecule hydrogen-bonded to the 228-amide, but Leu-226, compared with Gln-226, is pushed back from the sialic acid by more than 0.5Å, so that the lower methyl group of Leu-226 in the 220 loop can fit next to Ala-138 in the 130 loop (gray in Fig. 4). In the human H3 HA, both Ser-228 and Leu-226 are “pushed” back from the sialic acid, with a hydrogen bond from the Ser-228 OH group to the 9-OH of sialic acid replacing the water-mediated hydrogen bond (green in Fig. 4). A related difference was described earlier between the 1968 human H3 HA and a single residue variant selected for decreased α2,6 binding in which Leu-226 was replaced by Gln (L226Q) (22, 32). In that mutant, Ser-228 is located as in the wild-type human H3 HA and Gln-226 is located as in the avian H5 HA.
Figure 4.
Swine H9 and human H3 HA binding sites preferring α2,6 linkages are wider than avian H5 preferring α2,3. The H9 swine (gray), H3 human (green), and H5 avian (white) 226/228 loops are superimposed, showing that the H5 avian 220s loop (Gln-226/Gly-228) is closer to the opposing 130s loop than the H9 swine (Leu-226/Gly-228) or H3 human (Leu-226/Ser-228). Contact between Ala-138 and the lower methyl group of Leu-226 requires a more “open” site. The glycosidic oxygen of sialic acid (atom colors) is labeled with an asterisk. A water molecule (red sphere) mediates interactions between the amide group of Gly-228 and the 8- and 9-OHs of sialic acid in H9 swine and H5 avian HAs. The hydroxyl group of Ser-228 “replaces” the water molecule to form a hydrogen bond with 9-OH in the H3 human HA.
Evidently, the “closed” geometry of the avian H5 HA, which prefers α2,3 linkages, results from the Gln-226/Gly-228 pair. This geometry appears optimal for positioning Gln-226 to hydrogen-bond to the α2,3 trans motif composed of the 4-OH of Gal-2 and the glycosidic oxygen (Fig. 2c). The human H3 HA with the Leu-226/Ser-228 pair is at the opposite extreme, more “open” at both 228 and 226, which may be optimal for Leu-226 to make nonpolar contacts to α2,6 cis linkages. Swine H9 HA (Leu-226/Gly-228) and the L226Q variant of human H3 HA (Gln-226/Ser-228) appear to be intermediate, with partial avian and partial human character and the nonstandard Leu/Gly and Gln/Ser pairs (Fig. 2f).
However, these considerations do not apply to the HAs of the H1 subtype. H1 HAs isolated from humans prefer the α2,6 sialic acid linkage yet contain the Gln-226/Gly-228 pair characteristic of avian HAs (33–35). H1 HA structures will be required to understand the basis for these preferences.
Summary
We have compared how H5 avian, H9 swine, and H3 human virus HAs bind avian and human pentasaccharide sialoside receptor analogs by x-ray crystallography. Although differences in a few residues in the binding sites of the avian, swine, and human virus HAs cause small differences in the sialic acid position, the sialic acid makes a number of conserved contacts and hydrogen bonds (Fig. 1). The H5 avian HA recognizes the trans conformation of the α2,3 glycosidic linkage that creates a motif of the glycosidic oxygen and the axial 4-OH of Gal-2 that is complementary to the hydrogen-bonding capacity of Gln-226 (Fig. 2c). If α2,6 sialosides bind in their preferred cis conformation to the H5 avian HA, then nonpolar atoms of Gal-2 would cover Gln-226 (number 6 in Fig. 2d), unfavorably shielding it from making hydrogen bonds. (No saccharides beyond sialic acid were observed in this x-ray structure, presumably because the binding was so unfavorable (Fig. 2d).]
In contrast, the H9 swine and H3 human virus HAs recognize the cis conformation of the α2,6 and α2,3 glycosidic linkage, where the linkage carbon atoms of Gal-2 C6 in LSTc and Gal-2 C3 in LSTa and the Gal-2 ring of LSTa make nonpolar contacts with Leu-226 (Fig. 2 e and f and figure 2 in ref. 19) and the glycosidic oxygen points toward solution. α2,6 apparently is favored by the H9 swine HA (Fig. 3f) because only sialic acid and Gal-2 of the oligosaccharide are detected interacting with the HA in the α2,3 pentasaccharide sialoside complex (Fig. 3e). The width of avian virus-binding sites is narrower than the swine and human HA sites, which appears to optimize contacts with the preferred sialoside linkages. The H9 swine HA with Leu-226/Gly-228 may represent an evolutionary intermediate (33), with a binding site not yet as optimal for human cells as the human 1968 H3 site (Fig. 4).
Supplementary Material
Acknowledgments
We thank members of our laboratories for helpful discussions, the beamline staff at 14BMC at APS for assistance in data collection, and Dr. Chi-Huey Wong and colleagues for a helpful synthesis. This research is supported by National Institutes of Health Grant AI-13654, by a supplement to that grant for Expanded International Research on Emerging and Re-Emerging Diseases, by an International Partnership Research Award in Veterinary Epidemiology of the Wellcome Trust, by the Howard Hughes Medical Institute, and by the Medical Research Council. Y.H. is supported by the Jane Coffin Childs Memorial Fund for Medical Research. D.C.W. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- HA
hemagglutinin
- Sia
sialic acid
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1JSH, 1JSI, 1JSN, and 1JSO).
References
- 1.Who Memorandum. Bull W H O. 1980;58:585–591. [PMC free article] [PubMed] [Google Scholar]
- 2.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 3.Ito T, Couceiro J N, Kelm S, Baum L G, Krauss S, Castrucci M R, Donatelli I, Kida H, Paulson J C, Webster R G, et al. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers G N, D'Souza B L. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 5.Baum L G, Paulson J C. Acta Histochem Suppl. 1990;40:35–38. [PubMed] [Google Scholar]
- 6.Skehel J, Wiley D. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 7.Couceiro J N, Paulson J C, Baum L G. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 8.Pritchett T J, Paulson J C. J Biol Chem. 1989;264:9850–9858. [PubMed] [Google Scholar]
- 9.Matrosovich M, Gao P, Yoshihiro Kawaoka Y. J Virol. 1998;72:6373–6380. doi: 10.1128/jvi.72.8.6373-6380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subbarao K, Klimor A, Katz J, Reguery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, et al. Science. 1998;279:393–395. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 11.Claas E C, Osterhaus A D, van Beek R, De Jong J C, Rimmelzwaan G F, Seene D A, Krauss S, Shortridge K F, Webster R G. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 12.Suarez D L, Perdue M L, Cox N, Rowe T, Bender C, Huang J, Swayne D E. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bender C, Hall H, Huang J, Klimov A, Cox N, Hay A, Gregory V, Cameron K, Lim W, Subbarao K. Virology. 1999;254:115–123. doi: 10.1006/viro.1998.9529. [DOI] [PubMed] [Google Scholar]
- 14.Guan Y, Shortridge K F, Krauss S, Webster R G. Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peiris M, Yuen K Y, Leung C W, Chan K H, Ip P L S, Lai R W M, Orr W K, Shortridge K F. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y P, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, et al. Proc Natl Acad Sci USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y, Li J, Cheng X, Wang M, Zhou Y, Li X H, Cai F, Miao H L, Zhang H, Guo F. Chin J Exp Clin Virol. 1999;13:105–108. [Google Scholar]
- 18.Bridges C B, Katz J M, Seto W H, Chan P K S, Tsang D, Ho W, Mak K H, Lim W, Tam J S, Clarke M, et al. J Infect Dis. 2000;181:344–348. doi: 10.1086/315213. [DOI] [PubMed] [Google Scholar]
- 19.Eisen M B, Sabesan S, Skehel J J, Wiley D C. Virology. 1997;232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Brünger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:901–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Nature (London) 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 23.Sauter N K, Hanson J E, Glick G D, Brown J H, Crowther R L, Park S J, Skehel J J, Wiley D C. Biochemistry. 1992;31:9609–9621. doi: 10.1021/bi00155a013. [DOI] [PubMed] [Google Scholar]
- 24.Watowich S, Skehel J J, Wiley D C. Structure. 1994;2:719–731. doi: 10.1016/s0969-2126(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 25.Matrosovich M, Zhou N, Kawaoka Y, Webster R. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breg J, Kroon-Batenburg L M, Strecker G, Montreuil J, Vliegenthart J F. Eur J Biochem. 1989;178:727–739. doi: 10.1111/j.1432-1033.1989.tb14504.x. [DOI] [PubMed] [Google Scholar]
- 27.Poppe L, Stuike-Prill R, Meyer B, van Halbeek H. J Biomol NMR. 1992;2:109–136. doi: 10.1007/BF01875524. [DOI] [PubMed] [Google Scholar]
- 28.Sabesan S, Bock K, Paulson J C. Carbohydr Res. 1991;218:27–54. doi: 10.1016/0008-6215(91)84084-r. [DOI] [PubMed] [Google Scholar]
- 29.Stehle T, Harrison S C. Structure. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 30.Wright C S, Jaeger J. J Mol Biol. 1993;232:620–638. doi: 10.1006/jmbi.1993.1415. [DOI] [PubMed] [Google Scholar]
- 31.Merritt E A, Sarfaty S, van den Akker F, L'Hoir C, Martial J A, Hol W G. Protein Sci. 1994;3:166–175. doi: 10.1002/pro.5560030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers G N, Paulson J C, Daniels R S, Skehel J J, Wilson I A, Wiley D C. Nature (London) 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 33.Matrosovich M N, Gambaryan A S, Teneberg S, Piskarev V E, Yamnikova S S, Lvov D K, Robertson J S, Karlsson K-A. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 34.Reid A H, Fanning T G, Hultin J V, Taubenberger J K. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Kilmov A, Castrucci M R, Donatelli I, Kawaoka Y. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.