Abstract
The LhrC family of small regulatory RNAs (sRNAs) is known to be induced when the foodborne pathogen Listeria monocytogenes is exposed to infection-relevant conditions, such as human blood. Here we demonstrate that excess heme, the core component of hemoglobin in blood, leads to a strong induction of the LhrC family members LhrC1–5. The heme-dependent activation of lhrC1–5 relies on the response regulator LisR, which is known to play a role in virulence and stress tolerance. Importantly, our studies revealed that LhrC1–5 and LisR contribute to the adaptation of L. monocytogenes to excess heme. Regarding the regulatory function of the sRNAs, we demonstrate that LhrC1–5 act to down-regulate the expression of known LhrC target genes under heme-rich conditions: oppA, tcsA, and lapB, encoding surface exposed proteins with virulence functions. These genes were originally identified as targets for LhrC-mediated control under cell envelope stress conditions, suggesting a link between the response to heme toxicity and cell envelope stress in L. monocytogenes. We also investigated the role of LhrC1–5 in controlling the expression of genes involved in heme uptake and utilization: lmo2186 and lmo2185, encoding the heme-binding proteins Hbp1 and Hbp2, respectively, and lmo0484, encoding a heme oxygenase-like protein. Using in vitro binding assays, we demonstrated that the LhrC family member LhrC4 interacts with mRNAs encoded from lmo2186, lmo2185, and lmo0484. For lmo0484, we furthermore show that LhrC4 uses a CU-rich loop for basepairing to the AG-rich Shine–Dalgarno region of the mRNA. The presence of a link between the response to heme toxicity and cell envelope stress was further underlined by the observation that LhrC1–5 down-regulate the expression of lmo0484 in response to the cell wall-acting antibiotic cefuroxime. Collectively, this study suggests a role for the LisR-regulated sRNAs LhrC1–5 in a coordinated response to excess heme and cell envelope stress in L. monocytogenes.
Keywords: Listeria monocytogenes, heme toxicity, cell envelope stress, sRNA, two-component system, target mRNA
Introduction
Listeria monocytogenes is a Gram-positive, foodborne pathogen and the causative agent of listeriosis (Vazquez-Boland et al., 2001). To successfully establish an infection, this facultative intracellular pathogen must overcome several obstacles, such as low iron availability in the host (Chipperfield and Ratledge, 2000; Stojiljkovic and Perkins-Balding, 2002; Lungu et al., 2009). Upon infection, bacterial pathogens face a significant challenge in accessing iron, as it is mostly complexed to iron-binding proteins, such as hemoglobin, ferritin, lactoferrin, and transferrin (Hentze et al., 2004; Hammer and Skaar, 2011; Huang and Wilks, 2017). Bacterial pathogens are known to overcome this phenomenon, called nutritional immunity (Kochan, 1973; Wakeman and Skaar, 2012), through the activation of diverse mechanisms that allow iron acquisition from the iron-binding proteins in the host. Two-thirds of the total iron in the human body are sequestered in erythrocytes as heme bound to hemoglobin (Cassat and Skaar, 2013). In order to use heme as a source for iron, the pathogens first need to lyse erythrocytes, bind hemoglobin or other host heme proteins, and then extract and import the heme molecule for intracellular degradation to liberate free iron (Choby and Skaar, 2016). The iron acquisition system in L. monocytogenes has been the focus of diverse studies that have identified several specific iron transport and storage proteins (McLaughlin et al., 2011; Lechowicz and Krawczyk-Balska, 2015), such as Fhu, involved in the uptake of ferrichrome siderophores, and HupDGC, involved in the uptake of hemoglobin and hemin (i.e., the oxidized version of heme) (Jin et al., 2006; Xiao et al., 2011). Notably, HupDGC in L. monocytogenes is homologous to the well-studied iron-regulated surface determinant system (Isd) from Staphylococcus aureus (Skaar and Schneewind, 2004; Reniere et al., 2007). When the heme concentration in the environment is below 50 nM, heme acquisition in L. monocytogenes occurs with the participation of the heme-binding proteins 1 and 2, Hbp1 and Hbp2 (encoded by lmo2186 and lmo2185, respectively), which are anchored in the cell wall by Sortase B (Xiao et al., 2011). While a role for Hbp1 is still unclear, Hbp2 is known to scavenge for heme and hemoglobin and facilitate the transport of heme through the cell wall (Xiao et al., 2011; Klebba et al., 2012; Malmirchegini et al., 2014). Heme can then cross the membrane through the HupDGC ABC transporter (Jin et al., 2006; Xiao et al., 2011). At higher heme concentrations, free heme molecules are predicted to diffuse through the porous structure of peptidoglycan. Then, they are bound by HupD anchored to the cytoplasmic membrane and transported into the cell (Klebba et al., 2012; Lechowicz and Krawczyk-Balska, 2015). Once inside the cell, heme may be used, for example, as a cofactor for several enzymes, such as catalases and peroxidases, as a respiratory cofactor for oxygen transport and storage, or as a catalyst of electron transfer (Chiabrando et al., 2014). Alternatively, heme can be broken down by heme oxygenases, like the characterized Isd-type heme-degradation enzyme Isd-LmHde and/or the IsdG homolog Lmo0484, to liberate free iron (Wu et al., 2005; Travaglini-Allocatelli, 2013; Duong et al., 2014). To maintain intracellular iron homeostasis, L. monocytogenes possesses an iron-binding protein, Fur (ferric uptake regulator), which negatively regulates several genes under iron-replete conditions, including the genes encoding Hbp1 and Hbp2 (Ledala et al., 2010). Fur boxes have also been identified upstream from other genes coding for proteins involved in the heme uptake and utilization system, such as HupDGC and IsdG/Lmo0484 (McLaughlin et al., 2012), suggesting that a tight regulatory connection between the iron and heme uptake/utilization systems is crucial for L. monocytogenes. Conversely, while heme can be an essential source of iron for L. monocytogenes upon infection, during certain pathological states, severe hemolysis may occur, resulting in high levels of free heme (up to 20 μM) (Arruda et al., 2004). As heme is a highly reactive lipophilic molecule, the cells must protect themselves against the potential damaging effects of heme under heme-rich conditions, such as in the bloodstream and blood-rich organs (McLaughlin et al., 2011; Choby and Skaar, 2016; Huang and Wilks, 2017). For L. monocytogenes, the mechanism by which this pathogen senses and responds to excess heme is yet to be characterized.
The LhrC family in L. monocytogenes consists of seven sibling sRNAs with regulatory roles under infection-relevant conditions (Thorsing et al., 2017). The family includes the highly homologous sRNAs LhrC1–5, as well as Rli22 and Rli33-1, which share lower homology. rli22 and lhrC1–5 are positively regulated by the two-component system (TCS) LisRK that responds to cell envelope stress, whereas the general stress sigma factor σB controls the expression of rli33-1 (Sievers et al., 2014; Mollerup et al., 2016). The seven siblings are induced under various stress conditions and act to modulate the expression of specific target genes by an antisense mechanism. So far, three genes have been shown to be negatively regulated by the LhrCs at the post-transcriptional level: lapB, encoding a cell wall anchored virulence adhesin; oppA, encoding a substrate-binding protein of an oligopeptide transporter; and tcsA, encoding a CD4+ T cell-stimulating antigen (Sievers et al., 2014, 2015). For the mRNA targets characterized so far, the LhrC sRNAs act by basepairing to the Shine–Dalgarno (SD) region, leading to inhibition of translation initiation and/or decreased mRNA levels (Sievers et al., 2014, 2015; Mollerup et al., 2016). Strikingly, all seven sRNAs are highly induced when L. monocytogenes is exposed to human blood, suggesting an important regulatory role for the LhrC family in this host environment (Toledo-Arana et al., 2009). Yet, the specific component(s) in blood leading to induction of the LhrCs are presently unknown.
In this study, we aimed to investigate if the induction of the LhrC sRNAs by human blood could be linked to the increasing levels of heme in the bloodstream upon infection. Indeed, L. monocytogenes has the ability to secrete listeriolysin O (LLO) that facilitates the release of hemoglobin through erythrocyte lysis (Parrisius et al., 1986; Geoffroy et al., 1987; Foller et al., 2007). Similar to the LhrC family, hly, the gene encoding LLO, is also highly expressed in human blood, which supports the hypothesis that L. monocytogenes possibly encounters increasing levels of heme after host invasion (Toledo-Arana et al., 2009). Here, we demonstrate the LisR-dependent induction of LhrC family members, in particular LhrC1–5, by hemin, and show their capacity to regulate their known target genes under hemin stress conditions. In addition, we provide evidence that LhrC1–5 and LisR contribute to the adaptation of L. monocytogenes to excess hemin. Finally, we propose a role for LhrC1–5 in the post-transcriptional control of the heme uptake and utilization genes lmo0484, lmo2185, and lmo2186 in response to cell envelope stress and excess hemin.
Materials and Methods
Bacterial Strains and Growth Conditions
The wild-type strain used in this study was L. monocytogenes serotype 1/2c strain LO28 (Vazquez-Boland et al., 1992). The isogenic mutant derivatives LO28ΔlhrC1–5 and LO28ΔlisR were constructed in previous work (Kallipolitis et al., 2003; Sievers et al., 2014). The remaining isogenic mutant derivatives of this strain were constructed as previously described (Christiansen et al., 2004) by using the temperature-sensitive shuttle vector pAUL-A (Schaferkordt and Chakraborty, 1995). Primers used for in-frame deletions are listed in Supplementary Table S1. All strains used in this study are listed in Supplementary Table S2. L. monocytogenes was routinely grown at 37°C with aeration in brain heart infusion broth (BHI, Oxoid) unless stated otherwise. When appropriate, cultures were supplemented with kanamycin (50 μg/mL) or erythromycin (5 μg/mL). For induction of sRNA expression, cultures were supplemented with cefuroxime (4 μg/mL, corresponding to 9 μM) or various concentrations of hemin (Sigma). Hemin is the commercially available version of heme, which contains the oxidized Fe3+ ferric form instead of the reduced Fe2+ ferrous form. For experimental purposes, we will refer to hemin, while for the discussion we will refer to heme instead. Hemin was dissolved in 1.4 M NaOH and stock solutions were prepared fresh every time. In stress tolerance assays, overnight cultures were diluted to OD600 = 0.002 into BHI adjusted with various concentrations of hemin, or into BHI, when hemin was added to exponentially growing cells (OD600 = 0.2); growth was monitored until strains reached stationary phase. For cloning of plasmid vectors, Escherichia coli TOP10 cells (Invitrogen) were grown with aeration in Luria-Bertani broth supplemented with kanamycin (50 μg/mL) or erythromycin (150 μg/mL), when appropriate.
Plasmid Constructions and β-Galactosidase Assays
To study the transcriptional activity of lhrC1–5, rli22, and rli33-1, we used the promoter-less lacZ transcriptional fusion vector pTCV-lac (Poyart and Trieu-Cuot, 1997) fused to the promoter regions of the seven sRNA-encoding genes constructed previously (Sievers et al., 2014; Mollerup et al., 2016). Post-transcriptional regulation of LhrC target genes was monitored using in-frame translational lacZ fusions of lapB constructed in previous work (Sievers et al., 2014), and of lmo0484 constructed in the present study. Briefly, DNA fragments encoding a moderate promoter (Sievers et al., 2014) as well as a region spanning the SD region of lmo0484 (-48 to +47, relative to translation start site) were ligated into pCK-lac (Nielsen et al., 2010). For the β-galactosidase assay, L. monocytogenes strains carrying the plasmids were grown overnight, diluted to OD600 = 0.02 into fresh BHI and grown to OD600 = 0.2 (for strains with translational fusions) or OD600 = 0.35 (for strains with transcriptional fusions). Cultures were then split and stressed with either 9 μM cefuroxime or 8 μM hemin for 1 and/or 2 h. Samples (1 mL) were withdrawn prior to stress and at the indicated time points after the subjected stress. β-galactosidase assays were conducted as previously described (Christiansen et al., 2004). As the applied stress conditions resulted in impaired growth relative to the non-stress control condition, a direct comparison between the stressed and non-stressed cultures was not possible. However, the growth of the wild-type and mutant strains were comparable under each of the conditions tested (i.e., control, cefuroxime or hemin stress, respectively). Therefore, the β-galactosidase activities of wild-type and mutant strains were analyzed for each of the conditions using two-tailed Student’s t-test (i.e., wild-type, stressed vs. mutant, stressed). Only differences with at least 95% confidence were reported as statistically significant.
RNA Isolation and Purification
For primer extension and northern blot analysis, L. monocytogenes was grown to OD600 = 0.35. Cultures where then split, stressed with the indicated stressor concentration and samples were taken (10 mL) at the indicated time points. Cells were harvested by centrifugation at 11,000 rcf for 3 min at 4°C, and snap-frozen in liquid nitrogen. Cells were disrupted by the FastPrep instrument (Bio101, Thermo Scientific Corporation) and total RNA was extracted using TRI Reagent (Molecular Research Center, Inc.) as previously described (Nielsen et al., 2010). The integrity, concentration and purity of the RNA were confirmed by agarose gel electrophoresis, and NanoDrop 2000 or DeNovix DS-11 Fx+.
Primer Extension
Primer extension experiments were performed as previously described (Christiansen et al., 2004). The 32P-labeled, single-stranded primers used for detection of lmo0484 and lmo2186 transcription start sites are listed in Supplementary Table S1.
Northern Blotting
Total RNA (10 μg) was resolved on a 6% or 8% polyacrylamide-8 M urea gel as previously described (Nielsen et al., 2010); alternatively, 20 μg of total RNA was separated on a formaldehyde agarose gel for 3 h and 15 min prior to capillarity blotting on a Zeta-Probe membrane (Bio-Rad) (Sheehan et al., 1995). Membranes were hybridized with 32P-labeled DNA single-stranded probes listed in Supplementary Table S1. RNA bands were visualized using a Typhoon Trio or a Typhoon FLA9000 (GE Healthcare) and analyzed with IQTL 8.0 quantification software (GE Healthcare).
Reverse Transcriptase-Quantitative Polymerase Chain Reaction (RT-qPCR)
Fifty μg of total RNA was DNase-treated according to the manufacturer (Roche), and further purified with phenol-chloroform extraction. Three μg of DNA-free RNA was used for cDNA synthesis using the Maxima First Strand cDNA Synthesis Kit (Fermentas), following the manufacturer’s recommendations. RT-qPCR was performed using SYBR Green PCR Master Mix (Fermentas) and specific primer sets for the gene of interest (Supplementary Table S1). The samples were run on a MX3000 quantitative PCR thermocycler (Stratagene) with an initial step at 95°C for 10 min, 40 cycles of 15 s at 95°C, 30 s at 60°C and 30 s at 72°C. Data was analyzed using the Relative Expression Software Tool – Multiple Condition Solver REST-MCS version 2 (Pfaffl, 2001; Pfaffl et al., 2002). The tpi and rpoB genes served as reference genes. The experiment was carried out in three biological replicates, each in technical duplicates. Statistical differences were analyzed with a randomization test provided in the REST software. Only differences with at least 95% confidence were reported as statistically significant.
In Silico Predictions
The IntaRNA software (Busch et al., 2008; Wright et al., 2014; Mann et al., 2017) was used for predicting interactions between target mRNAs and sRNAs. Full length sequences of sRNAs and truncated versions of the targets were employed. Secondary structure predictions were obtained through Mfold (Zuker, 2003).
Electrophoretic Mobility Shift Assays (EMSAs)
Templates for in vitro transcription carried a T7 RNA polymerase binding site at their 5′-end and were generated by PCR. Templates for lmo0484 were obtained by PCR from chromosomal DNA and sRNA transcripts were made using overlapping primers (Supplementary Table S1). In vitro transcription, RNA purification, de-phosphorylation and labeling were performed as described previously (Sievers et al., 2014). EMSAs were performed as previously described (Nielsen et al., 2010). Briefly, 40 fmol of 5′-end labeled RNA was incubated with excess unlabeled RNA in a total volume of 10 μL in the presence of non-specific competitor (tRNA) for 1 h at 37°C followed by 10 min on ice. Samples were separated on a 5% non-denaturing gel at 4°C. RNA bands were visualized and analyzed as described for the northern blotting experiments.
In Vitro Enzymatic Structure Probing
5′-end labeled transcripts were prepared as described for the EMSAs. The enzymatic probing was carried out as previously described (Sievers et al., 2014), with some deviations. Briefly, for the alkaline hydrolysis ladder, 0.2 pmol of labeled RNA was mixed with alkaline hydrolysis buffer (Ambion) and 10 μg of yeast tRNA (Ambion) in a total volume of 10 μL and incubated at 95°C for 5 min; for T1 control sample, 0.2 pmol of labeled RNA was denatured and incubated with 0.01 U of T1 RNase (Ambion) for 5 min. Structure probing RNA interactions were incubated at 37°C for 1 h before treating the samples with the indicated cleaving agent: 0.01 U T1 RNase for 5 min and 0.0015 U V1 RNase (Ambion) for 2 min. Control samples were prepared likewise (except for the cleaving agents) and incubated at 37°C for the duration of the experiment. Samples were placed on ice and mixed with 2× loading buffer type II (Ambion). Five μL of each sample was separated on an 8% denaturing polyacrylamide gel. RNA bands were visualized and analyzed as described for the northern blotting experiments.
Results
LhrC1–5 Are Highly Induced by Hemin in a LisR-Dependent Manner
To investigate if LhrC1–5 are induced in response to hemin exposure, the sRNA levels were determined via northern blot analysis on total RNA purified from L. monocytogenes LO28 wild-type cells subjected to various concentrations of hemin for 1 h. As a control, wild-type cells were exposed to a sub-inhibitory concentration of the cell wall-acting antibiotic cefuroxime (9 μM), which is already known as an inducer of LhrC1–5 (Sievers et al., 2014). As seen in Figure 1A, LhrC1–5 levels were strongly induced with increasing concentrations of hemin (0, 1, 2, 4, 8, and 16 μM) and this induction was not caused by the hemin dissolvent NaOH (C1 and C2). In addition, exposure to 8 μM hemin induced LhrC1–5 to the same extent as 9 μM cefuroxime (Cef). The novel members of the LhrC family, Rli22 and Rli33-1, were also investigated through the same means to assess their induction under hemin stress conditions (Supplementary Figure S1A). Rli22 appeared to be induced when L. monocytogenes was exposed to the highest concentrations of hemin (8 and 16 μM), whereas the expression of Rli33-1 remained constant under all conditions tested.
FIGURE 1.
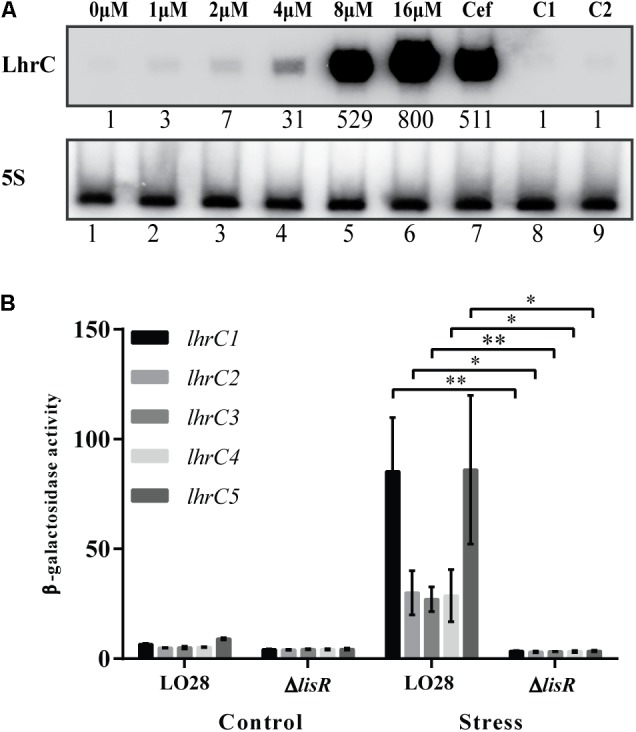
Induction of LhrC1–5 during hemin stress. (A) Northern blot analysis of LhrC1–5 expression. Samples were taken from Listeria monocytogenes LO28 wild-type cultures stressed with increasing concentrations of hemin (lanes 1–6), with a sub-inhibitory concentration of cefuroxime (9 μM) (lane 7) or with the hemin dissolvent NaOH (the same volume used to dissolve 8 and 16 μM hemin – lanes 8 and 9, respectively). Northern blot was probed for LhrC1–5 and 5S rRNA as a loading control. Relative levels of LhrC1–5 (normalized to 5S) are shown below each lane. (B) Transcriptional reporter gene fusions of lhrC promoters. Plasmids containing each of the five lhrC promoter regions fused to lacZ (Sievers et al., 2014) were transformed into LO28 wild-type and ΔlisR. The resulting strains were grown up to OD600 = 0.35 and stressed with hemin (8 μM), after control samples had been taken (Control). Further samples for a following β-galactosidase assay were withdrawn after 2 h (Stress). Results are the average of three biological replicates, each carried out in technical duplicates. After 2 h of stress, a significant difference between the ΔlisR mutant and wild-type cells was observed (∗p < 0.05, ∗∗p < 0.005).
To further investigate the induction of LhrC1–5 by hemin, the promoter activity of the five lhrC copies was determined using transcriptional fusions of each promoter to the reporter gene lacZ in the vector pTCV-lac (Sievers et al., 2014). As the TCS LisRK has been shown to play a role in the activation of lhrC1–5 under cell envelope stress conditions (Sievers et al., 2014), both the wild-type strain and a mutant strain lacking the response regulator LisR (ΔlisR) were transformed with the promoter-lacZ plasmids. The β-galactosidase activity was determined 2 h after subjecting the cultures to hemin stress (8 μM) and non-stressed cultures were included as controls (Figure 1B). In line with our previous observations, the promoter-lacZ constructs gave rise to β-galactosidase activity close to background levels under non-stress conditions (Sievers et al., 2014). However, after 2 h of hemin stress, a significant increase in the β-galactosidase activity was observed in the wild-type strain carrying the lhrC-lacZ fusion plasmids, while no increase in activity was detected in the ΔlisR cells. The activity of rli22 and rli33-1 promoters fused to lacZ was also determined in wild-type cells (Supplementary Figure S1B). Notably, the β-galactosidase activity in non-stressed and stressed cultures remained similar, showing that these promoters were not significantly activated by the addition of hemin. Overall, the results confirmed the induction of LhrC1–5 by hemin exposure and the requirement of LisR in the regulation of lhrC1–5 when L. monocytogenes faces excess concentrations of hemin. In contrast, the effect of hemin on rli22 and rli33-1 appeared to be negligible, and for that reason the following experiments will only focus on LhrC1–5.
LhrC1–5 and LisR Play a Role in the Adaptation to Hemin Toxicity
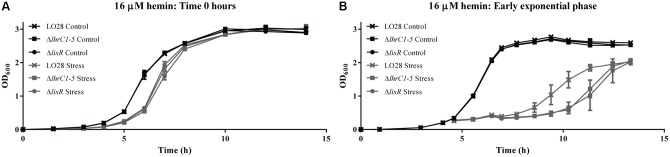
To investigate whether LhrC1–5 and/or the response regulator LisR contribute to the adaptation of L. monocytogenes to hemin stress, growth of the wild-type strain and strains lacking LhrC1–5 or LisR was compared when these cultures were exposed to 16 μM hemin. Hemin was either added to the cultures at the beginning of the growth experiment (Figure 2A), or when the cells reached the early exponential phase (Figure 2B). No difference in growth was observed between the wild-type and the two mutant strains when hemin was added at time 0 h (Figure 2A). However, when hemin was added to exponentially growing cells, the strains clearly responded differently. As seen in Figure 2B, both mutant strains struggled to adapt to the stress condition compared to the wild-type, suggesting the involvement of LhrC1–5 and LisR in the adaptation of L. monocytogenes to excess hemin.
FIGURE 2.
Stress tolerance assay. (A) LO28 wild-type, ΔlhrC1–5 and ΔlisR strains were grown in BHI (Control) and BHI containing 16 μM hemin (Stress). (B) LO28 wild-type, ΔlhrC1–5 and ΔlisR were grown in BHI to OD600 = 0.2. Then, the cultures were split in two; one half was stressed with 16 μM hemin (Stress) and the other half kept unstressed (Control). Bacterial growth was monitored until all cultures reached stationary phase. For each assay, the average of three biological replicates is shown.
LhrC1–5 Down-Regulate Their Known Target Genes Under Hemin Stress
Three genes (lapB, oppA, and tcsA) were previously identified as direct targets of LhrC1–5 (Sievers et al., 2014, 2015). More specifically, LhrC1–5 down-regulate their expression at the post-transcriptional level in response to cefuroxime stress. This raises the question whether LhrC1–5 regulate the same set of targets, no matter the stress factor causing their induction, or if LhrC1–5 have different targets under different stress conditions. To investigate if LhrC1–5 control the expression of oppA and tcsA under hemin stress, we made use of northern blot analysis to assess the oppA and tcsA mRNA levels in wild-type and ΔlhrC1–5 strains. The cultures were grown to early exponential phase and subjected to 8 μM hemin for 1 h; non-stressed cultures were included as controls (Figure 3A). In wild-type cells, hemin exposure resulted in a minor decrease in oppA mRNA (less than 1.5-fold), whereas for the lhrC1–5 mutant strain, the oppA mRNA level was 4-fold higher, when comparing hemin stressed and non-stressed cells. For tcsA, a threefold decrease was seen when wild-type cells were subjected to hemin stress, whereas no major changes were observed when comparing the levels of tcsA mRNA in stressed and non-stressed cultures of the ΔlhrC1–5 strain. Altogether, the northern blot analysis demonstrated that the induction of LhrC1–5 by hemin diminishes the expression of oppA and causes down-regulation of tcsA (Figure 3A). Strikingly, the regulatory effect observed for LhrC1–5 on oppA and tcsA is comparable to the one obtained when using cefuroxime as an inducer of LhrC1–5 (Sievers et al., 2015). Clearly, LhrC1–5 down-regulate the mRNA levels of oppA and tcsA to a similar extent in response to both cefuroxime and hemin stresses.
FIGURE 3.
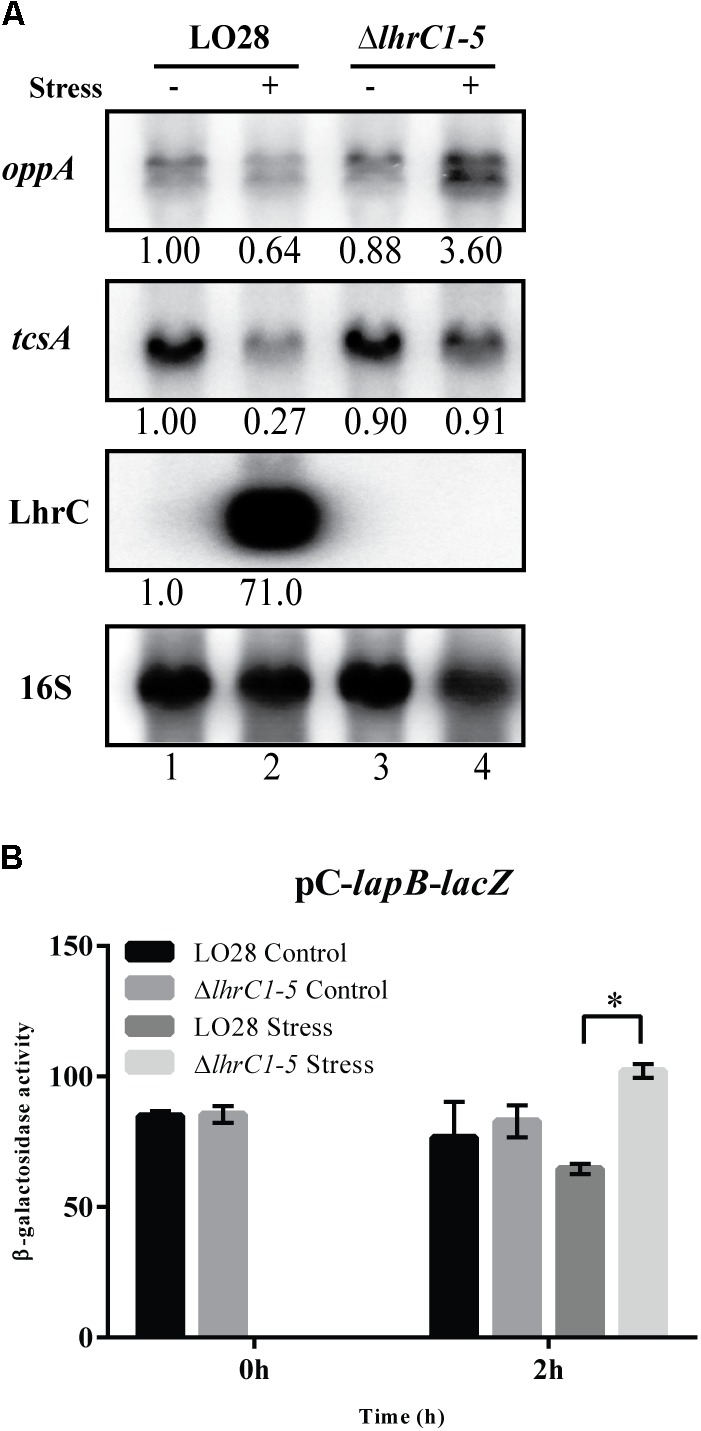
LhrC-mediated down-regulation of the known target genes oppA, tcsA and lapB. (A) Northern blot analysis of oppA mRNA, tcsA mRNA and LhrC1–5. Samples were taken from LO28 wild-type and ΔlhrC1–5 cultures exposed to 8 μM hemin stress for 1 h (+) as well as from non-stressed cultures (–). Northern blots were probed for oppA mRNA, tcsA mRNA, LhrC1–5 and 16S rRNA (loading control). Relative levels of oppA mRNA, tcsA mRNA and LhrC1–5 (normalized to 16S) are shown below each lane. (B) β-galactosidase assay of LO28 wild-type and ΔlhrC1–5 strains carrying a translational reporter gene fusion of lapB to lacZ in the vector pCK-lac (Sievers et al., 2014). β-galactosidase activities of wild-type and mutant cells were measured at the indicated time points under non-stress conditions (Control) and after exposure to 8 μM hemin (Stress). The results are the average of three biological replicates, each carried out in technical duplicates. After 2 h of stress, a significant difference (asterisk) between the mutant and wild-type cells was observed (p < 0.0001).
As lapB is part of a large operon, a reporter gene fusion strategy was used to assess the effect of hemin-induced LhrC sRNAs on this target gene (Sievers et al., 2014). In the pC-lapB-lacZ construct, a sequence including the 5′-untranslated region and the first codons of lapB’s coding region was fused downstream of a moderate promoter and inserted in-frame to lacZ in vector pCK-lac (Sievers et al., 2014). Notably, the moderate promoter was not affected by LhrC1–5 under hemin stress (Supplementary Figure S2A). The β-galactosidase activities were measured at time 0 and 2 h relative to the onset of hemin stress (8 μM), and non-stressed cultures were included as controls (Figure 3B and Supplementary Figure S2B). After hemin exposure, the ΔlhrC1–5 cells containing pC-lapB-lacZ showed higher β-galactosidase activity relative to the wild-type cells, whereas no difference in activity was observed under non-stress (control) conditions. These results demonstrate that LhrC1–5 down-regulate this target gene at the post-transcriptional level in response to hemin stress.
Proteins Related to Heme Uptake and Utilization Are Affected by LhrC1–5 Under Cefuroxime Stress
In a previous study, Sievers et al. (2015) performed transcriptomic and proteomic analyses of LO28 wild-type and ΔlhrC1–5 cells to identify genes controlled by LhrC1–5. For the transcriptome study, the cultures were subjected to cefuroxime stress for 30 min to induce LhrC1–5 regulation, whereas 1 h of cefuroxime stress was chosen for the proteomic analysis. These studies generated two lists of genes that were significantly up- or down-regulated in the lhrC1–5 mutant strain relative to the wild-type at the RNA and protein levels, respectively. By then, tight parameters were employed to select the genes to be further characterized as potential targets of LhrC1–5. Thus, only genes that were regulated at least 1.5-fold by LhrC1–5 at the RNA level and 2.0-fold at the protein level in all three biological replicates were chosen for further investigation (Sievers et al., 2015). Only three genes passed these strict criteria, including oppA and tcsA. Finding that LhrC1–5 also act as regulatory sRNAs in response to hemin stress prompted us to search these lists for potential targets related to heme uptake and utilization. Indeed, when searching through the data obtained from the proteomic analysis, we found that Lmo0484 was more than threefold up-regulated in ΔlhrC1–5 relative to the wild-type strain in all three biological replicates (Sievers et al., 2015). Lmo0484 is a homolog of the IsdG heme oxygenase from S. aureus, which degrades exogenous heme in the cytoplasm, leading to the release of free iron to be used as a nutrient source (Wu et al., 2005). In addition, we found that in two of the three biological replicates, the proteins Lmo2186 and Lmo2185 were more than three and twofold up-regulated, respectively, in the ΔlhrC1–5 strain relative to wild-type (Sievers et al., 2015). Lmo2186 and Lmo2185 are homologs of IsdC from S. aureus (Newton et al., 2005) and were previously characterized as heme-binding proteins 1 and 2 (Hbp1 and Hbp2), respectively (Xiao et al., 2011). Even though none of these genes were significantly affected by LhrC1–5 at the RNA level following 30 min of cefuroxime stress (Sievers et al., 2015), the results obtained from the proteomic analysis made it relevant to further investigate the regulatory effect of LhrC1–5 on lmo0484, lmo2186, and lmo2185.
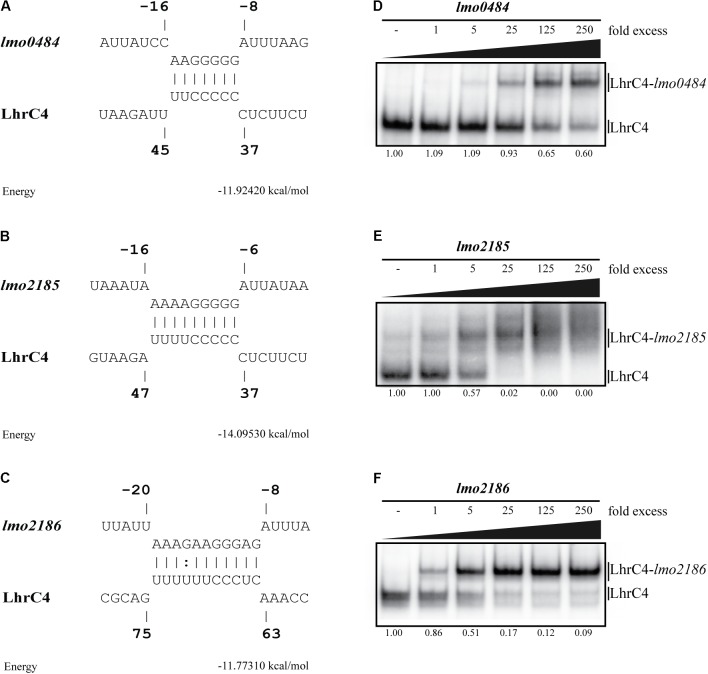
In regard to the known mode of action of LhrC1–5, we hypothesized that the sRNAs might act on lmo0484, lmo2186, and lmo2185 via direct binding to the mRNAs, leading to inhibition of translation initiation. To investigate this assumption, we first performed in silico analyses of the potential basepairing between the sRNAs and mRNAs. Using IntaRNA (Busch et al., 2008; Wright et al., 2014; Mann et al., 2017), we predicted that LhrC1–5 could bind to the SD region of lmo0484, lmo2185, and lmo2186 mRNAs (Figures 4A–C and Supplementary Figure S3). To verify experimentally the binding of the LhrCs to these mRNAs, EMSAs were performed, where LhrC4 was used as a representative of the LhrC family (Figures 4D–F). 5′-end labeled LhrC4 was visibly able to bind all three RNAs, where a single shifted band appeared with increasing concentrations of unlabeled lmo0484, lmo2185, or lmo2186 RNA. These results clearly demonstrate that LhrC4 interacts with these mRNAs (Figure 4).
FIGURE 4.
Analyzing the sRNA-mRNA interaction between LhrC4 and lmo0484, lmo2185, or lmo2186. (A–C) In silico prediction of sRNA-mRNA interactions. According to the IntaRNA Software (Busch et al., 2008; Wright et al., 2014; Mann et al., 2017), the loop A of LhrC4 binds to (A) lmo0484 and (B) lmo2185 mRNAs, and the single-stranded stretch binds to (C) lmo2186 mRNA. All the interactions are predicted to block the SD sequence of the mRNAs. LhrC4 is shown as a representative of the five LhrC copies. The nucleotides of lmo0484, lmo2185, and lmo2186 are numbered relative to the translation start site, and the nucleotides of LhrC4 are numbered relative to the 5′-end of the sRNA. (D–F) Testing the formation of sRNA-mRNA complexes by EMSAs. Labeled LhrC4 was shifted with increasing concentrations of unlabeled (D) lmo0484 RNA, (E) lmo2185 RNA or (F) lmo2186 RNA. Fold excess refers to the amount of unlabeled mRNA added to each sample, relative to the amount of labeled LhrC4. The fraction of unbound LhrC4 is shown below each lane.
LhrC4 Loop A Binds to the SD Region of lmo0484 mRNA
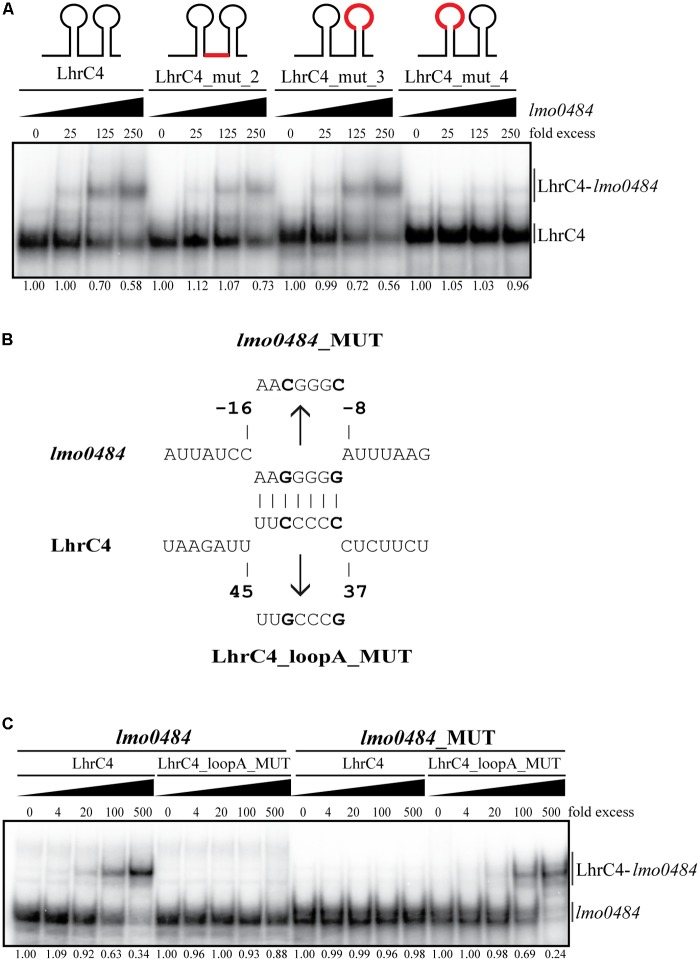
Based on the results obtained from the proteomic analysis and the in vitro binding studies, we decided to further analyze the basepairing between the sRNA LhrC4 and lmo0484 mRNA. All five LhrCs hold three single-stranded CU-rich regions known to interact with their target mRNAs (Sievers et al., 2014): loop A, a single-stranded stretch and the terminator loop. To investigate the importance of the CU-rich regions for the interaction with lmo0484 mRNA, mutant versions of LhrC4, where the entire CU-rich region is mutated, were tested for their ability to bind wild-type lmo0484 in an EMSA [for details about the mutations, see Sievers et al. (2014)]. The LhrC4 mutant derivatives were labeled and mixed with increasing concentrations of lmo0484 RNA (Figure 5A). The results revealed that mutations in the single-stranded stretch (LhrC4_mut_2) and in the terminator loop (LhrC4_mut_3) did not reduce the interaction between the two RNAs, whereas the loop A mutation (LhrC4_mut_4) abolished the basepairing, suggesting that this region is crucial for the binding of LhrC4 to lmo0484 (Figure 5A). To investigate if the SD region of lmo0484 is responsible for basepairing to LhrC4 loop A, lmo0484 was mutated in two of the seven nucleotides in the predicted LhrC4 binding region (Figure 5B). Interestingly, the mutated version of lmo0484 (lmo0484_MUT) was not able to bind the wild-type version of LhrC4 (Figure 5C). Finally, we created a mutant version of LhrC4 containing the complementary nucleotide mutations of lmo0484_MUT in loop A (LhrC4_loopA_MUT) (Figure 5B). As expected, the LhrC4_loopA_MUT variant was unable to bind the wild-type version of lmo0484, but when testing the binding of LhrC4_loopA_MUT to lmo0484_MUT, the interaction was restored (Figure 5C), confirming the importance of loop A for basepairing to the SD region of lmo0484 mRNA.
FIGURE 5.
Electrophoretic mobility shift assays (EMSAs) of the interaction between LhrC4 and lmo0484 mRNA. (A) LhrC4 mutant screening of loop A, single-stranded stretch, and the terminator loop. Labeled LhrC4 and the mutant derivatives were tested for their ability to interact with unlabeled lmo0484 RNA. The mutated regions are shown in red in the sRNA sketches. LhrC4: wild-type LhrC4; LhrC4_mut_2: mutation in single-stranded stretch; LhrC4_mut_3: mutation in terminator loop; LhrC4_mut_4: mutation in loop A. The fraction of unbound LhrC4 is shown below each lane. (B) Predicted basepairing between the SD region of lmo0484 mRNA and loop A of LhrC4. The mutated nucleotides are shown in bold and the sequences of the minimal mutant variants lmo0484_MUT and LhrC4_loopA_MUT are indicated. (C) Labeled lmo0484 RNA and lmo0484_MUT were each incubated with increasing concentrations of unlabeled LhrC4 or the mutant variant LhrC4_loopA_MUT. The fraction of unbound lmo0484 RNA is shown below each lane.
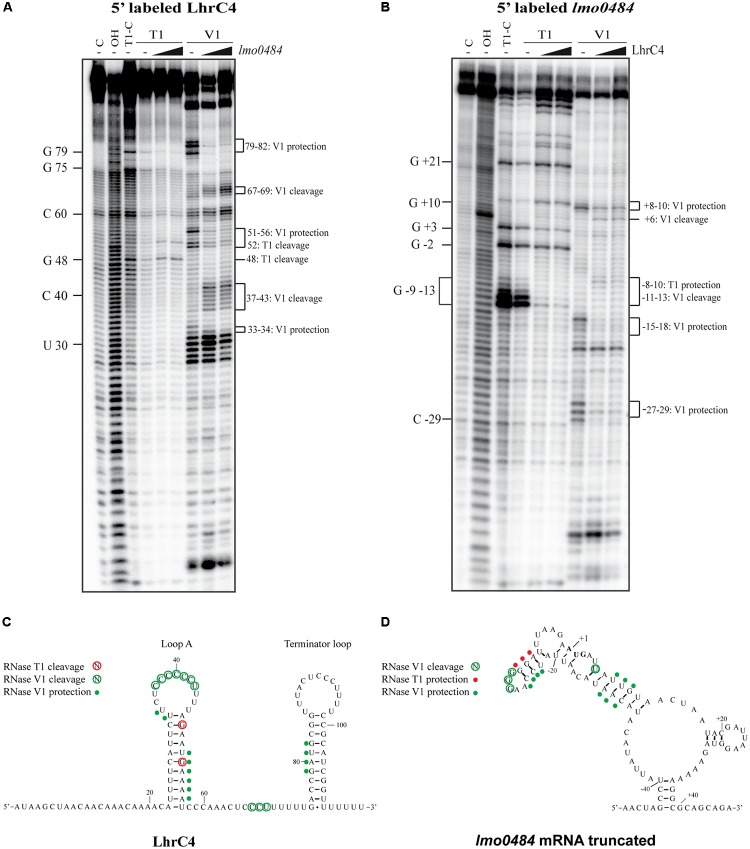
To study the structural implications of the interaction between the two RNAs, structural probing was employed. First, 5′-end labeled LhrC4 was subjected to RNase hydrolysis in the absence and presence of unlabeled lmo0484 RNA (Figure 6A). The enzymes RNase T1 and RNase V1 were used to cleave the RNAs, where the former is specific for unpaired guanine and the latter for double-stranded regions. The region from nucleotide 37 to 43, which resides in loop A, presented an increased V1 cleavage when LhrC4 interacts with lmo0484 (Figures 6A,C). Thus, the nucleotides residing in loop A went from single-stranded to double-stranded upon binding to lmo0484, confirming the importance of loop A for the sRNA-mRNA interaction. The V1 protection of nucleotides 51–56 and the increased T1 cleavage of G48 and G52 suggest that binding of loop A to lmo0484 disrupts the double-stranded structure of stem A. An increased V1 cleavage was also seen at nucleotides 67–69, corresponding to the single-stranded stretch in LhrC4, indicating that this CU-rich region may play a role in the basepairing of LhrC4 to lmo0484 as well, but to a lesser extent than loop A. Similarly, the region from nucleotides 79–82, residing in the double-stranded region of the terminator loop, obtained a more single-stranded conformation (V1 protection) in the presence of lmo0484, suggesting an opening of the terminator stem upon basepairing between the target mRNA and the single-stranded stretch (Figures 6A,C). To corroborate these results, 5′-end labeled lmo0484 RNA was subjected to the action of the same RNases in the absence and presence of unlabeled LhrC4 (Figure 6B). The nucleotides from -8 to -13, corresponding to the putative SD region of lmo0484, changed from single-stranded to double-stranded conformation (from -8 to -10: increased cleaving by V1; from -11 to -13: protection from T1) (Figures 6B,D). This result clearly supports that the SD region is bound by LhrC4. Additionally, some nucleotides shifted from double-stranded to single-stranded conformations (-27 to -29, -15 to -18 and +8 to +10), most likely reflecting changes in the secondary structure of lmo0484 occurring as a consequence of LhrC4 basepairing to the SD region (Figures 6B,D). Collectively, the results of the EMSAs and structure binding experiments support that the CU-rich region of loop A, and to a lesser extent the single-stranded stretch, binds to the SD region of lmo0484 mRNA.
FIGURE 6.
Structure probing of LhrC4 and lmo0484 mRNA interaction. (A) 5′-end labeled LhrC4 was treated with RNase T1 or RNase V1, either in the absence (–) or in the presence of 25- or 250-fold excess of unlabeled lmo0484 RNA. As a control, untreated LhrC4 was separated in the first lane (C), an alkaline ladder (OH) is shown in the second lane and an RNase T1 ladder (T1-C) was separated in the third lane. For an overview, selected nucleotides are labeled on the left side. Nucleotides showing structural changes upon lmo0484 binding are marked on the right side of the gel. (B) Partial digestion of 5′-end labeled lmo0484 RNA with RNases T1 and V1. Untreated lmo0484 RNA (C), an alkaline ladder (OH) and an RNase T1 ladder (T1-C) are shown in the first three lanes. Some of the cleaved G residues are labeled along the left side of the gel. The lmo0484 nucleotides, that were protected or cleaved in the presence of LhrC4, are indicated on the right side of the gel. (C) Secondary structure of LhrC4 illustrating the cleavage pattern upon binding to lmo0484 mRNA. Residues cleaved by RNase T1 (red) or RNase V1 (green) are encircled. Residues of LhrC4, that appeared to be protected by lmo0484 RNA, are indicated by green dots. The nucleotides of LhrC4 are numbered relative to the 5′-end of the sRNA. (D) Secondary structure of the truncated version of lmo0484 mRNA showing an altered cleavage pattern upon the addition of LhrC4. Residues cleaved by RNase V1 (green) are encircled. Residues of lmo0484 RNA that were protected in the presence of LhrC4 are indicated by red dots (T1 protection) or green dots (V1 protection). The start codon is indicated in bold. The nucleotides of lmo0484 are numbered relative to the translation start site (+1).
LhrC1–5 Control the Expression of lmo0484 at the Post-transcriptional Level in Response to Cefuroxime Stress
After showing that LhrC4 basepairs to the SD region of lmo0484 mRNA in vitro, we aimed to test the mechanism by which LhrC1–5 regulate this target in vivo. To this end, we analyzed the regulatory effect of LhrC1–5 at the post-transcriptional level on lmo0484 by making use once again of the lacZ reporter strategy. First, the 5′-end of the lmo0484 transcript was mapped by primer extension analysis to position -48 relative to the translation start site (Supplementary Figure S4). Then, a sequence encoding the 5′-end of the lmo0484 transcript and additional 47 bp of lmo0484 coding region was fused downstream of a moderate promoter and fused in-frame to lacZ in the vector pCK-lac (Nielsen et al., 2010). The generated pC-lmo0484-lacZ construct was transformed into both wild-type and ΔlhrC1–5 cells. The resulting strains were then subjected to cefuroxime stress for 1 or 2 h, and the β-galactosidase activity was measured. During cefuroxime stress, higher activity levels were obtained in ΔlhrC1–5 cells compared to wild-type cells, confirming that LhrC1–5 down-regulate the expression of lmo0484 at the post-transcriptional level in response to cefuroxime exposure (Figure 7A and Supplementary Figure S2C).
FIGURE 7.
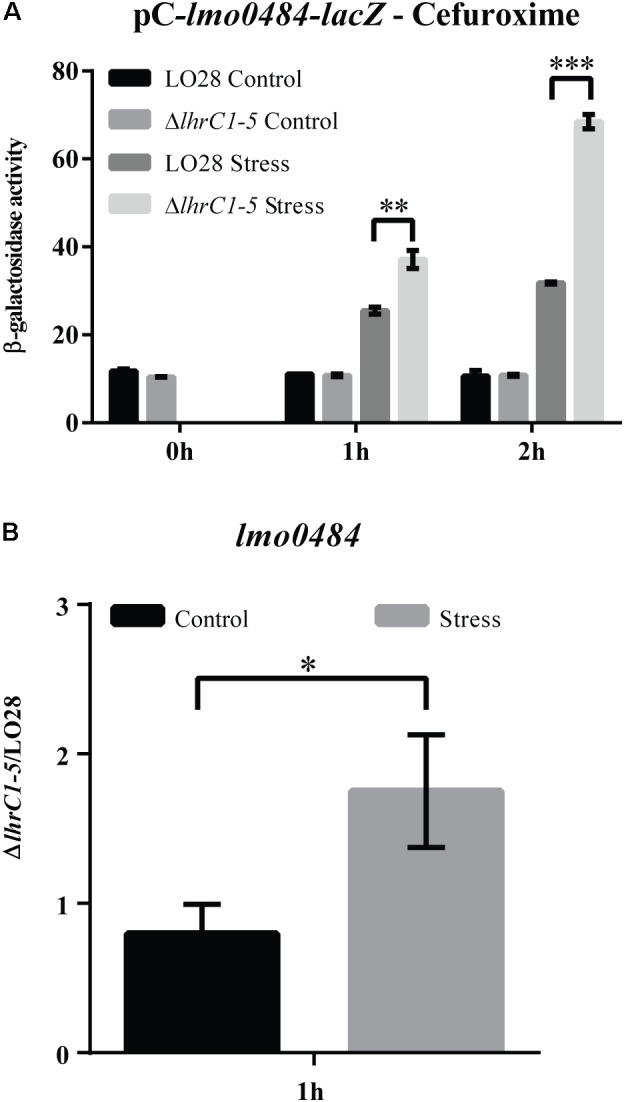
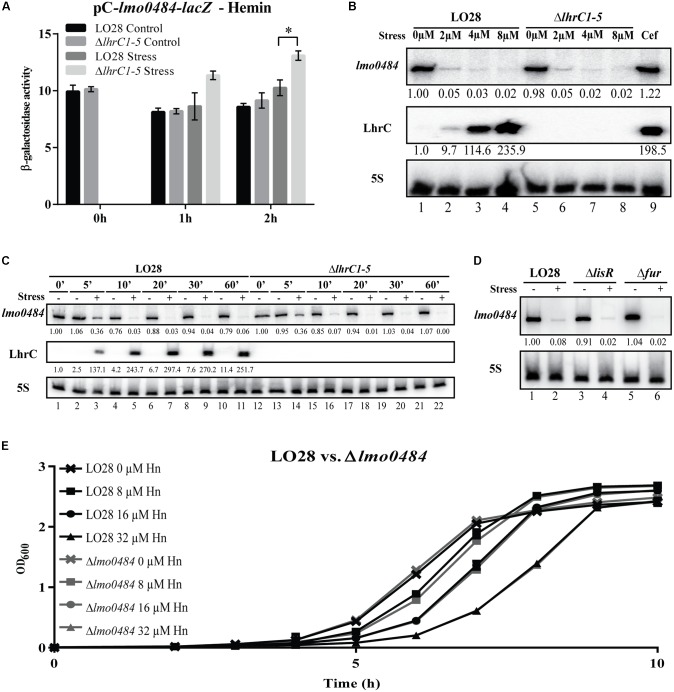
Assessing the effect of LhrC1–5 on the novel target gene lmo0484 upon cefuroxime exposure. (A) β-galactosidase assay of LO28 wild-type and ΔlhrC1–5 strains carrying a translational reporter gene fusion of lmo0484 to lacZ in the vector pCK-lac. β-galactosidase activities of LO28 wild-type and mutant cells were measured at the indicated time points under non-stress conditions (Control) and after exposure to 9 μM cefuroxime (Stress). The results are the average of three biological replicates, each carried out in technical duplicates. After 1 and 2 h of stress, a significant difference between the mutant and wild-type cells was observed (∗∗p < 0.001; ∗∗∗p < 0.0001). (B) Quantification of lmo0484 mRNA in ΔlhrC1–5 relative to LO28 wild-type by RT-qPCR. The ratio of ΔlhrC1–5/LO28 was determined at the 1 h time point for both non-stressed (Control) and cefuroxime-exposed samples (Stress). The result shown is the average of three biological replicates. The asterisk indicates a significant increase of the ratio under stress conditions compared to the control with p < 0.05.
As lmo0484 was not significantly affected by LhrC1–5 at the RNA level after 30 min of cefuroxime stress (Sievers et al., 2015), we performed RT-qPCR on total RNA purified from cells exposed to cefuroxime for 1 h, where the level of LhrC1–5 induction is known to be at its highest (Sievers et al., 2014); non-stressed samples were included as controls. The lmo0484 mRNA level was quantified in ΔlhrC1–5 cells relative to wild-type cells under non-stress (control) and stress conditions (Figure 7B). In the control samples, the mRNA ratio was approximately 1, showing that there was no difference in lmo0484 expression in the wild-type and mutant strains grown under non-stress conditions. In contrast, lmo0484 mRNA expression was 1.75-fold higher in ΔlhrC1–5 compared to wild-type after 1 h of cefuroxime stress. Collectively, the results of the β-galactosidase assay and the RT-qPCR experiment demonstrate that LhrC1–5 down-regulate lmo0484 expression at the post-transcriptional level upon subjecting L. monocytogenes to cefuroxime stress. Thus, lmo0484 is clearly a target for LhrC1–5-mediated control when L. monocytogenes is exposed to cell envelope stress conditions.
lmo0484 Is Strongly Repressed in Response to Hemin Exposure and Is Dispensable for Growth Under Hemin Stress Conditions
Based on the results obtained so far, we reasoned that the LhrCs could down-regulate the expression of lmo0484 in response to hemin stress. To test this hypothesis, wild-type and ΔlhrC1–5 cells containing the pC-lmo0484-lacZ plasmid were exposed to hemin stress for 1 and 2 h, and non-stressed cultures were used as controls. Upon the addition of hemin, the β-galactosidase activity in the wild-type and ΔlhrC1–5 cells remained at relatively low levels (Figure 8A and Supplementary Figure S2D), which is in stark contrast to the increase in lmo0484-lacZ expression observed upon cefuroxime exposure (Figure 7A). After 2 h of hemin stress, the lmo0484-lacZ expression was only slightly, but significantly higher in the ΔlhrC1–5 cells relative to the wild-type cells (Figure 8A). These results prompted us to investigate the effect of hemin stress on lmo0484 at the RNA level. First, the expression of lmo0484 was investigated in wild-type and ΔlhrC1–5 cells subjected to increasing concentrations of hemin for 1 h (Figure 8B). The results of the northern blot experiment clearly demonstrated a strong repression of lmo0484 transcript levels in the wild-type strain already at the lowest concentration of hemin tested. For comparison, the lmo0484 mRNA level in the wild-type strain remained unaffected by 9 μM cefuroxime (Cef). When comparing the wild-type and ΔlhrC1–5 cells after 1 h of hemin exposure, a regulatory effect of LhrC1–5 on lmo0484 was not evident, although LhrC1–5 was clearly expressed (Figure 8B).
FIGURE 8.
Assessing the effect of LhrC1–5 on the novel target gene lmo0484 under hemin stress. (A) β-galactosidase assay of LO28 wild-type and ΔlhrC1–5 strains carrying a translational reporter gene fusion of lmo0484 to lacZ in the vector pCK-lac. β-galactosidase activities of wild-type and mutant cells were measured at the indicated time points under non-stress conditions (Control) and after exposure to 8 μM hemin (Stress). The results are the average of three biological replicates, each carried out in technical duplicates. After 2 h of stress, a significant difference between the mutant and wild-type cells was observed (p < 0.005). (B) Northern blot analysis of lmo0484 mRNA after exposure to various concentrations of hemin. Samples were taken from LO28 wild-type and ΔlhrC1–5 cultures exposed to 1 h of 0, 2, 4, or 8 μM hemin stress. A sample from LO28 wild-type culture exposed to 9 μM cefuroxime was used as a control. Northern blots were probed for lmo0484 mRNA, LhrC1–5 and 5S rRNA (loading control). Relative levels of lmo0484 mRNA and LhrC1–5 (normalized to 5S) are shown below each lane. (C) LhrC induction profile and time-dependent regulation of lmo0484 mRNA in response to hemin stress. At time = 0, corresponding to OD600 = 0.35, LO28 wild-type and ΔlhrC1–5 cells were split in two, and one half of the culture was treated with 8 μM hemin (+), whereas the other half was left untreated (–). Samples were harvested at several time points (0, 5, 10, 20, 30, and 60 min relative to the addition of hemin) and total RNA was prepared for northern blot analysis. The blot was probed for lmo0484 mRNA, LhrC1–5 and 5S rRNA (loading control). Relative levels of lmo0484 mRNA and LhrC1–5 were normalized to 5S and are shown below each lane. (D) Testing the role of LisR and Fur in the heme-dependent repression of lmo0484. Samples were taken from LO28 wild-type, ΔlisR and Δfur cultures exposed for 1 h to 8 μM hemin stress (+) as well as from non-stressed cultures (–). Northern blots were probed for lmo0484 mRNA and 5S rRNA (loading control). Relative levels of lmo0484 mRNA normalized to 5S are shown below each lane. (E) Stress tolerance assay of the Δlmo0484 strain compared to LO28 wild-type. Wild-type and Δlmo0484 strains were grown in BHI containing 0, 8, 16, or 32 μM hemin. Bacterial growth was monitored until all cultures reached stationary phase. The average of three biological replicates is shown.
To further explore the response of L. monocytogenes to excess hemin, a time course experiment was performed. Using northern blot analysis, the levels of LhrC1–5 and lmo0484 mRNA were determined in wild-type and ΔlhrC1–5 cells at various time points after hemin exposure (Figure 8C). In the wild-type background, LhrC1–5 were clearly detected just 5 min after the addition of hemin, reaching a peak after 20 min of stress. A strong decrease in the transcript level of lmo0484 was observed after 5 min of stress, demonstrating that L. monocytogenes responds very quickly to excess hemin by shutting down the expression of lmo0484 mRNA. Once again, no major differences were observed when comparing the lmo0484 mRNA levels in the wild-type and ΔlhrC1–5 strains, suggesting that additional regulatory factors are involved in mediating the repression of lmo0484 (Figure 8C). Indeed, a Fur box is present upstream from lmo0484 (Supplementary Figure S4), suggesting that the iron-responsive regulator Fur could be a negative regulator of lmo0484 expression. To investigate this, the lmo0484 levels were tested in a mutant strain lacking the Fur regulator (Figure 8D). The results clearly demonstrate that Fur is not responsible for down-regulating the expression of lmo0484 in response to hemin stress, and LisR seems not to be required either. Curiously, lmo2185 and lmo2186 transcript levels in the wild-type strain were also strongly repressed by the lowest concentration of hemin tested, and similarly, Fur was not responsible for the down-regulation observed (Supplementary Figures S4, S5).
In Gram-positive bacteria, members of the IsdG family are known to be required for the utilization of heme as an iron source (Reniere et al., 2010; Sheldon and Heinrichs, 2015). In addition, IsdG in Bacillus anthracis also acts to protect the bacterium against heme-mediated toxicity (Skaar et al., 2006). To assess the role of lmo0484 during growth of L. monocytogenes under hemin stress conditions, a mutant strain lacking lmo0484 was constructed. No significant difference was observed when comparing the growth of the Δlmo0484 and wild-type strains under hemin stress (Figure 8E).
To summarize, the mRNA level of lmo0484 is quickly diminished upon hemin exposure by an unknown mechanism. Likewise, the level of lmo2186-2185 mRNA is strongly reduced under hemin-rich conditions. It is possible that LhrC1–5 act to ensure an efficient shut-down of the translation of the few lmo0484 and lmo2186-2185 mRNAs remaining after the onset of hemin exposure. In accordance with the expression profile of lmo0484 upon hemin exposure, this IsdG-encoding gene does not contribute to growth under hemin stress conditions.
Discussion
Bacteria are known to utilize heme as an iron source during infection, however, in heme-rich environments, such as in the bloodstream and blood-rich organs, successful pathogens must defend themselves against the harmful effects of heme toxicity (Choby and Skaar, 2016; Joubert et al., 2017). Indeed, during severe hemolysis, free heme may reach concentrations up to 20 μM (Arruda et al., 2004). Information on how L. monocytogenes maintains heme homeostasis is relatively limited and biased toward studies on the acquisition of hemoglobin and heme under iron-limited conditions (Klebba et al., 2012; Lechowicz and Krawczyk-Balska, 2015). In this study, we focused on how L. monocytogenes senses and responds to excess heme. We report that heme generates a signal that stimulates LisR-mediated activation of members of the LhrC family of sRNAs. Furthermore, we show that LisR and LhrC1–5 contribute to the adaptation of L. monocytogenes to growth under conditions of excess heme. LisRK has previously been found to be important for infection in mice (Cotter et al., 1999; Kallipolitis and Ingmer, 2001), and, together with LhrC1–5, LisR contributes to the intracellular replication of L. monocytogenes in macrophage-like cells (Sievers et al., 2015). The expression of LhrC1–5 is known to be highly induced when L. monocytogenes is exposed to whole human blood (Toledo-Arana et al., 2009), which corresponds well with a role for the LisR-regulated sRNAs in the adaptation to heme-rich environments.
We have previously shown that LhrC1–5 are induced in a LisR-dependent manner under cell envelope stress conditions, such as exposure to the cell wall-acting antibiotic cefuroxime, and that LhrC1–5 down-regulate the expression of cell envelope-associated proteins with virulence functions (Sievers et al., 2014, 2015). In the present study, we show that LhrC1–5 also repress the expression of the three known target genes in response to heme toxicity, suggesting that a fine-tuning of the levels of the cell envelope-associated proteins TcsA, OppA, and LapB is beneficial for L. monocytogenes under heme-rich conditions. Notably, whole human blood contains multiple components participating in host immunity, and surface exposed proteins are readily recognized by the immune system (Sievers et al., 2015). Thus, in heme-rich conditions, such as the human blood, LhrC-mediated down-regulation of surface exposed proteins may be viewed as an attempt by L. monocytogenes to evade detection by the immune system. Interestingly, the present study suggests that genes encoding cell envelope-associated proteins with known functions in heme acquisition belong to the LhrC regulon as well. The LhrC4 sRNA bound readily to the SD regions of lmo2186-lmo2185 mRNA, encoding the heme uptake proteins Hbp1 and Hbp2, respectively. Additionally, the lmo0484 gene, encoding a heme oxygenase, was found to be a target for LhrC regulation. The LhrC sRNAs were shown to down-regulate the expression of lmo0484 in response to cell envelope stress and LhrC4 was found to bind specifically to the SD region of lmo0484 mRNA using one of its three CU-rich regions. These observations suggest that the sRNAs act to inhibit translation initiation of lmo0484 under LhrC-inducing conditions, such as upon exposure to cefuroxime or heme. Whereas LhrC-mediated down-regulation of lmo0484 expression was clearly detected in response to cefuroxime stress, the regulatory effect of LhrC1–5 on lmo0484 was less pronounced under heme stress due to a rapid decrease in lmo0484 mRNA levels following the addition of excess heme to the growth medium. Interestingly, a similar decrease was observed when testing the level of lmo2186-lmo2185 mRNA upon heme exposure, suggesting a common mechanism for down-regulation of genes involved in heme uptake and utilization in response to heme stress. Although potential Fur boxes were found in the promoter regions upstream from lmo2186-lmo2185 and lmo0484, the Fur regulator was not required for the heme-mediated repression. Thus, the mechanism underlying the repressive effect of excess heme on the heme uptake and utilization genes in L. monocytogenes remains elusive. Considering the potential damaging effects of heme, an instant shut-down of the heme uptake genes lmo2186 and lmo2185 in response to excess heme seems logical, whereas the repression of the heme oxygenase-encoding gene lmo0484 is more ambiguous. In other organisms, including the bacterial pathogen B. anthracis, heme oxygenases have been shown to contribute to the heme detoxification process (Skaar et al., 2006), but in L. monocytogenes, the heme oxygenase encoded by lmo0484 appears to be dispensable in heme-rich environments. Indeed, L. monocytogenes wild-type and Δlmo0484 strains were found to grow equally well under heme stress conditions (Figure 8E). Whether the second heme oxygenase encoded by L. monocytogenes (Isd-LmHde) contributes to the heme detoxification process remains to be investigated.
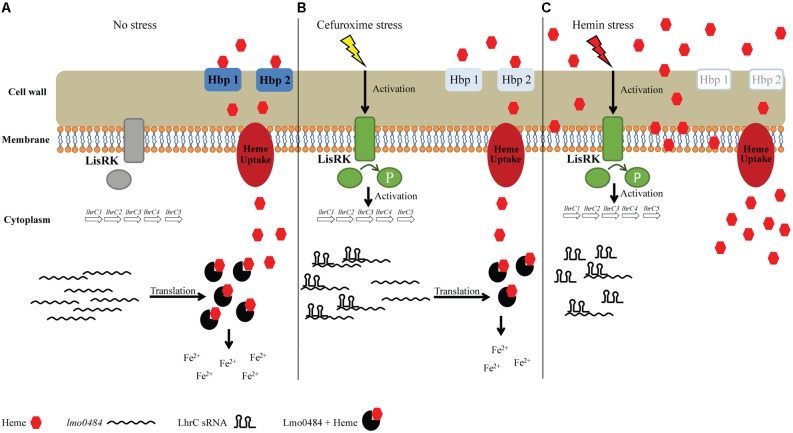
Based on our findings, we propose the following model for LhrC-mediated control of the heme uptake and utilization genes lmo0484, lmo2186, and lmo2185 under cefuroxime stress and heme stress (see Figure 9): under non-stress conditions, these genes are clearly expressed (Figures 8B–D and Supplementary Figure S5), indicating that L. monocytogenes is using the heme already present in the BHI medium as a source of iron (Figure 9A); in response to cefuroxime stress, LhrC1–5 fine-tune the expression of lmo0484, and most likely also lmo2186 and lmo2185, suggesting that L. monocytogenes is employing sRNA-mediated control to maintain heme homeostasis under cell envelope stress conditions (Figure 9B); upon exposure to excess heme, the levels of lmo0484 and lmo2186-lmo2185 mRNAs are strongly reduced by an unknown mechanism (e.g., heme-induced transcriptional repression and/or mRNA degradation) (Figure 9C). In this case, we speculate that the role of LhrC1–5 is to prevent translation of the remaining lmo0484 mRNA, and possibly also lmo2186-lmo2185 mRNA, following heme exposure. Notably, LhrC1–5 were induced more than 50-fold after just 5 min of heme stress, showing that the LhrC sRNAs are available for basepairing to target mRNAs within a few minutes (Figure 8C). Together, these regulatory mechanisms act to repress the expression of genes involved in heme uptake and utilization, which allows L. monocytogenes to adapt very quickly to heme-rich conditions. Collectively, this study shows that the detected outcome of LhrC-mediated control relies not only on the presence of the sRNAs, but also on the availability of the partner mRNAs, which may be subject to control by other regulatory mechanisms under specific stress conditions (Figure 9C).
FIGURE 9.
Proposed model of the regulatory effect of LhrC1–5 on heme uptake and utilization genes. (A) During growth in BHI medium, there is no activation of the TCS LisRK and subsequently LhrC1–5 are not induced. Under this condition, lmo0484, lmo2186, and lmo2185 are expressed and heme-uptake from the BHI medium is taking place according to the cells’ needs. The lmo0484 mRNA is translated into a IsdG-like heme oxygenase, which will catalyze the degradation of heme to obtain free iron. (B) When the cells are subjected to cefuroxime stress, LisRK will be activated. LhrC1–5 are then induced and can bind lmo0484 mRNAs. Binding of LhrC sRNA to the SD region of lmo0484 mRNA leads to inhibition of translation. Under cefuroxime stress, LhrC1–5 most likely act to control the expression of the heme uptake genes lmo2186 and lmo2185 in a similar fashion (not shown). Thus, when exposed to cefuroxime, LhrC1–5 act to fine-tune the expression of heme uptake and utilization genes, suggesting a need for L. monocytogenes to slightly reduce the uptake of heme when the cell envelope is being stressed. (C) Under hemin stress, LhrC1–5 are strongly induced as well in a LisR-dependent manner. Furthermore, when the cell faces high concentrations of heme, the mRNA levels of lmo0484 are almost immediately diminished by an unknown mechanism. In this situation, LhrC1–5 will bind the remaining lmo0484 mRNAs in the cell and act as ‘vacuum cleaners’ to avoid translation of the residual lmo0484 mRNA. The heme uptake genes lmo2185 and lmo2186 are most likely controlled by heme and LhrC1–5 in a similar fashion (not shown). Consequently, the expression of heme acquisition genes is quickly reduced in response to heme stress to avoid further uptake of heme and liberation of iron.
The results obtained in this study clearly demonstrate a role for LisRK in the response to excess heme. In addition to LisRK, at least one more TCS is expected to contribute to the adaptation of L. monocytogenes to heme-rich conditions. In Gram-positive pathogens, the TCS HssRS has been shown to play a major role in the response to heme stress (Choby and Skaar, 2016). In S. aureus and B. anthracis, HssRS controls the expression of genes encoding a heme-regulated transporter, HrtAB, which protects the bacteria from heme toxicity by exporting heme (Torres et al., 2007; Stauff and Skaar, 2009). The HssRS system and HrtAB exporter are conserved in L. monocytogenes as well (Torres et al., 2007), however, their roles in dealing with heme toxicity remains to be clarified. Curiously, the HssRS system of B. anthracis was recently found to interact with another TCS, HitRS, which responds to compounds that affect the cell envelope integrity (Mike et al., 2014). HssRS and HitRS both act to stimulate hrtAB expression in response to heme and cell envelope stress, respectively. Furthermore, the histidine kinase HssS was shown to cross-phosphorylate the response regulator HitR to activate the expression of the hitPQRS operon upon heme exposure (Mike et al., 2014). This operon encodes components of an unstudied ABC transporter and the HitRS system. The cross-regulation between HssRS and HitRS is thought to ensure a coordinated response to heme and cell envelope stress in B. anthracis, which may enable this pathogen to better adapt and survive during infection (Mike et al., 2014). We found that heme stimulates LisR-dependent activation of LhrC1–5 as efficiently as the cell-wall acting antibiotic cefuroxime, revealing a link between the response to heme toxicity and cell envelope stress in L. monocytogenes as well. Future studies should focus on clarifying the interconnections (if any) between LisRK and the putative heme-responsive HssRS system in L. monocytogenes. Most importantly, it should be investigated if LisRK cross-phosphorylates with HssRS, as shown for HitRS and HssRS in B. anthracis (Mike et al., 2014).
In addition to the LhrCs in L. monocytogenes, a heme-responsive sRNA has also been described in Pseudomonas aeruginosa. The sRNA PrrH is encoded from the prrF locus in P. aeruginosa and overlaps with two iron-regulated sRNAs, PrrF1, and PrrF2 (Oglesby-Sherrouse and Vasil, 2010). In contrast to LhrC1–5, which are highly induced in response to excess heme, the PrrH sRNA is repressed by heme via an unknown mechanism. Furthermore, PrrH is repressed by iron, most likely via the Fur protein in P. aeruginosa. Although PrrH has been predicted to regulate genes involved in heme homeostasis and virulence through the unique sequence derived from the prrF1-prrF2 intergenic region (Oglesby-Sherrouse and Vasil, 2010; Reinhart et al., 2015), a recent study showed that PrrH is not required for acute murine lung infection (Reinhart et al., 2017). Notably, the region unique to PrrH is highly conserved across P. aeruginosa strains of clinical origin, suggesting that a role for PrrH could be found using alternative infection models (Reinhart et al., 2017). The finding of heme-regulated sRNAs in both L. monocytogenes and P. aeruginosa raises the possibility of a more widespread role for sRNA-mediated control in the response of bacterial pathogens to heme.
Author Contributions
PdS, PM-G, DS, and BK: conceived and designed the experiments. PdS, PM-G, DS, J-HC, MB, and EL: performed the experiments. PdS, PM-G, DS, J-HC, MB, EL, and BK: analyzed the data. PdS and BK: wrote the paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement No. 641984, the Novo Nordisk Foundation, and a “La Caixa” Fellowship Grant for Post-graduate Studies, “La Caixa” Banking Foundation, Barcelona, Spain to PM-G.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00599/full#supplementary-material
References
- Arruda M. A., Rossi A. G., de Freitas M. S., Barja-Fidalgo C., Graca-Souza A. V. (2004). Heme inhibits human neutrophil apoptosis: involvement of phosphoinositide 3-kinase, MAPK, and NF-kappaB. J. Immunol. 173 2023–2030. 10.4049/jimmunol.173.3.2023 [DOI] [PubMed] [Google Scholar]
- Busch A., Richter A. S., Backofen R. (2008). IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24 2849–2856. 10.1093/bioinformatics/btn544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat J. E., Skaar E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13 509–519. 10.1016/j.chom.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiabrando D., Vinchi F., Fiorito V., Mercurio S., Tolosano E. (2014). Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 5:61. 10.3389/fphar.2014.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipperfield J. R., Ratledge C. (2000). Salicylic acid is not a bacterial siderophore: a theoretical study. Biometals 13 165–168. 10.1023/A:1009227206890 [DOI] [PubMed] [Google Scholar]
- Choby J. E., Skaar E. P. (2016). Heme synthesis and acquisition in bacterial pathogens. J. Mol. Biol. 428 3408–3428. 10.1016/j.jmb.2016.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J. K., Larsen M. H., Ingmer H., Sogaard-Andersen L., Kallipolitis B. H. (2004). The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186 3355–3362. 10.1128/JB.186.11.3355-3362.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. D., Emerson N., Gahan C. G., Hill C. (1999). Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181 6840–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T., Park K., Kim T., Kang S. W., Hahn M. J., Hwang H. Y., et al. (2014). Structural and functional characterization of an Isd-type haem-degradation enzyme from Listeria monocytogenes. Acta Crystallogr. D Biol. Crystallogr. 70(Pt 3), 615–626. 10.1107/S1399004713030794 [DOI] [PubMed] [Google Scholar]
- Foller M., Shumilina E., Lam R., Mohamed W., Kasinathan R., Huber S., et al. (2007). Induction of suicidal erythrocyte death by listeriolysin from Listeria monocytogenes. Cell Physiol. Biochem. 20 1051–1060. 10.1159/000110715 [DOI] [PubMed] [Google Scholar]
- Geoffroy C., Gaillard J. L., Alouf J. E., Berche P. (1987). Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 55 1641–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer N. D., Skaar E. P. (2011). Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 65 129–147. 10.1146/annurev-micro-090110-102851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M. W., Muckenthaler M. U., Andrews N. C. (2004). Balancing acts: molecular control of mammalian iron metabolism. Cell 117 285–297. 10.1016/S0092-8674(04)00343-5 [DOI] [PubMed] [Google Scholar]
- Huang W., Wilks A. (2017). Extracellular heme uptake and the challenge of bacterial cell membranes. Annu. Rev. Biochem. 86 799–823. 10.1146/annurev-biochem-060815-014214 [DOI] [PubMed] [Google Scholar]
- Jin B., Newton S. M., Shao Y., Jiang X., Charbit A., Klebba P. E. (2006). Iron acquisition systems for ferric hydroxamates, haemin and haemoglobin in Listeria monocytogenes. Mol. Microbiol. 59 1185–1198. 10.1111/j.1365-2958.2005.05015.x [DOI] [PubMed] [Google Scholar]
- Joubert L., Dagieu J. B., Fernandez A., Derre-Bobillot A., Borezee-Durant E., Fleurot I., et al. (2017). Visualization of the role of host heme on the virulence of the heme auxotroph Streptococcus agalactiae. Sci. Rep. 7:40435. 10.1038/srep40435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallipolitis B. H., Ingmer H. (2001). Listeria monocytogenes response regulators important for stress tolerance and pathogenesis. FEMS Microbiol. Lett. 204 111–115. 10.1111/j.1574-6968.2001.tb10872.x [DOI] [PubMed] [Google Scholar]
- Kallipolitis B. H., Ingmer H., Gahan C. G., Hill C., Sogaard-Andersen L. (2003). CesRK, a two-component signal transduction system in Listeria monocytogenes, responds to the presence of cell wall-acting antibiotics and affects beta-lactam resistance. Antimicrob. Agents Chemother. 47 3421–3429. 10.1128/AAC.47.11.3421-3429.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba P. E., Charbit A., Xiao Q., Jiang X., Newton S. M. (2012). Mechanisms of iron and haem transport by Listeria monocytogenes. Mol. Membr. Biol. 29 69–86. 10.3109/09687688.2012.694485 [DOI] [PubMed] [Google Scholar]
- Kochan I. (1973). The role of iron in bacterial infections, with special consideration of host-tubercle bacillus interaction. Curr. Top. Microbiol. Immunol. 60 1–30. 10.1007/978-3-642-65502-9_1 [DOI] [PubMed] [Google Scholar]
- Lechowicz J., Krawczyk-Balska A. (2015). An update on the transport and metabolism of iron in Listeria monocytogenes: the role of proteins involved in pathogenicity. Biometals 28 587–603. 10.1007/s10534-015-9849-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledala N., Sengupta M., Muthaiyan A., Wilkinson B. J., Jayaswal R. K. (2010). Transcriptomic response of Listeria monocytogenes to iron limitation and Fur mutation. Appl. Environ. Microbiol. 76 406–416. 10.1128/AEM.01389-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lungu B., Ricke S. C., Johnson M. G. (2009). Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions: a review. Anaerobe 15 7–17. 10.1016/j.anaerobe.2008.08.001 [DOI] [PubMed] [Google Scholar]
- Malmirchegini G. R., Sjodt M., Shnitkind S., Sawaya M. R., Rosinski J., Newton S. M., et al. (2014). Novel mechanism of hemin capture by Hbp2, the hemoglobin-binding hemophore from Listeria monocytogenes. J. Biol. Chem. 289 34886–34899. 10.1074/jbc.M114.583013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M., Wright P. R., Backofen R. (2017). IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 45(W1), W435–W439. 10.1093/nar/gkx279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin H. P., Hill C., Gahan C. G. (2011). The impact of iron on Listeria monocytogenes; inside and outside the host. Curr. Opin. Biotechnol. 22 194–199. 10.1016/j.copbio.2010.10.005 [DOI] [PubMed] [Google Scholar]
- McLaughlin H. P., Xiao Q., Rea R. B., Pi H., Casey P. G., Darby T., et al. (2012). A putative P-type ATPase required for virulence and resistance to haem toxicity in Listeria monocytogenes. PLoS One 7:e30928. 10.1371/journal.pone.0030928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mike L. A., Choby J. E., Brinkman P. R., Olive L. Q., Dutter B. F., Ivan S. J., et al. (2014). Two-component system cross-regulation integrates Bacillus anthracis response to heme and cell envelope stress. PLoS Pathog. 10:e1004044. 10.1371/journal.ppat.1004044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollerup M. S., Ross J. A., Helfer A. C., Meistrup K., Romby P., Kallipolitis B. H. (2016). Two novel members of the LhrC family of small RNAs in Listeria monocytogenes with overlapping regulatory functions but distinctive expression profiles. RNA Biol. 13 895–915. 10.1080/15476286.2016.1208332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton S. M., Klebba P. E., Raynaud C., Shao Y., Jiang X., Dubail I., et al. (2005). The svpA-srtB locus of Listeria monocytogenes: fur-mediated iron regulation and effect on virulence. Mol. Microbiol. 55 927–940. 10.1111/j.1365-2958.2004.04436.x [DOI] [PubMed] [Google Scholar]
- Nielsen J. S., Lei L. K., Ebersbach T., Olsen A. S., Klitgaard J. K., Valentin-Hansen P., et al. (2010). Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 38 907–919. 10.1093/nar/gkp1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby-Sherrouse A. G., Vasil M. L. (2010). Characterization of a heme-regulated non-coding RNA encoded by the prrF locus of Pseudomonas aeruginosa. PLoS One 5:e9930. 10.1371/journal.pone.0009930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrisius J., Bhakdi S., Roth M., Tranum-Jensen J., Goebel W., Seeliger H. P. (1986). Production of listeriolysin by beta-hemolytic strains of Listeria monocytogenes. Infect. Immun. 51 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart C., Trieu-Cuot P. (1997). A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156 193–198. 10.1016/S0378-1097(97)00423-0 [DOI] [PubMed] [Google Scholar]
- Reinhart A. A., Nguyen A. T., Brewer L. K., Bevere J., Jones J. W., Kane M. A., et al. (2017). The Pseudomonas aeruginosa PrrF small RNAs regulate iron homeostasis during acute murine lung infection. Infect. Immun. 85:e00764-16. 10.1128/IAI.00764-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart A. A., Powell D. A., Nguyen A. T., O’Neill M., Djapgne L., Wilks A., et al. (2015). The prrF-encoded small regulatory RNAs are required for iron homeostasis and virulence of Pseudomonas aeruginosa. Infect. Immun. 83 863–875. 10.1128/IAI.02707-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere M. L., Torres V. J., Skaar E. P. (2007). Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals 20 333–345. 10.1007/s10534-006-9032-0 [DOI] [PubMed] [Google Scholar]
- Reniere M. L., Ukpabi G. N., Harry S. R., Stec D. F., Krull R., Wright D. W., et al. (2010). The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol. 75 1529–1538. 10.1111/j.1365-2958.2010.07076.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaferkordt S., Chakraborty T. (1995). Vector plasmid for insertional mutagenesis and directional cloning in Listeria spp. Biotechniques 720-722 724–725. [PubMed] [Google Scholar]
- Sheehan B., Klarsfeld A., Msadek T., Cossart P. (1995). Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 177 6469–6476. 10.1128/jb.177.22.6469-6476.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon J. R., Heinrichs D. E. (2015). Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol. Rev. 39 592–630. 10.1093/femsre/fuv009 [DOI] [PubMed] [Google Scholar]
- Sievers S., Lund A., Menendez-Gil P., Nielsen A., Storm Mollerup M., Lambert Nielsen S., et al. (2015). The multicopy sRNA LhrC controls expression of the oligopeptide-binding protein OppA in Listeria monocytogenes. RNA Biol. 12 985–997. 10.1080/15476286.2015.1071011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers S., Sternkopf Lillebaek E. M., Jacobsen K., Lund A., Mollerup M. S., Nielsen P. K., et al. (2014). A multicopy sRNA of Listeria monocytogenes regulates expression of the virulence adhesin LapB. Nucleic Acids Res. 42 9383–9398. 10.1093/nar/gku630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar E. P., Gaspar A. H., Schneewind O. (2006). Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 188 1071–1080. 10.1128/JB.188.3.1071-1080.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar E. P., Schneewind O. (2004). Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6 390–397. 10.1016/j.micinf.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Stauff D. L., Skaar E. P. (2009). Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol. Microbiol. 72 763–778. 10.1111/j.1365-2958.2009.06684.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I., Perkins-Balding D. (2002). Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21 281–295. 10.1089/104454902753759708 [DOI] [PubMed] [Google Scholar]
- Thorsing M., Dos Santos P. T., Kallipolitis B. H. (2017). Small RNAs in major foodborne pathogens: from novel regulatory activities to future applications. Curr. Opin. Biotechnol. 49 120–128. 10.1016/j.copbio.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459 950–956. 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- Torres V. J., Stauff D. L., Pishchany G., Bezbradica J. S., Gordy L. E., Iturregui J., et al. (2007). A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1 109–119. 10.1016/j.chom.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini-Allocatelli C. (2013). Protein machineries involved in the attachment of heme to cytochrome c: protein structures and molecular mechanisms. Scientifica 2013 505714. 10.1155/2013/505714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland J. A., Kocks C., Dramsi S., Ohayon H., Geoffroy C., Mengaud J., et al. (1992). Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland J. A., Kuhn M., Berche P., Chakraborty T., Dominguez-Bernal G., Goebel W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14 584–640. 10.1128/CMR.14.3.584-640.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman C. A., Skaar E. P. (2012). Metalloregulation of Gram-positive pathogen physiology. Curr. Opin. Microbiol. 15 169–174. 10.1016/j.mib.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P. R., Georg J., Mann M., Sorescu D. A., Richter A. S., Lott S., et al. (2014). CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 42 W119–W123. 10.1093/nar/gku359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Skaar E. P., Zhang R., Joachimiak G., Gornicki P., Schneewind O., et al. (2005). Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 280 2840–2846. 10.1074/jbc.M409526200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Jiang X., Moore K. J., Shao Y., Pi H., Dubail I., et al. (2011). Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes. Mol. Microbiol. 80 1581–1597. 10.1111/j.1365-2958.2011.07667.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.