Abstract
Dietary supplementation of docosahexaenoic acid (DHA)-enriched products inhibits the final step of biohydrogenation in the adult rumen, resulting in the accumulation of 18:1 isomers, particularly of trans(t)-11 18:1. Occasionally, a shift toward the formation of t10 intermediates at the expense of t11 intermediates can be triggered. However, whether similar impact would occur when supplementing DHA-enriched products during pregnancy or early life remains unknown. Therefore, the current in vivo study aimed to investigate the effect of a nutritional intervention with DHA in the early life of goat kids on rumen biohydrogenation and microbial community. Delivery of DHA was achieved by supplementing DHA-enriched microalgae (DHA Gold) either to the maternal diet during pregnancy (prenatal) or to the diet of the young offspring (postnatal). At the age of 12 weeks, rumen fluid was sampled for analysis of long-chain fatty acids and microbial community based on bacterial 16S rRNA amplicon sequencing. Postnatal supplementation with DHA-enriched microalgae inhibited the final biohydrogenation step, as observed in adult animals. This resulted particularly in increased ruminal proportions of t11 18:1 rather than a shift to t10 intermediates, suggesting that both young and adult goats might be less prone to dietary induced shifts toward the formation of t10 intermediates, in comparison with cows. Although Butyrivibrio species have been identified as the most important biohydrogenating bacteria, this genus was more abundant when complete biohydrogenation, i.e. 18:0 formation, was inhibited. Blautia abundance was positively correlated with 18:0 accumulation, whereas Lactobacillus spp. Dialister spp. and Bifidobacterium spp. were more abundant in situations with greater t10 accumulation. Extensive comparisons made between current results and literature data indicate that current associations between biohydrogenation intermediates and rumen bacteria in young goats align with former observations in adult ruminants.
Keywords: docosahexaenoic acid, early life, goat, microalgae, rumen biohydrogenation, rumen microbiome
Introduction
Ruminant diets commonly contain forages and concentrates. Fats are often incorporated to increase the energy density in diets for high yielding dairy cattle. Moreover, fat sources have been included in ruminant diets to increase concentrations of human-health promoting n-3 fatty acids (FA) and bioactive conjugated linoleic acids (CLA) in milk and meat. The majority of the dietary FA are 18-carbon unsaturated FA (linolenic acid, 18:3n-3; linoleic acid, 18:2n-6 and oleic acid, cis-9 18:1; Ferlay et al., 2017). However, following lipolysis, dietary unsaturated FA are converted to saturated FA in the rumen by bacteria (Buccioni et al., 2012). This process is called biohydrogenation and involves several consecutive conversions via various pathways, resulting in a plethora of FA isomers. Nevertheless, the predominant biohydrogenation pathway of 18:2n-6 is through its isomerization to cis-9, trans-11 CLA (c9, t11 CLA) which is further hydrogenated to t11 18:1 and ultimately to 18:0 (Bauman and Griinari, 2003; Harvatine et al., 2009). 18:3n-3 is mainly isomerized to c9, t11, c15 conjugated linolenic acid (CLnA) followed by a hydrogenation to t11, c15 18:2, t11 18:1, and ultimately to 18:0 (Shingfield and Wallace, 2014).
Regarding the potential benefits for human health, several studies investigated the effect of supplementation of poly-unsaturated FA (PUFA) in the ruminant diet (Wasowska et al., 2006; Shingfield et al., 2012; Toral et al., 2016), of which some contain docosahexaenoic acid (DHA, 22:6n-3). In monogastric animals and humans, feeding of such DHA-enriched supplements during pregnancy or in the early life has gained interest, as they are claimed to support vitality and growth at this young stage (Tanghe and De Smet, 2013; Brenna and Carlson, 2014). However, those supplements modify rumen biohydrogenation of 18:2n-6 and 18:3n-3 (Klein and Jenkins, 2011) as well as the rumen microbial population in adult ruminants (Boeckaert et al., 2008; Shingfield et al., 2012). Docosahexaenoic acid inhibits the final step of biohydrogenation to 18:0, which results in the accumulation of 18:1 isomers (Boeckaert et al., 2007; Zhao et al., 2016), mainly t11 18:1 (e.g., Shingfield et al., 2012; Zhao et al., 2016; Zhu et al., 2016). Some studies also observed an increase of t10 18:1 after dietary supplementation of DHA (Boeckaert et al., 2007; Shingfield et al., 2012; Zhu et al., 2016) which could indicate a shift from the main biohydrogenation pathway toward the formation of t10 intermediates at the expense of t11 intermediates in the rumen. In lactating ruminants, these intermediates (e.g., t10, c12 CLA) could inhibit milk fat synthesis in the mammary gland (Harvatine et al., 2009).
It is unknown whether similar changes occur when supplementing DHA-enriched products to ruminants at young age, during the period of rumen microbial colonization, as to our knowledge, there are no studies investigating the effect of DHA on rumen biohydrogenation using young ruminants. Therefore, the current in vivo study aimed to investigate the effect of a nutritional intervention with DHA in the early life of goat kids on rumen biohydrogenation and microbial community. Delivery of DHA to young animals can be either through the maternal diet (either prenatal during pregnancy or through the dam's milk) or directly through the diet of the young animal (postnatal). In the current study, we investigated the effect of prenatal and/or postnatal supplementation of DHA-enriched microalgae, which we hypothesized to induce changes in rumen microbial community and biohydrogenation, similar as in adult animals. As the microbial community in young ruminants may be less complex, it was hypothesized that the results of this experiment also could highlight a potential role of particular bacterial species in different rumen biohydrogenation steps.
Materials and methods
Animals, diets, and experimental design
All experimental procedures involving animals were approved by the Ethical Committee of the Faculty of Veterinary Medicine and Bioscience Engineering of Ghent University (EC2015/148). One hundred and eight Saanen dairy goats (46 multiparous and 62 primiparous goats) at similar pregnancy stage (insemination between 02/09/2015 and 25/09/2015) were selected during the last 6 weeks of pregnancy. All animals were housed in group pens and were fed, according to their maintenance requirements, grass silage (first 4 weeks) or a roughage mixture (% on DM basis: 24/60/16 of grass silage/maize silage/fodder beet; last 2 weeks) ad libitum supplemented with a standard concentrate (1 kg DM/day) which contained rapeseed (18.2 g/kg of fresh product). During the last 3 weeks of pregnancy, the animals were randomly divided into two experimental groups. One group (D−) received the standard concentrate during this period. The other group (D+, prenatal treatment) was supplemented with a DHA-enriched microalgae product, DHA Gold, replacing rapeseed (18.2 g/kg of fresh product; DHAgoldTM, DSM Nutritional Products, Deinze, Belgium; FA composition (g/100 g fresh material): 14:0, 3.07; 16:0, 8.38; 18:0, 0.20; 18:1, 0.10; 18:2n-6, < 0.01; 22:5n-6, 5.40; 22:6n-3, 14.84). Both concentrates were formulated to be isoenergetic and isoproteic (Table 1). Diets were offered as two equal meals at 09h00 and 15h00. Animals had free access to fresh water.
Table 1.
Ingredients and fatty acid composition (g/kg of fresh product) of concentrates of does.
| Ingredient | Concentrate | |
|---|---|---|
| Soybean meal | 260 | |
| Rapeseed (D−) or DHA Gold (D+)1 | 18.2 | |
| Maize | 231.8 | |
| Beet pulp | 200 | |
| Wheat | 150 | |
| Molasses | 50 | |
| Lignosulphonate | 20 | |
| Vitamin premix AD2 | 16 | |
| Vitamin E2 | 1 | |
| Feed phosphate | 16 | |
| Trace elements3 | 16 | |
| Salt | 10 | |
| MgO 93% | 7 | |
| Calcium carbonate | 4 | |
| DVE4 | 155 | |
| VEM5 | 920 | |
| Fatty acid composition | D− | D+ |
| 14:0 | n.d. | 0.9 |
| 16:0 | 3.3 | 5.1 |
| 18:0 | 0.6 | 0.5 |
| 18:1 | 8.9 | 3.8 |
| 18:2n-6 | 9.6 | 8.3 |
| 22:5n-6 | n.d. | 1.1 |
| 22:6n-3 | < 0.1 | 2.8 |
n.d.: not detected.
D, doe; +, supplemented daily with DHA Gold during the last 3 weeks of gestation (0.28 g per kg BW); −, no DHA Gold supplementation.
VIT A, D and E (UI/g): A (312), D (62.5), and E (6,000).
Premix of trace elements and minerals (mg/g): Fe (281), Cu (62.5), Mn (116), Co (1.4), Zn (175), I (4.8), Ca (244.3), P (157), K (248), Mg (73.8), and Na (8.1).
DVE, true protein digested in the small intestine (Tamminga et al., 1994).
VEM, feed unit milk (1,000 VEM = 6.9 MJ; Van Es, 1978).
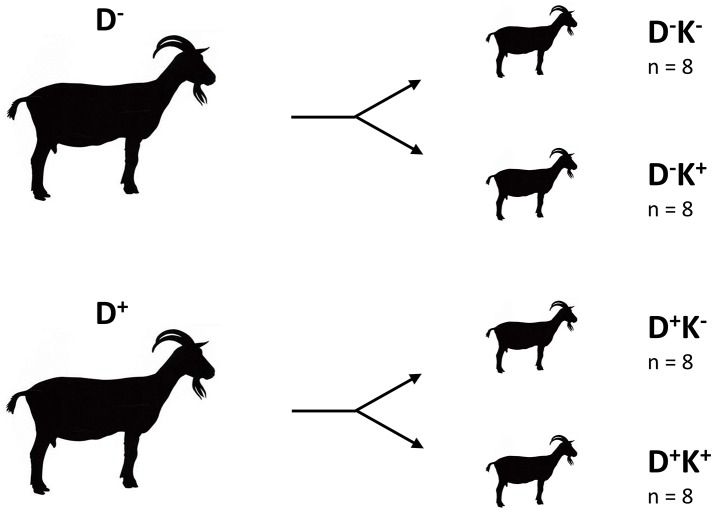
After birth, only male twin goat kids were further used in the experiment. From each group of does (D− and D+), eight male twins were selected. The twins were immediately separated from their mother and each randomly allocated to one of two experimental groups. One group (K+, postnatal treatment) was supplemented daily with 0.28 g DHA Gold per kg of body weight (BW) whereas the other group (K−) was not. This resulted in four experimental groups of eight kids per group (D−K−, D−K+, D+K− and D+K+) as illustrated in Figure 1. Kids were housed in pairs of the same experimental condition in pens equipped with rubber mats and bedded with straw (1.8 × 2.2 m). The pens were constructed to avoid physical contact between the neighboring kids during the first weeks of life. Treatment (K+) started immediately after birth until the age of 85 ± 2 days (ca. 12 weeks old). A DHA Gold emulsion in water (0.333 g/mL) was prepared fresh and was administered orally before the morning and afternoon feeding with a 10-mL syringe. All kids received colostrum during the first 2 days after birth, which was replaced by goat milk powder from day 3 until weaning at 9 weeks. All kids received hay ad libitum and a standard concentrate (maximum 0.5 kg/day per pen) from week 3 onwards. From week 6 until weaning, milk powder gradually decreased and the amount of concentrate (maximum 1 kg/day per pen) increased. Diets were divided into two meals at 8h30 and 16h30. All kids had constant access to fresh water.
Figure 1.
Experimental groups. D, doe; K, kid; +, supplemented with DHA Gold (0.28 g per kg BW); −, no DHA Gold supplementation.
Rumen sample collection
At the age of 85 ± 2 days (ca. 12 weeks old), rumen fluid was collected by stomach tube before the morning feeding after an overnight period without access to concentrate and hay. Briefly, a flexible plastic tube (15 mm i.d.) with 3 holes (± 5 mm i.d.) in the probe head was warmed up using hot water and was inserted in the rumen via the esophagus. Rumen samples were obtained using an electric vacuum pump. This method has been validated as a feasible alternative to surgical rumen cannulation in goats to examine dietary effects on the rumen FA profile and the rumen bacterial community (Ramos-Morales et al., 2014). Treated kids (K+) received their last supplement of DHA Gold 2 h before the sampling. An aliquot of 50 mL rumen fluid was filtered through a sieve with a pore size of 1 mm. Subsamples for analysis of long-chain FA (LCFA; 2 mL homogenized rumen fluid) were collected in glass tubes, stored at −20°C and freeze-dried prior to LCFA analysis. Subsamples for bacterial community analysis (3 mL homogenized rumen fluid) were collected in cryovials and stored at −80°C until gDNA extraction. For practical reasons, rumen sample collection was performed on two different days. On both sampling days, animals from each experimental condition were included.
Long-chain fatty acid composition
Fatty acids were methylated as described by Vlaeminck et al. (2014). Briefly, toluene (2 mL) containing the internal standard (21:0; Sigma Aldrich, Diegem, Belgium) and methanolic NaOH (2 mL) were added and the mixture was incubated at 70°C for 60 min. This was followed by 30 min at 50°C after addition of methanolic HCl (3 mL), prepared by dissolving acetyl chloride in methanol (5/1, v/v). Fatty acid methyl esters (FAME) were extracted with hexane. Analysis of FAME was carried out using a gas chromatograph (HP7890A; Agilent Technologies, Diegem, Belgium) equipped with a SP-2560 capillary column (75 m × 0.18 mm i.d. × 0.14 μm thickness; Supelco Analytical, Bellefonte, PA, USA) and a flame ionization detector. The temperature program was as follows: initially 70°C for 2 min, increasing by 15°C/min to 150°C, followed by a second increase at 1°C/min up to 165°C and holding for 12 min, followed by a third increase at 2°C/min to 170°C, held at 170°C for 5 min, increased at 5°C/min to 215°C and held at 215°C for 20 min. Inlet and detector temperatures were 250°C and 255°C, respectively. The split ratio was 50:1. Hydrogen was used as the carrier gas at a flow rate of 1 mL/min. Identities of peaks were determined using mixtures of methyl ester standards (22:5n-6 and GLC463, Nu-Chek-Prep, Elysian, MN, USA; c9, t11 CLA and t10, c12 CLA, Larodan 279, Fine Chemicals AB, Malmö, Sweden). Quantification of FA was based on the area of the internal standard and on the conversion of peak areas to the weight of FA by a theoretical response factor for each FA (Ackman and Sipos, 1964; Wolff et al., 1995).
Statistical analysis
Data were analyzed using the MIXED procedure of SAS (version Enterprise Guide 7.1; SAS Institute Inc., Cary, NC, USA) by the following model: Yijkl = μ + Di + Kj + Tk + DOl + Di × Kj + εijkl, with Di the fixed effect of prenatal treatment (i = D− or D+), Kj the fixed effect of postnatal treatment (j = K− or K+), Tk the random effect of sampling day (k = day 1 or 2), DOl the random effect of doe (l = doe 1 until 16), Di × Kj the interaction between prenatal and postnatal treatment and εijkl the residual error term. Least square means are reported with treatment effects declared significant at P < 0.05. Tukey-Kramer multiple comparison test was used to evaluate significant differences.
Bacterial community analysis
DNA extraction
gDNA extraction was performed using the repeated bead beating plus column purification (RBB+C) method as described by Yu and Morrison (2004). The yield and quality of extracted DNA were determined using a NanoDrop spectrophotometer (VWR International BVBA, Leuven, Belgium).
Illumina library generation and data mining
Bacterial 16S rRNA amplicon sequencing (V3-V4 region) using Illumina MiSeq technology (2 × 300 bp) was performed by Macrogen Sequencing Service (Macrogen Korea, Seoul, Rep. of Korea). Preparation of the amplicon barcoded library (primers: 344F and 806R; Klindworth et al., 2013) was based on the 16S metagenomic sequencing library preparation protocol provided by the manufacturer (Illumina, https://support.illumina.com).
The amplicon sequencing dataset was demultiplexed and barcodes were clipped off by the sequence provider. Forward and reverse reads were merged using the fastq-join method (Aronesty, 2011) after which primer removal and quality filtering was performed using the open-source software package QIIME (v1.9.1; Caporaso et al., 2010). This resulted in 43 459 ± 5812 reads per sample. Rarefaction analyses were performed using the QIIME software package (Caporaso et al., 2010) indicating that the sequencing depth was sufficient to analyze the bacterial communities in all samples (data not shown). The subsequent analysis, picking Operational Taxonomic Units (OTU), assigning taxonomy, inferring phylogeny and creating OTU tables, were also performed by QIIME software (Caporaso et al., 2010). The sequences were clustered into OTU using the open-reference OTU picking workflow with a 97 % similarity threshold using UCLUST (Edgar, 2010) and chimeras were removed using UCHIME (Edgar, 2010). Representative sequences from each OTU were aligned using PyNAST (Caporaso et al., 2010) and a taxonomy identity was assigned to each representative sequence using the method UCLUST (Edgar, 2010) and the GreenGenes database for reference (v13_8; DeSantis et al., 2006). OTU with <0.005% of the total number of sequences were removed. To ensure the comparability of the species diversity between the samples, normalized/rarefied OTU sets were used for further analysis.
Alpha diversity indices (Chao1 index, Observed OTU, PD whole tree) were calculated and significant differences between experimental groups were determined by the non-parametric Kruskal-Wallis test in QIIME (Caporaso et al., 2010). Beta diversity indices between samples were determined in QIIME (Caporaso et al., 2010) based on Bray-Curtis dissimilarity (Bray and Curtis, 1957) and Unweighted UniFrac metric (Lozupone and Knight, 2005). The non-parametric permutational MANOVA-based statistical test ANOSIM was used in QIIME (Caporaso et al., 2010) to determine differences in microbial community between experimental groups. Differences in relative abundance of the different taxa (at genus level) between treatments were determined using the MIXED procedure of SAS (version Enterprise Guide 7.1; SAS Institute Inc., Cary, NC, USA) by the following model: Yijkl = μ + Di + Kj + Tk + DOl + Di × Kj + εijkl, with Di the fixed effect of prenatal treatment (i = D− or D+), Kj the fixed effect of postnatal treatment (j = K− or K+), Tk the random effect of sampling day (k = day 1 or 2), DOl the random effect of doe (l = doe 1 until 16), Di × Kj the interaction between prenatal and postnatal treatment and εijkl the residual error term. Least square means are reported with treatment effects declared significant at P < 0.05 and with a trend toward significance at 0.05 ≤ P < 0.10. Significant differences were evaluated with the Tukey-Kramer multiple comparison test. In addition, Spearman Rank correlation was used to check the correlation between different FA and different taxa (at genus level) using QIIME (Caporaso et al., 2010).
Sequence data have been deposited in the National Center for Biotechnology Information (NCBI) database under accession number PRJNA414378.
Multivariate statistical analysis
A bipartite network was inferred using a similarity matrix obtained from a regularized canonical correlation analysis (rCCA), using the package mixOmics (v6.2.0; Lê Cao et al., 2011) in R (v3.4.1; Kurtz et al., 2015). In this analysis, the correlation values between the relative abundances of bacterial taxa (at genus level) and each LCFA were computed to calculate a similarity matrix. Then, these values were projected onto the space spanned by the first components retained in the analysis. Three relevant components were obtained setting a threshold to R = 0.40. Relevance networks are a robust approach to highlight functional relationships, because they simultaneously represent positive and negative correlations, which are missed by methods using Euclidian distances. Another advantage of the rCCA is its ability to represent correlations across disparate biological measures, such as the bacterial relative abundances and metabolic information (De Weirdt et al., 2017).
Results
Rumen fatty acid composition
Postnatal treatment with 0.28 g DHA Gold per kg BW increased the rumen proportions of 14:0 (P = 0.003), 16:0 (P < 0.001), 22:5n-6 (P < 0.001) and 22:6n-3 (P < 0.001, Table 2). This may be associated with the high amounts of these FA in the algal cell biomass. In contrast, prenatal treatment did not affect the rumen proportion of any of these FA (P > 0.05, Table 2).
Table 2.
Effect of prenatal and/or postnatal treatment of goat kids with DHA Gold on proportions of long-chain fatty acids (g/100 g fatty acids) in rumen fluid.
| Fatty acid3 | Experimental group1 | SEM2 | P-value | |||||
|---|---|---|---|---|---|---|---|---|
| D−K− (n = 8) | D−K+ (n = 8) | D+K− (n = 8) | D+K+ (n = 8) | Prenatal treatment (D) | Postnatal treatment (K) | D × K | ||
| 14:0 | 1.58 | 3.33 | 2.59 | 3.35 | 0.478 | 0.203 | 0.003 | 0.178 |
| 16:0 | 15.31 | 17.59 | 16.01 | 17.38 | 0.667 | 0.570 | <0.001 | 0.247 |
| 18:0 | 43.46 | 32.05 | 40.43 | 32.32 | 2.710 | 0.633 | 0.001 | 0.503 |
| t6 18:1 + t7 18:1 + t8 18:1* | 0.21 | 0.51 | 0.16 | 0.38 | 0.089 | 0.132 | 0.002 | 0.474 |
| t9 18:1 | 0.05 | 0.31 | 0.06 | 0.23 | 0.046 | 0.489 | <0.001 | 0.356 |
| t10 18:1* | 0.70 | 1.09 | 0.56 | 1.61 | 0.597 | 0.403 | 0.320 | 0.731 |
| t11 18:1* | 0.64 | 1.62 | 0.81 | 1.37 | 0.323 | 0.921 | 0.002 | 0.232 |
| t12 18:1 | 0.15 | 0.54 | 0.10 | 0.32 | 0.078 | 0.106 | 0.002 | 0.272 |
| c9 18:1 + t13 18:1 + t14 18:1 | 2.39 | 2.85 | 2.37 | 2.43 | 0.335 | 0.476 | 0.372 | 0.493 |
| c11 18:1 + t15 18:1 | 1.23 | 2.03 | 1.58 | 1.59 | 0.246 | 0.850 | 0.123 | 0.131 |
| c12 18:1 | 0.41 | 0.29 | 0.39 | 0.27 | 0.054 | 0.706 | 0.037 | 0.968 |
| c13 18:1 | 0.10 | 0.32 | 0.27 | 0.38 | 0.207 | 0.430 | 0.015 | 0.334 |
| c14 18:1 + t16 18:1 | 0.33 | 0.49 | 0.13 | 0.25 | 0.181 | 0.174 | 0.059 | 0.801 |
| Sum 18:1* | 6.31 | 10.14 | 6.35 | 8.74 | 1.271 | 0.380 | 0.018 | 0.372 |
| c9, t11 CLA | 0.07 | 0.19 | 0.12 | 0.17 | 0.058 | 0.873 | 0.164 | 0.574 |
| t10, c12 CLA | 0.01 | 0.01 | <0.01 | <0.01 | 0.010 | 0.179 | 0.969 | 0.969 |
| 18:2n-6 | 2.67 | 1.97 | 2.57 | 1.71 | 0.347 | 0.615 | 0.028 | 0.812 |
| c9, t11 CLA + t11 18:1* | 0.72 | 1.81 | 0.92 | 1.53 | 0.334 | 0.980 | 0.003 | 0.227 |
| t10, c12 CLA + t10 18:1* | 0.71 | 1.11 | 0.56 | 1.61 | 0.597 | 0.363 | 0.312 | 0.719 |
| 18:3n-3 | 0.59 | 0.43 | 0.70 | 0.56 | 0.075 | 0.157 | 0.038 | 0.884 |
| c9, t11, c15 CLnA | 0.09 | 0.15 | 0.10 | 0.06 | 0.034 | 0.350 | 0.673 | 0.058 |
| 22:5n-6 | <0.01 | 1.43 | 0.12 | 1.75 | 0.293 | 0.140 | <0.001 | 0.654 |
| 22:6n-3 | <0.01 | 3.81 | 0.34 | 4.63 | 0.817 | 0.137 | <0.001 | 0.716 |
D, doe; K, kid; +, supplemented with DHA Gold (0.28 g per kg BW); −, no DHA Gold supplementation.
SEM, standard error of the mean.
t, trans; c, cis; CLA, conjugated linoleic acid; CLnA, conjugated linolenic acid.
Reported P-values are the P-values from the logarithm.
Prenatal treatment with DHA Gold did not affect the rumen proportions of 18:2n-6, 18:3n-3 and their biohydrogenation intermediates (P > 0.05, Table 2), whereas postnatal treatment decreased the proportions of 18:2n-6 (P = 0.028) and 18:3n-3 (P = 0.038) in the rumen. However, when expressed as μg per mL of rumen fluid (Supplementary Table 1), no significant effect of postnatal treatment on 18:2n-6 or 18:3n-3 was observed (P > 0.05). The rumen proportions of t6 18:1 + t7 18:1 + t8 18:1 (P = 0.002), t9 18:1 (P < 0.001), t11 18:1 (P = 0.002), t12 18:1 (P = 0.002), and c13 18:1 (P = 0.015) increased upon postnatal DHA Gold supplementation (Table 2). This resulted in a higher proportion of total 18:1 FA (P = 0.018), which was accompanied by a decrease in 18:0 (P = 0.001). In contrast to the other 18:1 FA, c12 18:1 decreased (P = 0.037) after postnatal treatment with DHA Gold. No difference between treatments was observed for c9, t11 CLA (P = 0.164), t10, c12 CLA (P = 0.969) and c9, t11, c15 CLnA (P = 0.673). In none of the treatments, t10, c12, c15 CLnA was detected in the rumen (data not shown).
Neither prenatal nor postnatal treatment with DHA Gold induced a shift toward the formation of t10 intermediates at the expense of t11 intermediates. Nevertheless, both t10 18:1 as well as t11 18:1 increased upon postnatal DHA Gold supplementation. However, the increase in t10 intermediates was not significant due to large inter-animal variation ( ± SD = 1.00 ± 1.66 g/100 g FA; SEM = 0.597 g/100 g FA).
Rumen microbiome
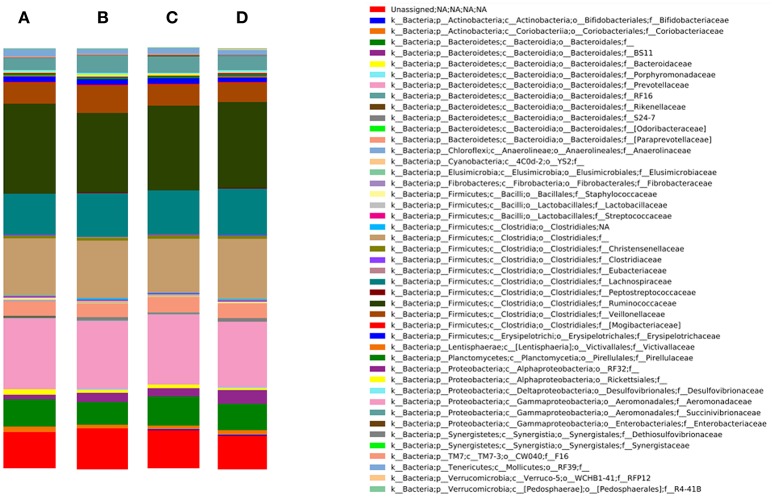
Figure 2 represents the bacterial community composition in the rumen of goats from the different experimental groups. The 3 most abundant phyla were Firmicutes (52.8% of relative abundance), Bacteroidetes (29.9% of relative abundance) and Proteobacteria (4.6% of relative abundance). Within the Firmicutes, the families Veillonellaceae, Ruminococcaceae (predominant genus: Ruminococcus) and Lachnospiraceae and undefined families within the order of the Clostridiales dominated the rumen. The predominant families of the Bacteroidetes were Prevotellaceae (predominant genus: Prevotella) and Paraprevotellaceae, whereas the Proteobacteria mainly consisted of undefined genera within the family Succinivibrionaceae.
Figure 2.
Bacterial community composition on family level in the rumen of the different experimental groups: (A) D−K−, (B) D−K+, (C) D+K−, and (D) D+K+ (n = 8). D, doe; K, kid; +, supplemented with DHA Gold (0.28 g per kg BW); −, no DHA Gold supplementation.
Neither prenatal nor postnatal treatment influenced bacterial species richness as expressed by different alpha diversity indices (P > 0.10; Supplementary Figure 1). Nevertheless, the statistical test ANOSIM revealed differences in microbial community between experimental groups (PBray−Curtis = 0.011; PUnweightedUniFrac = 0.016). Taxa with significant differences or with a trend toward significance upon prenatal and/or postnatal treatment with DHA Gold are shown in Table 3. Prenatal treatment increased or tended to increase the relative abundance of the families RF16 (P = 0.020) and BS11 (P = 0.075) within the order of the Bacteroidales and the genera Clostridium (P = 0.009) and Butyrivibrio (P = 0.079) within the order of the Clostridiales. Postnatal treatment increased the relative abundance of the families BS11 (P = 0.038) and S24-7 (P = 0.006) within the order of the Bacteroidales and of Butyrivibrio species (P = 0.007) and the family Veillonellaceae (P = 0.011; order Clostridiales) whereas it decreased the relative abundance of BF311 (P = 0.033; order Bacteroidales), Blautia species (P = 0.028; order Clostridiales), RF39 (P = 0.016) and YS2 (P = 0.025). Besides this, interaction effects between prenatal and postnatal treatment were observed for some taxa. Postnatal treatment altered (P < 0.05) the relative abundance of some taxa, but only when does were not treated with DHA Gold (YRC22, Ruminococcaceae, Acidaminococcus, Selenomonas, Succiniclasticum, L7A_E11).
Table 3.
Average relative abundance (%) of different bacterial taxa in the rumen of goat kids supplemented pre- and/or postnatally with DHA Gold.
| Taxon: order/family/genus3 | Experimental group1 | SEM2 | P-value | |||||
|---|---|---|---|---|---|---|---|---|
| D−K− (n = 8) | D−K+ (n = 8) | D+K− (n = 8) | D+K+ (n = 8) | Prenatal treatment (D) | Postnatal treatment (K) | D × K | ||
| Aeromonadales/Succinivibrionaceae | 2.28B | 3.59A | 3.44AB | 2.93AB | 0.506 | 0.624 | 0.449 | 0.093 |
| Anaerolineales/Anaerolinaceae/SHD-231 | <0.01 | 0.06 | 0.03 | 0.10 | 0.032 | 0.334 | 0.060 | 0.910 |
| Bacteroidales/Bacteroidaceae/BF311 | 0.48 | 0.28 | 0.34 | 0.21 | 0.069 | 0.147 | 0.033 | 0.608 |
| Bacteroidales/BS11 | 0.87 | 2.13 | 1.94 | 3.01 | 0.508 | 0.075 | 0.038 | 0.854 |
| Bacteroidales/Paraprevotellaceae/YRC22 | 0.37b | 0.72a | 0.55ab | 0.45ab | 0.147 | 0.654 | 0.189 | 0.027 |
| Bacteroidales/RF16 | 0.08 | 0.02 | 0.18 | 0.16 | 0.060 | 0.020 | 0.233 | 0.632 |
| Bacteroidales/S24-7 | 0.28 | 0.49 | 0.39 | 0.72 | 0.111 | 0.111 | 0.006 | 0.490 |
| Clostridiales | 0.40 | 0.49 | 0.30 | 0.41 | 0.054 | 0.138 | 0.091 | 0.876 |
| Clostridiales/Clostridiaceae/Clostridium | 0.03 | <0.01 | 0.18 | 0.21 | 0.152 | 0.009 | 0.836 | 0.538 |
| Clostridiales/Lachnospiraceae/Blautia | 0.45 | 0.21 | 0.37 | 0.27 | 0.071 | 0.862 | 0.028 | 0.368 |
| Clostridiales/Lachnospiraceae/Butyrivibrio | 1.88 | 2.67 | 2.38 | 3.17 | 0.258 | 0.079 | 0.007 | 0.996 |
| Clostridiales/Ruminococcaceae | 15.70a | 13.85b | 13.71b | 14.12b | 0.664 | 0.075 | 0.089 | 0.013 |
| Clostridiales/Veillonellaceae | 2.15 | 3.26 | 2.02 | 2.21 | 0.323 | 0.168 | 0.011 | 0.055 |
| Clostridiales/Veillonellaceae/Acidaminococcus | 0.31a | 0.17b | 0.13b | 0.18ab | 0.059 | 0.210 | 0.082 | 0.004 |
| Clostridiales/Veillonellaceae/Selenomonas | 0.02b | 0.12a | 0.02b | 0.05b | 0.019 | 0.135 | <!0.001 | 0.048 |
| Clostridiales/Veillonellaceae/Succiniclasticum | 0.79b | 1.25a | 1.17a | 1.02ab | 0.098 | 0.462 | 0.127 | 0.007 |
| Desulfovibrionales/Desulfovibrionaceae | 0.04 | 0.08 | 0.03 | 0.07 | 0.022 | 0.795 | 0.079 | 0.862 |
| Enterobacteriales/Enterobacteriaceae/NA | 0.01b | 0.02ab | 0.05a | <0.01b | 0.011 | 0.435 | 0.100 | 0.023 |
| Erysipelotrichales/Erysipelotrichaceae/L7A_E11 | 0.04b | 0.12a | 0.10a | 0.10a | 0.037 | 0.504 | 0.041 | 0.042 |
| Pirellulales/Pirellulaceae | 0.43A | 0.32B | 0.32AB | 0.40AB | 0.047 | 0.717 | 0.540 | 0.067 |
| RF39 | 1.73 | 1.08 | 1.33 | 1.15 | 0.186 | 0.388 | 0.016 | 0.141 |
| Synergistales/Synergistaceae/NA | 0.09 | 0.04 | 0.02 | <0.01 | 0.028 | 0.191 | 0.031 | 0.296 |
| YS2 | 0.35 | 0.21 | 0.39 | 0.17 | 0.073 | 0.984 | 0.025 | 0.567 |
Only taxa with significant differences (P < 0.05) or with a trend toward significance (0.05 ≤ P < 0.10) are shown.
D, doe; K, kid; +, supplemented with DHA Gold (0.28 g per kg BW); −, no DHA Gold supplementation.
SEM, standard error of the mean.
NA, not assigned.
Means annotated with a different letter differ (P < 0.05) between experimental groups.
Means annotated with a different capital letter tend to differ (0.05 ≤ P < 0.10) between experimental groups.
Correlation between important 18-carbon fatty acids and microbial population in the rumen
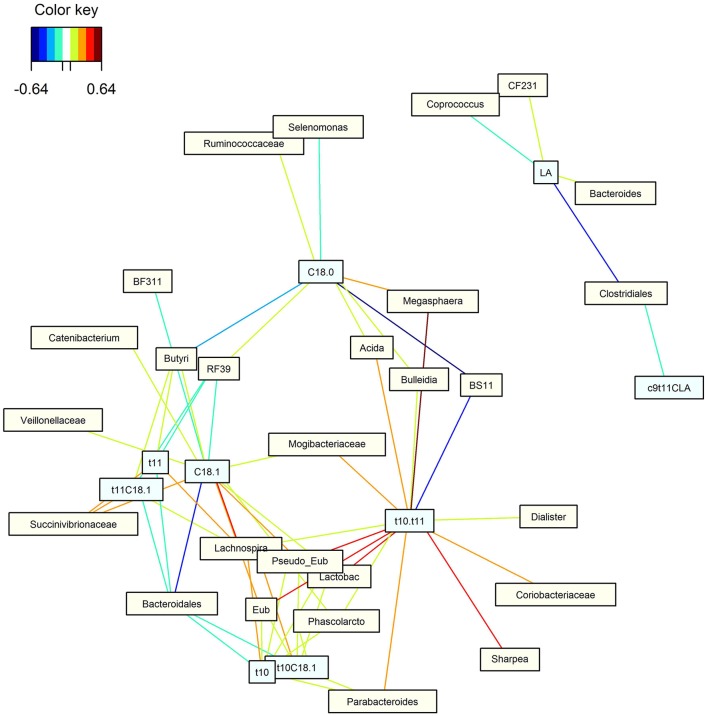
Two different approaches were used to investigate correlations between 18:2n-6, 18:3n-3 or their biohydrogenation intermediates and bacterial taxa. Spearman Rank correlations were calculated in QIIME and are presented in Table 4. Secondly, a bipartite network was inferred using a similarity matrix obtained from a regularized canonical correlation analysis (rCCA; Figure 3). Although most of the significant correlations were rather weak (|R| < 0.50), some stronger correlations were found. Undefined genera of the order Clostridiales were found to be negatively correlated with 18:2n-6 (Figure 3) whereas the family S24-7 within the order of the Bacteroidales was found to be negatively correlated with 18:3n-3 (Table 4). Acidaminococcus, RF16, Ruminococcaceae and BF311 were correlated with t11 intermediates based on Spearman Rank correlation (Table 4), however no strong correlations with these taxa were observed in Figure 3. Bifidobacterium (Table 4), Dialister (Table 4), Megasphaera (Table 4, Figure 3), Coriobacteriaceae (Table 4, Figure 3), Lactobacillus (Table 4, Figure 3), Sharpea (Figure 3), Pseudoramibacter eubacterium (Figure 3) and Eubacterium (Figure 3) were positively correlated with t10 intermediates or the ratio of t10 to t11 intermediates whereas Pirellulaceae (Table 4), Rickettsiales (Table 4) and BS11 (Figure 3) correlated negatively with these intermediates. The family Succinivibrionaceae within the order of the Aeromonadales (Table 4, Figure 3) and the genus Lachnospira (Figure 3) correlated positively with 18:1 FA whereas unknown genera within the order of the Bacteroidales correlated negatively with 18:1 FA (Figure 3). Besides this, BS11 as well as Butyrivibrio correlated negatively with 18:0 (Table 4, Figure 3).
Table 4.
Ruminal bacterial taxa (97% sequence similarity) correlated with 18:2n-6, 18:3n-3 or related biohydrogenation intermediates.
| Taxon: order/family/genus3 | R1,2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 18:2n-6 | 18:3n-3 | c9, t11, c15 CLnA | c9, t11 CLA | t11 18:1 | t10 18:1 | t10:t11 | Sum 18:1 | 18:0 | |
| Aeromonadales/Succinivibrionaceae | 0.33$ | 0.54# | |||||||
| Aeromonadales/Succinivibrionaceae/Succinivibrio | 0.31$ | ||||||||
| Bacteroidales | 0.37# | −0.38# | −0.39# | ||||||
| Bacteroidales/Bacteroidaceae/Bacteroides | 0.31$ | 0.35$ | 0.35$ | ||||||
| Bacteroidales/Bacteroidaceae/BF311 | 0.44# | 0.42# | −0.53# | −0.31$ | −0.45# | ||||
| Bacteroidales/BS11 | −0.49# | −0.51# | |||||||
| Bacteroidales/Odoribacteraceae/Butyricimonas | 0.42# | 0.38# | |||||||
| Bacteroidales/Porphyromonadaceae/Parabacteroides | 0.32$ | 0.40# | 0.30$ | 0.40# | |||||
| Bacteroidales/Paraprevotellaceae | −0.49# | ||||||||
| Bacteroidales/Paraprevotellaceae/CF231 | −0.33$ | ||||||||
| Bacteroidales/Paraprevotellaceae/Prevotella | 0.43# | ||||||||
| Bacteroidales/Paraprevotellaceae/YRC22 | 0.30$ | ||||||||
| Bacteroidales/Prevotellaceae | −0.49# | 0.35$ | |||||||
| Bacteroidales/Prevotellaceae/Prevotella | 0.37# | ||||||||
| Bacteroidales/RF16 | −0.51# | ||||||||
| Bacteroidales/Rikenellaceae | 0.36# | ||||||||
| Bacteroidales/S24-7 | −0.50# | 0.38# | |||||||
| Bifidobacteriales/Bifidobacteriaceae/Bifidobacterium | 0.47# | 0.50# | 0.43# | 0.41# | |||||
| Clostridiales | −0.35# | ||||||||
| Clostridiales/Christensenellaceae | 0.47# | −0.31$ | |||||||
| Clostridiales/Christensenellaceae/Christensenella | 0.41# | ||||||||
| Clostridiales/Clostridiaceae/Clostridium | 0.31$ | ||||||||
| Clostridiales/Eubacteriaceae/Pseudoramibacter eubacterium | −0.42# | −0.30$ | 0.47# | 0.47# | |||||
| Clostridiales/Lachnospiraceae | 0.34$ | ||||||||
| Clostridiales/Lachnospiraceae/Anaerostipes | 0.44# | 0.44# | |||||||
| Clostridiales/Lachnospiraceae/Blautia | −0.31$ | 0.42# | |||||||
| Clostridiales/Lachnospiraceae/Butyrivibrio | 0.41# | −0.57# | |||||||
| Clostridiales/Lachnospiraceae/Coprococcus | −0.43# | ||||||||
| Clostridiales/Lachnospiraceae/Lachnobacterium | 0.34$ | 0.35# | |||||||
| Clostridiales/Lachnospiraceae/Lachnospira | −0.32$ | −0.44# | 0.33$ | 0.43# | 0.35# | 0.47# | 0.49# | ||
| Clostridiales/Lachnospiraceae/Pseudobutyrivibrio | |||||||||
| Clostridiales/Mogibacteriaceae | −0.37# | 0.36# | |||||||
| Clostridiales/NA/NA | −0.36# | 0.44# | |||||||
| Clostridiales/Ruminococcaceae | 0.32$ | −0.34$ | −0.58# | 0.32$ | |||||
| Clostridiales/Ruminococcaceae/NA | NS | 0.31$ | |||||||
| Clostridiales/Ruminococcaceae/Ruminococcus | −0.34$ | ||||||||
| Clostridiales/Veillonellaceae | 0.45# | 0.37# | |||||||
| Clostridiales/Veillonellaceae/NA | −0.33$ | 0.41# | 0.44# | 0.42# | 0.36# | ||||
| Clostridiales/Veillonellaceae/Acidaminococcus | 0.53# | 0.43# | 0.32$ | ||||||
| Clostridiales/Veillonellaceae/Dialister | 0.35$ | 0.63# | 0.56# | 0.38# | |||||
| Clostridiales/Veillonellaceae/Megasphaera | −0.31$ | 0.45# | 0.34$ | 0.48# | 0.51# | 0.49# | |||
| Clostridiales/Veillonellaceae/Phascolarctobacterium | 0.47# | 0.34$ | 0.33$ | 0.44# | |||||
| Clostridiales/Veillonellaceae/Selenomonas | 0.40# | ||||||||
| Clostridiales/Veillonellaceae/Succiniclasticum | −0.39# | 0.32$ | |||||||
| Coriobacteriales/Coriobacteriaceae | 0.48# | 0.65# | |||||||
| CW040/F16 | 0.34$ | −0.32$ | |||||||
| Desulfovibrionales/Desulfovibrionaceae/Desulfovibrio | −0.31$ | ||||||||
| Elusimicrobiales/Elusimicrobiaceae | −0.30$ | ||||||||
| Enterobacteriales/Enterobacteriaceae | 0.38# | ||||||||
| Erysipelotrichales/Erysipelotrichaceae/Allobaculum | 0.38# | 0.32$ | 0.31$ | −0.32$ | |||||
| Erysipelotrichales/Erysipelotrichaceae/Bulleidia | −0.39# | 0.32$ | 0.51# | ||||||
| Erysipelotrichales/Erysipelotrichaceae/Catenibacterium | 0.30$ | ||||||||
| Erysipelotrichales/Erysipelotrichaceae/Eubacterium | −0.45# | 0.45# | 0.45# | ||||||
| Erysipelotrichales/Erysipelotrichaceae/L7A_E11 | 0.30$ | ||||||||
| Erysipelotrichales/Erysipelotrichaceae/p-75-a5 | −0.43# | −0.46# | |||||||
| Erysipelotrichales/Erysipelotrichaceae/Sharpea | 0.31$ | 0.34$ | |||||||
| Lactobacillales/Lactobacillaceae/Lactobacillus | 0.33$ | 0.50# | 0.57# | ||||||
| Lactobacillales/Streptococcaceae/Streptococcus | |||||||||
| Pedosphaerales/R4-41B | −0.43# | −0.41# | −0.45# | −0.32$ | −0.41# | ||||
| Pirellulales/Pirellulaceae | −0.49# | −0.54# | −0.49# | −0.45# | |||||
| RF32 | 0.34$ | ||||||||
| RF39 | −0.43$ | −0.36# | 0.34$ | ||||||
| Rickettsiales | −0.51# | ||||||||
| Synergistales/Synergistaceae/NA | |||||||||
| YS2 | 0.30$ | ||||||||
Significances are based on Spearman P-values.
P < 0.05;
,0.05 ≤ P < 0.10; gray fields, P ≥ 0.10.
t, trans; c, cis; CLA, conjugated linoleic acid; CLnA, conjugated linolenic acid.
NA, not assigned.
Figure 3.
Relationships among clusters of bacterial genera and 18-carbon fatty acids irrespective of experimental group. This bipartite network was based on the regularized canonical correlations between relative bacterial abundances and relative concentrations of 18-carbon fatty acids. Interactions have been filtered for an absolute correlation above 0.4 and are colored following the key shown. This representation uncovers potentially functional populations. LA, 18:2n-6; c9t11CLA, cis-9, trans-11 conjugated linoleic acid; t11C18:1, trans-11 18:1; t11, total trans-11 fatty acids; C18.1, total 18:1 fatty acids; t10, total trans-10 fatty acids; t10C18:1, trans-10 18:1; t10.t11, ratio of trans-10 to trans-11 intermediates; C18.0, 18:0. Acida, Acidaminococcus, Butyri, Butyrivibrio, Eub, Eubacterium, Lactobac, Lactobacillus, Phascolarcto, Phascolarctobacterium, Pseudo_Eub, Pseudoramibacter eubacterium. No correlations were found with 18:3n-3 and cis-9, trans-11, cis-15 conjugated linolenic acid.
Discussion
Dietary supplementation of products containing DHA previously showed to inhibit the final step of biohydrogenation to 18:0 in the rumen of adult animals, resulting in the accumulation of 18:1 isomers (e.g., Shingfield et al., 2012; Toral et al., 2012; Szczechowiak et al., 2016). However, it is unknown whether similar effects on ruminal biohydrogenation would occur when supplementing DHA-enriched products to ruminants at young age, e.g., during pregnancy or early life. Postnatal supplementation of goat kids with DHA-enriched microalgae altered the rumen FA profile in a similar way as in adult animals, whereas prenatal treatment did not change the rumen FA profile. The ruminal proportion of total 18:1 FA increased from 6.33 to 9.44 g/100 g FA (+3.11 g/100 g FA, +49%) whereas 18:0 decreased from 41.95 to 32.19 g/100 g FA (−9.76 g/100 g FA, −23%) upon postnatal administration of DHA-enriched microalgae. In absolute proportions, the increase in total 18:1 was mostly caused by increased ruminal proportion of t11 18:1 (from 0.73 to 1.50 g/100 g FA, +0.77 g/100 g FA, +107%) concurring with other studies (e.g., Shingfield et al., 2012; Toral et al., 2012; Szczechowiak et al., 2016). As opposed to previous reports (e.g., Boeckaert et al., 2007, 2008; Zhu et al., 2016), the absolute increase in t10 18:1 of +0.72 g/100 g FA (from 0.63 to 1.35 g/100 g FA, +114%) upon postnatal DHA Gold supplementation was not significant. This could be due to large inter-animal variation ( ± SD = 1.00 ± 1.66 g/100 g FA; SEM = 0.597 g/100 g FA) which was also observed in other studies (Kim et al., 2008; Toral et al., 2012). In addition, the amount of DHA supplemented (0.28 g DHA Gold per kg BW corresponding to 0.04 g DHA per kg BW) might have been too limited to induce a general increase in t10 18:1. Indeed, supplementation of algae in the rumen of goats induced an increase in ruminal t10 18:1 at a dose of 0.19 g DHA per kg BW whereas a dose of 0.06 g DHA per kg BW did not (Zhu et al., 2016). In contrast, supplementation of 0.05 g DHA per kg BW induced an increase in t10 18:1 in cows (Shingfield et al., 2012). This discrepancy between goat and cow trials could indicate that both young and adult goats are less prone to diet-induced shifts toward the formation of t10 intermediates, in comparison with cows (Toral et al., 2015). The increased ruminal proportion of c13 18:1 might indicate that supplementation of DHA-enriched products not only limited trans 18:1 but also cis 18:1 saturation (e.g., Shingfield et al., 2012; Toral et al., 2012, 2017).
The ruminal proportions of 18:2n-6 and 18:3n-3 were lower in the postnatally supplemented group, which might indicate that DHA does not cause a general slowdown of the ruminal metabolism of these PUFA. Similar results were observed in vitro (Toral et al., 2017) and in vivo with cows (Shingfield et al., 2012) and sheep (Toral et al., 2012). However, other in vivo studies observed either no differences in 18:2n-6, and 18:3n-3 (Boeckaert et al., 2007) or higher ruminal proportions of 18:2n-6 (Szczechowiak et al., 2016) when supplementing DHA-containing products to cows. Nevertheless, lower relative proportions of 18:2n-6 and 18:3n-3 can also partly reflect the increase in the total lipid content in the rumen due to supplementation of FA through DHA Gold (dilution effect). If the amounts of 18:2n-6 and 18:3n-3 are expressed as μg/mL rumen fluid (Supplementary Table 1), no significant difference was observed after postnatal DHA Gold supplementation.
As reported by other authors (Boeckaert et al., 2007; Shingfield et al., 2012; Toral et al., 2017), rumen proportions of c9, t11 CLA, t10, c12 CLA, or c9, t11, c15 CLnA were not significantly different between treatments. Others did observe an effect of DHA supplementation on rumen concentrations of CLA and CLnA (Kim et al., 2008; Toral et al., 2012; Szczechowiak et al., 2016). Discrepancies between studies might be related to the amount of DHA supplemented as CLA generally increased with increasing DHA supplementation level. In addition, the time of sampling might also explain the lack of a treatment effect in the current study. Indeed, CLA and CLnA isomers particularly accumulate within a short period after feeding, since thereafter the disappearance rate of these FA exceeds the rate of formation. In the current study, samples of rumen fluid were collected before the morning feeding after an overnight period without access to concentrate and hay.
Hence, postnatal supplementation of DHA-enriched microalgae to goat kids from birth until 12 weeks old affected rumen biohydrogenation in a similar way as in adult animals by inhibition of the final step of biohydrogenation to 18:0. This resulted particularly in increased proportions of t11 18:1 rather than a shift to t10 intermediates, which suggests that young goats might also be less prone to dietary induced shifts toward the formation of t10 intermediates.
Previous studies showed that delivery of FA through the maternal diet during pregnancy can affect ruminal protozoa population, ruminal bacterial community, methane production and reticulorumen weight of the offspring (De Barbieri et al., 2015a,b). Indeed, in the current study, supplementing the maternal diet with DHA-enriched microalgae during pregnancy affected the rumen microbiome of the offspring. In line with the effect of postnatal supplementation on rumen metabolism as discussed in the previous paragraphs, postnatal supplementation of DHA-enriched microalgae also affected the rumen microbiome. Nevertheless, with some taxa (e.g. Succiniclasticum, L7A_E11), postnatal treatment with DHA Gold only shifted the relative abundance when goats were not treated prenatally, which could indicate adaptation to a repeated treatment later in life.
The accumulation of different 18:1 isomers and the reduced ruminal proportion of 18:0 after postnatal DHA Gold supplementation may be associated with an inhibitory effect of DHA-enriched microalgae on the proliferation of rumen micro-organisms involved in the reduction of 18:1 FA. Butyrivibrio proteoclasticus is the only bacterial species identified to reduce 18:1 FA to 18:0 (Kemp et al., 1975; Wallace et al., 2006; McKain et al., 2010). Indeed, in vitro relative abundance of B. proteoclasticus decreased when reduction to 18:0 was inhibited by supplementation of a blend of fish oil and soybean oil (Szczechowiak et al., 2016). However, no decrease was observed in the in vivo part of that study in accordance with our results and other reports (Huws et al., 2010; Toral et al., 2012; Zhu et al., 2016). Noncultivated Butyrivibrio, Pseudobutyrivibrio and other unknown Lachnospiraceae strains might play a role in the final biohydrogenation step (Boeckaert et al., 2008). In the current study, the relative abundance of Blautia (family Lachnospiraceae) decreased upon postnatal supplementation of DHA Gold. Furthermore, there was a trend toward negative correlation with t11 18:1, potentially indicating that this genus is involved in ruminal 18:0 formation (Huws et al., 2011). In addition, the order RF39 also decreased after postnatal DHA Gold supplementation and was negatively correlated with 18:1 isomers and positively with 18:0. Therefore, genera within this order could also be involved in ruminal 18:0 formation. An alternative explanation is that supplementation of DHA-enriched microalgae reduced the metabolic and perhaps specifically the biohydrogenating activity of B. proteoclasticus instead of its proliferation. Metabolic activity may not be proportional to 16S rRNA gene concentration and thus, RNA should be targeted to investigate this hypothesis. However, the current experimental design, sampling at one single time point after overnight fasting, is not appropriate for this purpose. According to Jeyanathan et al. (2016), an alternative explanation is that B. proteoclasticus starts to hydrogenate DHA before converting 18:1 isomers to 18:0 because of the higher toxicity of DHA in comparison with 18:1 isomers.
Besides 18:0 as an end product, the biohydrogenation pathways of 18:2n-6 and 18:3n-3 involve plenty of 18:2 and 18:1 FA intermediates. 18:2n-6 is mainly isomerized to c9, t11 CLA which is further hydrogenated to t11 18:1 and ultimately to 18:0 (Bauman and Griinari, 2003; Harvatine et al., 2009). The main biohydrogenation pathway of 18:3n-3 involves c9, t11, c15 CLnA, t11, c15 18:2, and t11 18:1 as intermediates (Shingfield and Wallace, 2014). The first ruminal bacterial species reported to produce c9, t11 CLA was B. fibrisolvens (Kepler et al., 1966). According to Wallace et al. (2007), several bacterial species can convert 18:2n-6 to c9, t11 CLA. However, all members of the (Pseudo)butyrivibrio group form c9, t11 CLA from 18:2n-6 much more rapidly than do other species (Shingfield et al., 2012). B. fibrisolvens also further hydrogenates c9, t11 CLA to t11 18:1 (McKain et al., 2010). Previous in vitro experiments in our lab (L. Dewanckele, unpublished data) revealed that this bacterial species is also involved in the formation of c9, t11, c15 CLnA, t11, c15 18:2 and t11 18:1 when incubated with 18:3n-3. In the current study, Butyrivibrio only showed a weak positive correlation with t11 intermediates whereas Pseudobutyrivibrio showed no correlation with t11 intermediates. Together with the study of Zened et al. (2016) in which no correlation was found between Butyrivibrio and t11 FA, this questions the role of (Pseudo)butyrivibrio species in ruminal t11 formation. Other as yet uncultured bacteria belonging to genera Anaerovorax, Prevotella, Lachnospiraceae Incertae Sedis, Ruminococcus, Butyrivibrio, and Pseudobutyrivibrio, Tanerella and unclassified Bacteroidales, Clostridia and Clostridiales, Ruminococcaceae, Lachnospiraceae, Prevotellaceae and Porphyromonadaceae might be involved in ruminal t11 formation (Huws et al., 2011). This study confirmed this potential role for e.g., Parabacteroides (Porphyromonadaceae family), Prevotella, the families Rikenellaceae and S24-7 (Bacteroidales), Christensenella and Lachnospira (Clostridiales).
Dietary supplementation of DHA-enriched supplements could induce a shift from the main biohydrogenation pathway toward the formation of t10 intermediates (e.g., t10, c12 CLA, t10, c15 18:2, and t10 18:1). However, the ruminal bacteria involved in t10 formation remain unclear. In this study, Megasphaera correlated positively with t10 18:1 and with the ratio of t10 to t11 intermediates. M. elsdenii was found to convert in vitro 18:2n-6 to t10, c12 CLA (Kim et al., 2002). Nevertheless, Maia et al. (2007) observed no production of t10, c12 CLA by this bacterial species. In vitro studies by the group of Wallace (Wallace et al., 2007) further indicated that Propionibacterium acnes is a producer of t10, c12 CLA, which is the end product of its 18:2n-6 metabolism (McKain et al., 2010). This was confirmed in our previous in vitro experiments (Dewanckele et al., 2017). However, ruminal abundance of this species is very low (Shingfield et al., 2012) and in the current study, it was even not observed. Lactobacillus spp. have also been shown to produce t10, c12 CLA in vitro (Alonso et al., 2003; Renes et al., 2017), which was highlighted in the current study. However, to what extent this microorganism plays a role in ruminal biohydrogenation remains unclear. Moreover, we validated that Dialister was positively correlated with t10 FA (Zened et al., 2016). In vitro experiments with pure cultures are required to confirm the capacity of this genus to produce t10 isomers. Other genera positively correlated with ruminal t10 isomers according to this study are: Bifidobacterium, Sharpea, Pseudoramibacter eubacterium, Eubacterium, and undefined genera belonging to the family Coriobacteriaceae. However, none of them have been identified as major t10 FA producers (e.g., Devillard et al., 2007; McIntosh et al., 2009; Gorissen et al., 2010). Nevertheless, production of trans isomers is species- and strain-dependent (Gorissen et al., 2010).
Correlations between different biohydrogenation intermediates and rumen bacteria are comparable between our results, which were based on young goats, and other reports, which were based on adult animals. Hence, these extensive comparisons made between current results and literature data indicate that associations between biohydrogenation intermediates and rumen bacteria in goat kids of 12 weeks old align with former observations in adult ruminants.
Conclusion
Postnatal supplementation of goat kids from birth until 12 weeks old with DHA-enriched microalgae affected rumen biohydrogenation in a similar way as in adult animals by inhibition of the final step of biohydrogenation to 18:0. This resulted particularly in increased ruminal proportions of t11 18:1 rather than a shift to t10 intermediates, which suggests that young goats, just as adult ones, might be less prone to dietary induced shifts toward the formation of t10 intermediates, in comparison with cows. Higher abundance of Butyrivibrio when the reduction to 18:0 was inhibited is surprising as they have been identified as the most important biohydrogenating bacteria. Blautia abundance was positively correlated with 18:0 accumulation, whereas Lactobacillus spp. Dialister spp. and Bifidobacterium spp. were more abundant in situations with greater t10 accumulation. Extensive comparisons made between current results and literature data indicate that current associations between biohydrogenation intermediates and rumen bacteria in young goats align with former observations in adult ruminants.
Author contributions
VF, AR-G, and SD conceived and designed the animal experiment. LD performed the chemical analysis of LCFA. LD, JJ, and EH-S performed the bacterial community analysis. LD analyzed the data. LD, BV, and VF discussed and interpreted the obtained data. LD wrote the manuscript. BV and VF contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the goat farm where does were housed (‘t Leenhof, Zele, Belgium). We acknowledge Laura Rogge, Dries Vermander and Longhui Jing for their help with feeding the kids and with rumen sampling, and Charlotte Melis for assistance in the lab.
Glossary
Abbreviations
- BW
body weight
- c
cis
- CLA
conjugated linoleic acid
- CLnA
conjugated linolenic acid
- DHA
docosahexaenoic acid
- EPA
eicosapentaenoic acid
- FA
fatty acid
- FAME
fatty acid methyl ester
- LCFA
long-chain fatty acid
- OTU
operational taxonomic unit
- PUFA
poly-unsaturated fatty acid
- SD
standard deviation
- t
trans.
Footnotes
Funding. Ph.D. research of LD is financed by the Special Research Fund of Ghent University (BOF-Belgium) [BOF15/DOC/246 to LD]. BV was a Postdoctoral Fellow of the Fund for Scientific Research-Flanders (Belgium). EH-S is a Postdoctoral Fellow supported by the Flanders Innovation and Entrepreneurship agency (VLAIO-Belgium). AR-G receives a scholarship from Consejo Nacional de Ciencia y Tecnología (Conacyt, Mexico) [grant number: 401184] and SD receives a Ph.D. grant from the government agency Flanders Innovation & Entrepreneurship (VLAIO-Belgium) [grant number: IWT/SB-141326/SDB]. Post-doctoral fellowship of JJ is financed by the Fund for Scientific Research-Flanders (Belgium) and the Special Research Fund of Ghent University. Research is also partially financed by the Fund for Scientific Research-Flanders (FWO-Flanders, Belgium) and is partially financed within the frame of the FACCE JPI Multi-partner Call on Agricultural Greenhouse Gas Research (RumenStability).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00573/full#supplementary-material
References
- Ackman R. G., Sipos J. C. (1964). Application of specific response factors in gas chromatographic analysis of methyl esters of fatty acids with flame ionization detectors. J. Am. Oil. Chem. Soc. 41, 377–378. [Google Scholar]
- Alonso L., Cuesta E. P., Gilliland S. E. (2003). Production of free conjugated linoleic acid by Lactobacillus acidophilus and Lactobacillus casei of human intestinal origin. J. Dairy Sci. 86, 1941–1946. 10.3168/jds.S0022-0302(03)73781-3 [DOI] [PubMed] [Google Scholar]
- Aronesty E. (2011). ea-utils: Command-Line Tools for Processing Biological Sequencing Data. Durham, NC: Expression Analysis. [Google Scholar]
- Bauman D. E., Griinari J. M. (2003). Nutritional regulation of milk fat synthesis. Annu. Rev. Nutr. 23, 203–227. 10.1146/annurev.nutr.23.011702.073408 [DOI] [PubMed] [Google Scholar]
- Boeckaert C., Fievez V., Van Hecke D., Verstraete W., Boon N. (2007). Changes in rumen biohydrogenation intermediates and ciliate protozoa diversity after algae supplementation to dairy cattle. Eur. J. Lipid Sci. Technol. 109, 767–777. 10.1002/ejlt.200700052 [DOI] [Google Scholar]
- Boeckaert C., Vlaeminck B., Fievez V., Maignien L., Dijkstra J., Boon N. (2008). Accumulation of trans C18:1 fatty acids in the rumen after dietary algal supplementation is associated with changes in the Butyrivibrio community. Appl. Environ. Microb. 74, 6923–6930. 10.1128/AEM.01473-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray J. R., Curtis J. T. (1957). An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349. 10.2307/1942268 [DOI] [Google Scholar]
- Brenna J. T., Carlson S. E. (2014). Docosahexaenoic acid and human brain development: Evidence that a dietary supply is needed for optimal development. J. Hum. Evol. 77, 99–106. 10.1016/j.jhevol.2014.02.017 [DOI] [PubMed] [Google Scholar]
- Buccioni A., Decandia M., Minieri S., Molle G., Cabiddu A. (2012). Lipid metabolism in the rumen: New insights on lipolysis and biohydrogenation with an emphasis on the role of endogenous plant factors. Anim. Feed Sci. Tech. 174, 1–25. 10.1016/j.anifeedsci.2012.02.009 [DOI] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Barbieri I., Hegarty R. S., Silveira C., Gulino L. M., Oddy V. H., Gilbert R. A., et al. (2015b). Programming rumen bacterial communities in newborn Merino lambs. Small Ruminant Res. 129, 48–59. 10.1016/j.smallrumres.2015.05.015 [DOI] [Google Scholar]
- De Barbieri I., Hegarty R. S., Silveira C., Oddy V. H. (2015a). Positive consequences of maternal diet and post-natal rumen inoculation on rumen function and animal performance of Merino lambs. Small Ruminant Res. 129, 37–47. 10.1016/j.smallrumres.2015.05.017 [DOI] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microb. 72, 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devillard E., McIntosh F. M., Duncan S. H., Wallace R. J. (2007). Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J. Bacteriol. 189, 2566–2570. 10.1128/JB.01359-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewanckele L., Vlaeminck B., Jeyanathan J., Fievez V. (2017). Effect of pH and 22:6n-3 on in vitro biohydrogenation of 18:2n-6 by different ratios of Butyrivibrio fibrisolvens to Propionibacterium acnes. American Dairy Science Association, Proceedings of the ADSA Annual Meeting; 2017 June 25-28. (Pittsburgh, PA: ), Abstract 259. [Google Scholar]
- De Weirdt R., Hernandez-Sanabria E., Fievez V., Mees E., Geirnaert A., Van Herreweghen F., et al. (2017). Mucosa-associated biohydrogenating microbes protect the simulated colon microbiome from stress associated with high concentrations of poly-unsaturated fat. Environ. Microbiol. 19, 722–739. 10.1111/1462-2920.13622 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Ferlay A., Bernard L., Meynadier A., Malpuech-Brugère C. (2017). Production of trans and conjugated fatty acids in dairy ruminants and their putative effects on human health: a review. Biochimie 141, 107–120. 10.1016/j.biochi.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Gorissen L., Raes K., Weckx S., Dannenberger D., Leroy F., De Vuyst L., et al. (2010). Production of conjugated linoleic acid and conjugated linolenic acid isomers by Bifidobacterium species. Appl. Microbiol. Biotechnol. 87, 2257–2266. 10.1007/s00253-010-2713-1 [DOI] [PubMed] [Google Scholar]
- Harvatine K. J., Boisclair Y. R., Bauman D. E. (2009). Recent advances in the regulation of milk fat synthesis. Animal 3, 40–54. 10.1017/S1751731108003133 [DOI] [PubMed] [Google Scholar]
- Huws S. A., Kim E. J., Lee M. R. F., Scott M. B., Tweed J. K. S., Pinloche E., et al. (2011). As yet uncultured bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales and Ruminococcaceae may play a predominant role in ruminal biohydrogenation. Environ. Microbiol. 13, 1500–1512. 10.1111/j.1462-2920.2011.02452.x [DOI] [PubMed] [Google Scholar]
- Huws S. A., Lee M. R. F., Muetzel S. M., Scott M. B., Wallace R. J., Scollan N. D. (2010). Forage type and fish oil cause shifts in rumen bacterial diversity. FEMS Microbiol. Ecol. 73, 396–702. 10.1111/j.1574-6941.2010.00892.x [DOI] [PubMed] [Google Scholar]
- Jeyanathan J., Escobar M., Wallace R. J., Fievez V., Vlaeminck B. (2016). Biohydrogenation of 22:6n-3 by Butyrivibrio proteoclasticus P18. BMC Microbiol. 16:104. 10.1186/s12866-016-0720-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp P., White R. W., Lander D. J. (1975). The hydrogenation of unsaturated fatty acids by five bacterial isolates from the sheep rumen, including a new species. Microbiology 90, 100–114. 10.1099/00221287-90-1-100 [DOI] [PubMed] [Google Scholar]
- Kepler C. R., Hirons K. P., McNeill J. J., Tove S. B. (1966). Intermediates and products of the biohydrogenation of linoleic acid by Butyrivibrio fibrisolvens. J. Biol. Chem. 241, 1350–1354. [PubMed] [Google Scholar]
- Kim E. J., Huws S. A., Lee M. R. F., Wood J. D., Muetzel S. M., Wallace R. J., et al. (2008). Fish oil increases the duodenal flow of long chain polyunsaturated fatty acids and trans-11 18:1 and decreases 18:0 in steers via changes in the rumen bacterial community. J. Nutr. 138, 889–896. 10.1093/jn/138.5.889 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Liu R. H., Rychlik J. L., Russell J. B. (2002). The enrichment of a ruminal bacterium (Megasphaera elsdenii YJ-4) that produces the trans-10, cis-12 isomer of conjugated linoleic acid. J. Appl. Microbiol. 92, 976–982. 10.1046/j.1365-2672.2002.01610.x [DOI] [PubMed] [Google Scholar]
- Klein C. M., Jenkins T. C. (2011). Docosahexaenoic acid elevates trans-18:1 isomers but is not directly converted into trans-18:1 isomers in ruminal batch cultures. J. Dairy Sci. 94, 4676–4683. 10.3168/jds.2011-4344 [DOI] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz Z. D., Müller C. L., Miraldi E. R., Littman D. R., Blaser M. J., Bonneau R. A. (2015). Sparse and compositionally robust inference of microbial ecological networks. Plos Comput. Biol. 11:e1004226. 10.1371/journal.pcbi.1004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Cao K. A., Boitard S., Besse P. (2011). Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics. 12:253. 10.1186/1471-2105-12-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia M. R., Chaudhary L. C., Figueres L., Wallace R. J. (2007). Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie Van Leeuwenhoek 91, 303–314. 10.1007/s10482-006-9118-2 [DOI] [PubMed] [Google Scholar]
- McIntosh F. M., Shingfield K. J., Devillard E., Russell W. R., Wallace R. J. (2009). Mechanism of conjugated linoleic acid and vaccenic acid formation in human faecal suspensions and pure cultures of intestinal bacteria. Microbiology 155, 285–294. 10.1099/mic.0.022921-0 [DOI] [PubMed] [Google Scholar]
- McKain N., Shingfield K. J., Wallace R. J. (2010). Metabolism of conjugated linoleic acids and 18:1 fatty acids by ruminal bacteria: products and mechanisms. Microbiology 156, 579–588. 10.1099/mic.0.036442-0 [DOI] [PubMed] [Google Scholar]
- Ramos-Morales E., Arco-Pérez A., Martín-García A. I., Yáñez-Ruiz D. R., Frutos P., Hervás G. (2014). Use of stomach tubing as an alternative to rumen cannulation to study ruminal fermentation and microbiota in sheep and goats. Anim. Feed Sci. Tech. 198, 57–66. 10.1016/j.anifeedsci.2014.09.016 [DOI] [Google Scholar]
- Renes E., Linares D. M., González L., Fresno J. M., Tornadijo M. E., Stanton C. (2017). Study of the conjugated linoleic acid synthesis by Lactobacillus strains and by different co-cultures designed for this ability. J. Funct. Foods 35, 74–80. 10.1016/j.jff.2017.05.015 [DOI] [Google Scholar]
- Shingfield K. J., Kairenius P., Ärölä A., Paillard D., Muetzel S., Ahvenjärvi S., et al. (2012). Dietary fish oil supplements modify ruminal biohydrogenation, alter the flow of fatty acids at the omasum, and induce changes in the ruminal Butyrivibrio population in lactating cows. J. Nutr. 142, 1437–1448. 10.3945/jn.112.158576 [DOI] [PubMed] [Google Scholar]
- Shingfield K. J., Wallace R. J. (2014). Synthesis of Conjugated Linoleic Acid in Ruminants and Humans, in Conjugated Linoleic Acids and Conjugated Vegetable Oils, ed Sels B., Philippaerts A. 1–65. Available online at: http://pubs.rsc.org/en/Content/eBook/978-1-84973-900-9
- Szczechowiak J., Szumacher-Strabel M., El-Sherbiny M., Pers-Kamczyc E., Pawlak P., Cieslak A. (2016). Rumen fermentation, methane concentration and fatty acid proportion in the rumen and milk of dairy cows fed condensed tannin and/or fish-soybean oils blend. Anim. Feed Sci. Tech. 216, 93–107. 10.1016/j.anifeedsci.2016.03.014 [DOI] [Google Scholar]
- Tamminga S., Van Straalen W. M., Subnel A. P., Meijer R. G. M., Steg A., Wever C. G., et al. (1994). The Dutch protein evaluation system: the DVE/OEB-system. Livest. Prod. Sci. 40, 139–155. 10.1016/0301-6226(94)90043-4 [DOI] [Google Scholar]
- Tanghe S., De Smet S. (2013). Does sow reproduction and piglet performance benefit from the addition of n-3 polyunsaturated fatty acids to the maternal diet? Vet. J. 197, 560–569. 10.1016/j.tvjl.2013.03.051 [DOI] [PubMed] [Google Scholar]
- Toral P. G., Belenguer A., Shingfield K. J., Hervás G., Toivonen V., Frutos P. (2012). Fatty acid composition and bacterial community changes in the rumen fluid of lactating sheep fed sunflower oil plus incremental levels of marine algae. J. Dairy Sci. 95, 794–806. 10.3168/jds.2011-4561 [DOI] [PubMed] [Google Scholar]
- Toral P. G., Bernard L., Belenguer A., Rouel J., Hervás G., Chilliard Y., et al. (2016). Comparison of ruminal lipid metabolism in dairy cows and goats fed diets supplemented with starch, plant oil, or fish oil. J. Dairy Sci. 99, 301–316. 10.3168/jds.2015-10292 [DOI] [PubMed] [Google Scholar]
- Toral P. G., Chilliard Y., Rouel J., Leskinen H., Shingfield K. J., Bernard L. (2015). Comparison of the nutritional regulation of milk fat secretion and composition in cows and goats. J. Dairy Sci. 98, 7277–7297. 10.3168/jds.2015-9649 [DOI] [PubMed] [Google Scholar]
- Toral P. G., Hervás G., Carreño D., Leskinen H., Belenguer A., Shingfield K. J., et al. (2017). In vitro response to EPA, DPA, and DHA: comparison of effects on ruminal fermentation and biohydrogenation of 18-carbon fatty acids in cows and ewes. J. Dairy Sci. 100, 6187–6198. 10.3168/jds.2017-12638 [DOI] [PubMed] [Google Scholar]
- Van Es A. H. (1978). Feed evaluation for ruminants: I. The systems in use from May 1977 onwards in the Netherlands. Livest. Prod. Sci. 5, 331–345. 10.1016/0301-6226(78)90029-5 [DOI] [Google Scholar]
- Vlaeminck B., Braeckman T., Fievez V. (2014). Rumen metabolism of 22:6n-3 in vitro is dependent on its concentration and inoculum size, but less dependent on substrate carbohydrate composition. Lipids 49, 517–525. 10.1007/s11745-014-3905-8 [DOI] [PubMed] [Google Scholar]
- Wallace R. J., Chaudhary L. C., McKain N., McEwan N. R., Richardson A. J., Vercoe P. E., et al. (2006). Clostridium proteoclasticum: a ruminal bacterium that forms stearic acid from linoleic acid. FEMS Microbiol. Lett. 265, 195–201. 10.1111/j.1574-6968.2006.00487.x [DOI] [PubMed] [Google Scholar]
- Wallace R. J., McKain N., Shingfield K. J., Devillard E. (2007). Isomers of conjugated linoleic acids are synthesized via different mechanisms in ruminal digesta and bacteria. J. Lipid Res. 48, 2247–2254. 10.1194/jlr.M700271-JLR200 [DOI] [PubMed] [Google Scholar]
- Wasowska I., Maia M. R. G., Niedzwiedzka K. M., Czauderna M., Ribeiro J. M., Devillard E., et al. (2006). Influence of fish oil on ruminal biohydrogenation of C18 unsaturated fatty acids. Br. J. Nutr. 95, 1199–1211. 10.1079/BJN20061783 [DOI] [PubMed] [Google Scholar]
- Wolff R. L., Bayard C. C., Fabien R. J. (1995). Evaluation of sequential methods for the determination of butterfat fatty acid composition with emphasis on trans-18:1 acids. Application to the study of seasonal variations in french butters. J. Am. Oil Chem. Soc. 72, 1471–1483. 10.1007/BF02577840 [DOI] [Google Scholar]
- Yu Z., Morrison M. (2004). Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36, 808–812. Available online at: https://www.biotechniques.com/multimedia/archive/00036/BTN_A_04365ST04_O_36541a.pdf [DOI] [PubMed] [Google Scholar]
- Zened A., Meynadier A., Cauquil L., Mariette J., Klopp C., Dejean S., et al. (2016). Trans-11 to trans-10 shift of ruminal biohydrogenation of fatty acids is linked to changes in rumen microbiota. Gut Microbiology 2016, in Proceedings of the 10th Joint Symposium on Gut Microbiology, 2016 June 20-23 (Clermont-Ferrand: ), 150. [Google Scholar]
- Zhao T., Ma Y., Qu Y., Luo H., Liu K., Zuo Z., et al. (2016). Effect of dietary oil sources on fatty acid composition of ruminal digesta and populations of specific bacteria involved in hydrogenation of 18-carbon unsaturated fatty acid in finishing lambs. Small Ruminant Res. 144, 126–134. 10.1016/j.smallrumres.2016.06.012 [DOI] [Google Scholar]
- Zhu H., Fievez V., Mao S., He W., Zhu W. (2016). Dose and time response of ruminally infused algae on rumen fermentation characteristics, biohydrogenation and Butyrivibrio group bacteria in goats. J. Anim. Sci. Biotechnol. 7:22. 10.1186/s40104-016-0080-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.