ABSTRACT
The bacterium Rickettsia parkeri has been reported to infect ticks of the “Amblyomma maculatum species complex” in the New World, where it causes spotted fever illness in humans. In South America, three additional rickettsial strains, namely, Atlantic rainforest, NOD, and Parvitarsum, have been isolated from the ticks Amblyomma ovale, Amblyomma nodosum, and Amblyomma parvitarsum, respectively. These three strains are phylogenetically closely related to R. parkeri, Rickettsia africae, and Rickettsia sibirica. Herein, we performed a robust phylogenetic analysis encompassing 5 genes (gltA, ompA, virB4, dnaA, and dnaK) and 3 intergenic spacers (mppE-pur, rrl-rrf-ITS, and rpmE-tRNAfMet) from 41 rickettsial isolates, including different isolates of R. parkeri, R. africae, R. sibirica, Rickettsia conorii, and strains Atlantic rainforest, NOD, and Parvitarsum. In our phylogenetic analyses, all New World isolates grouped in a major clade distinct from the Old World Rickettsia species (R. conorii, R. sibirica, and R. africae). This New World clade was subdivided into the following 4 clades: the R. parkeri sensu stricto clade, comprising the type strain Maculatum 20 and all other isolates of R. parkeri from North and South America, associated with ticks of the A. maculatum species complex; the strain NOD clade, comprising two South American isolates from A. nodosum ticks; the Parvitarsum clade, comprising two South American isolates from A. parvitarsum ticks; and the strain Atlantic rainforest clade, comprising six South American isolates from the A. ovale species complex (A. ovale or Amblyomma aureolatum). Under such evidences, we propose that strains Atlantic rainforest, NOD, and Parvitarsum are South American strains of R. parkeri.
IMPORTANCE Since the description of Rickettsia parkeri infecting ticks of the “Amblyomma maculatum species complex” and humans in the New World, three novel phylogenetic close-related rickettsial isolates were reported in South America. Herein, we provide genetic evidence that these novel isolates, namely, strains Atlantic rainforest, NOD, and Parvitarsum, are South American strains of R. parkeri. Interestingly, each of these R. parkeri strains seems to be primarily associated with a tick species group, namely, R. parkeri sensu stricto with the “Amblyomma maculatum species group,” R. parkeri strain NOD with Amblyomma nodosum, R. parkeri strain Parvitarsum with Amblyomma parvitarsum, and R. parkeri strain Atlantic rainforest with the “Amblyomma ovale species group.” Such rickettsial strain-tick species specificity suggests a coevolution of each tick-strain association. Finally, because R. parkeri sensu stricto and R. parkeri strain Atlantic rainforest are human pathogens, the potential of R. parkeri strains NOD and Parvitarsum to be human pathogens cannot be discarded.
KEYWORDS: Amblyomma, New World, Rickettsia parkeri, spotted fever group, tick
INTRODUCTION
During the first half of the 20th century, a novel bacterial agent of the spotted fever group was isolated from Amblyomma maculatum ticks in the southern United States (1). The agent, shown to be mildly pathogenic for guinea pigs (2), was later described as Rickettsia parkeri (3). After almost 6 decades in which R. parkeri was known only from ticks, in 2004, there was the first description of a spotted fever clinical case in a human in the United States (4). This first case has been followed by a growing number of R. parkeri rickettsiosis cases in the United States, all linked to the transmission by Amblyomma maculatum (5, 6). More recently in Arizona, R. parkeri was reported to have infected Amblyomma triste ticks, which were the likely vector of the infection for two human clinical cases (7).
In South America, the first report of R. parkeri dates from 2004, when the agent was found to have infected A. triste ticks in southern Uruguay (8), an area where clinical cases of a tick-borne spotted fever clinically similar to Mediterranean spotted fever had been reported (9, 10). A subsequent study provided serological evidence for R. parkeri as the etiological agent of the Uruguayan spotted fever (11). In Argentina, R. parkeri was reported to have infected A. triste ticks in 2008 (12) and later shown to be the etiological agent of clinical cases of spotted fever (13). Yet, during the 21st century, R. parkeri was reported to have infected A. triste ticks in Brazil (14) and A. maculatum ticks in Peru (15), although the diseases caused by R. parkeri have never been confirmed in these two countries. Additional records of R. parkeri include the infection of Amblyomma tigrinum ticks in Uruguay (16), Bolivia (17), Argentina (18), and Brazil (19). In the latter two countries, A. tigrinum was epidemiologically associated with human clinical cases of spotted fever rickettsiosis, confirmed to be caused by R. parkeri at least in Argentina (18). Amblyomma maculatum, A. triste, and A. tigrinum are morphologically and genetically closely related tick species, forming the “A. maculatum species complex” (20). The above reports of R. parkeri indicate that R. parkeri sensu stricto is primarily associated with ticks of the A. maculatum species complex in the New World.
From 2010 to 2016, three clinical cases of spotted fever rickettsiosis were reported in Brazil (21–23). The cases were shown to be caused by a novel agent, named strain Atlantic rainforest, phylogenetically related to R. parkeri, Rickettsia africae, and Rickettsia sibirica (21). Subsequent studies showed that these clinical cases were epidemiologically associated with the tick Amblyomma ovale (24, 25), and also with Amblyomma aureolatum (26). These two tick species form the “A. ovale species complex” (27). A laboratory study showed that A. ovale is a competent vector of strain Atlantic rainforest (28). Additional studies reported strain Atlantic rainforest-infected A. ovale ticks in Colombia (29) and Belize (30). Recently, a unique North American strain of R. parkeri isolated from Dermacentor parumapertus ticks collected in Texas was determined genetically as nearly identical to Rickettsia sp. strain Atlantic rainforest, further supporting the relatedness of these taxa (31).
In 2009, a novel spotted fever group agent (strain NOD) was isolated from Amblyomma nodosum ticks in Brazil (32). More recently, another spotted fever group agent, named strain Parvitarsum, was isolated from Amblyomma parvitarsum ticks in Argentina and Chile (33). These two novel agents, known only from ticks, were shown to be phylogenetically related to R. parkeri, R. africae, and R. sibirica. While the distribution of R. parkeri in association with the A. maculatum species complex was shown to encompass North and South America, the taxonomic status of strains Atlantic rainforest, NOD, and Parvitarsum remains unresolved. Herein, we provide phylogenetic evidence for the classification of these strains as belonging to the species R. parkeri.
RESULTS
Partial sequences of 5 genes (gltA, ompA, virB4, dnaA, and dnaK) and 3 intergenic spacers (mppE-pur, rrl-rrf-ITS, and rpmE-tRNAfMet) were obtained for the 39 rickettsial isolates listed in Table 1 and used for alignment with corresponding sequences of R. africae strain ESF and R. sibirica sibirica strain 246 from GenBank. The maximum parsimony (MP) analyses revealed the segregation of Rickettsia species into three groups for the gltA gene, seven groups for the ompA gene, seven groups for the dnaA gene, four groups for the dnaK gene, nine groups for the virB4 gene, three groups for the mppA-purC intergenic spacer, five groups for the rrl-rrf-ITS intergenic spacer, and eight groups for the rpmE-tRNAfMet intergenic spacer (see Fig. S1 to S8 in the supplemental material). The divergence values were calculated for each of the eight molecular markers. The highest divergence was found for the ompA gene (1.61%) followed by the intergenic spacer rpmE-tRNAfMet (1.21%). The lowest values were for gltA (0.14%) and dnaA (0.31%) genes.
TABLE 1.
Rickettsial isolates used for DNA amplification in the present study
| No. | Rickettsia species or strain | Code | Source | Geographical locality | Country | Rickettsial collectiona | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Rickettsia parkeri | Maculatum 20T | Amblyomma maculatum | Liberty County, Texas | United States | CDC | 2 |
| 2 | R. parkeri | Tate's Hell | A. maculatum | Franklin County, Florida | United States | CDC | 51 |
| 3 | R. parkeri | Cash Bayou | A. maculatum | Franklin County, Florida | United States | CDC | 51 |
| 4 | R. parkeri | Oktibbeha | A. maculatum | Oktibbeha County, Mississippi | United States | CDC | 51 |
| 5 | R. parkeri | Moss Point | A. maculatum | Jackson County, Mississippi | United States | CDC | 51 |
| 6 | R. parkeri | Escatawpa | A. maculatum | Jackson County, Mississippi | United States | CDC | 51 |
| 7 | R. parkeri | NC-3 | A. maculatum | Mecklenburg County, North Carolina | United States | CDC | 52 |
| 8 | R. parkeri | NC-8 | A. maculatum | Mecklenburg County, North Carolina | United States | CDC | 52 |
| 9 | R. parkeri | NC-15 | A. maculatum | Mecklenburg County, North Carolina | United States | CDC | 52 |
| 10 | R. parkeri | Portsmouth | Human | Norfolk County, Virginia | United States | CDC | 4 |
| 11 | R. parkeri | Ft. Story | Human | Virginia Beach County, Virginia | United States | CDC | 53 |
| 12 | R. parkeri | Fairfax | A. maculatum | Fairfax County, Virginia | United States | CDC | 54 |
| 13 | R. parkeri | I-66 | A. maculatum | Fairfax County, Virginia | United States | CDC | 54 |
| 14 | R. parkeri | 45 | Amblyomma triste | Delta do Paraná, Buenos Aires Province | Argentina | From tick DNA | 12 |
| 15 | R. parkeri | 132 | A. triste | Delta do Paraná, Buenos Aires Province | Argentina | From tick DNA | 12 |
| 16 | R. parkeri | 136 | A. triste | Delta do Paraná, Buenos Aires Province | Argentina | From tick DNA | 12 |
| 17 | R. parkeri | 34 | A. triste | Delta do Paraná, Buenos Aires Province | Argentina | From tick DNA | 12 |
| 18 | R. parkeri | 218 | A. triste | Delta do Paraná, Buenos Aires Province | Argentina | From tick DNA | 12 |
| 19 | R. parkeri | At24 | A. triste | Paulicéia, São Paulo | Brazil | FMVZ/USP | 14 |
| 20 | R. parkeri | Corumbá | A. triste | Corumbá, Mato Grosso do Sul | Brazil | FMVZ/USP | Unpublished |
| 21 | R. parkeri | Água Clara | A. triste | Água Clara, Mato Grosso do Sul | Brazil | FMVZ/USP | 55 |
| 22 | R. parkeri | Pantanal At46 | A. triste | Poconé, Mato Grosso | Brazil | FMVZ/USP | 56 |
| 23 | R. parkeri | At5URG | A. triste | Toledo Chico, Canelones | Uruguay | FMVZ/USP | 57 |
| 24 | Strain Atlantic rainforest | P-240 | Amblyomma ovale | Peruíbe, São Paulo | Brazil | FMVZ/USP | 24 |
| 25 | Strain Atlantic rainforest | P-51 | A. ovale | Peruíbe, São Paulo | Brazil | FMVZ/USP | 24 |
| 26 | Strain Atlantic rainforest | Adrianópolis | A. ovale | Adrianópolis, Paraná | Brazil | FMVZ/USP | 25 |
| 27 | Strain Atlantic rainforest | Paty | A. ovale | Chapada Diamantina, Bahia | Brazil | FMVZ/USP | 25 |
| 28 | Strain Atlantic rainforest | Aa47 | Amblyomma aureolatum | Blumenau, Santa Catarina | Brazil | FMVZ/USP | 26 |
| 29 | Strain Atlantic rainforest | Aa46 | A. aureolatum | Blumenau, Santa Catarina | Brazil | FMVZ/USP | 26 |
| 30 | Strain NOD | NOD | Amblyomma nodosum | Pontal do Paranapanema | Brazil | FMVZ/USP | 32 |
| 31 | Strain NOD | Pantanal | A. nodosum | Nhecolândia, Mato Grosso do Sul | Brazil | FMVZ/USP | Unpublished |
| 32 | Strain Parvitarsum | Argentina | Amblyomma parvitarsum | Salta | Argentina | FMVZ/USP | 33 |
| 33 | Strain Parvitarsum | Chile | A. parvitarsum | Arica and Parinacota | Chile | FMVZ/USP | 33 |
| 34 | Rickettsia africae | Z8-Ah | Amblyomma hebraeum | South of the country | Zimbabwe | UTMB | 58 |
| 35 | R. africae | RaPele | Human | Hluhluwe-Imfolozi Park | South Africa | FMVZ/USP | Unpublished |
| 36 | Rickettsia conorii Israeli | PoHu16026 | Human | Beja, Alentejo region | Portugal | INSA | 59 |
| 37 | R. conorii Malish | PoHu10908 | Human | Faro, Algarve region | Portugal | INSA | 59 |
| 38 | R. conorii Malish | PoHu17458 | Human | Faro, Algarve region | Portugal | INSA | 59 |
| 39 | R. sibirica mongolitimonae | PoHu10991 | Human | Évora, Alentejo region | Portugal | INSA | 60 |
CDC, Rickettsial Zoonoses Branch, Centers for Disease Control and Prevention, Atlanta, GA, United States; FMVZ/USP, Faculty of Veterinary Medicine, University of São Paulo, Brazil; UTMB, University of Texas Medical Branch, Galveston, TX, United States. INSA, National Institute of Health Dr. Ricardo Jorge, Águas de Moura, Portugal.
In both the gltA and the mppA-purC trees, all New World isolates formed a single group with R. sibirica, which was separated from R. africae and R. conorii isolates (Fig. S1 and S6). In the dnaA tree, the A. maculatum-R. parkeri isolates (North America) formed a group separated from the A. triste-R. parkeri isolates (South America), which were separated from the remaining South American and Old World isolates (Fig. S4). In the ompA, virB4, dnaK, rrl-rrf-ITS, and rpmE-tRNAfMet trees, the 23 R. parkeri isolates from A. maculatum (North America) or A. triste (South America) formed a group separate from the remaining groups (Fig. S2, S3, S5, S7, and S8); in the case of the dnaK tree, this single group also included the R. sibirica isolates and the two isolates of strain NOD (NOD and Pantanal) (Fig. S5). In the ompA, virB4, dnaA, and rpmE-tRNAfMet trees, the six isolates of strain Atlantic rainforest formed a group with isolates of strain Parvitarsum (Fig. S2 to S4 and S8); in the dnaA tree, this group also included R. sibirica sibirica (Fig. S4). In the rrl-rrf-ITS tree, the six isolates of strain Atlantic rainforest formed a separate group (Fig. S7). The two isolates of strain NOD formed a single group in the ompA, virB4, dnaA, and rpmE-tRNAfMet trees (Fig. S2 to S4 and S8); in the rrl-rrf-ITS tree, these isolates grouped with isolates of strain Parvitarsum and R. sibirica (Fig. S7). All New World isolates (R. parkeri, strain Atlantic rainforest, strain NOD, and strain Parvitarsum) regardless of separation, remained sisters to each other, well separated from the clade containing strains of R. conorii and, most of the time, from the different strains of R. africae and R. sibirica.
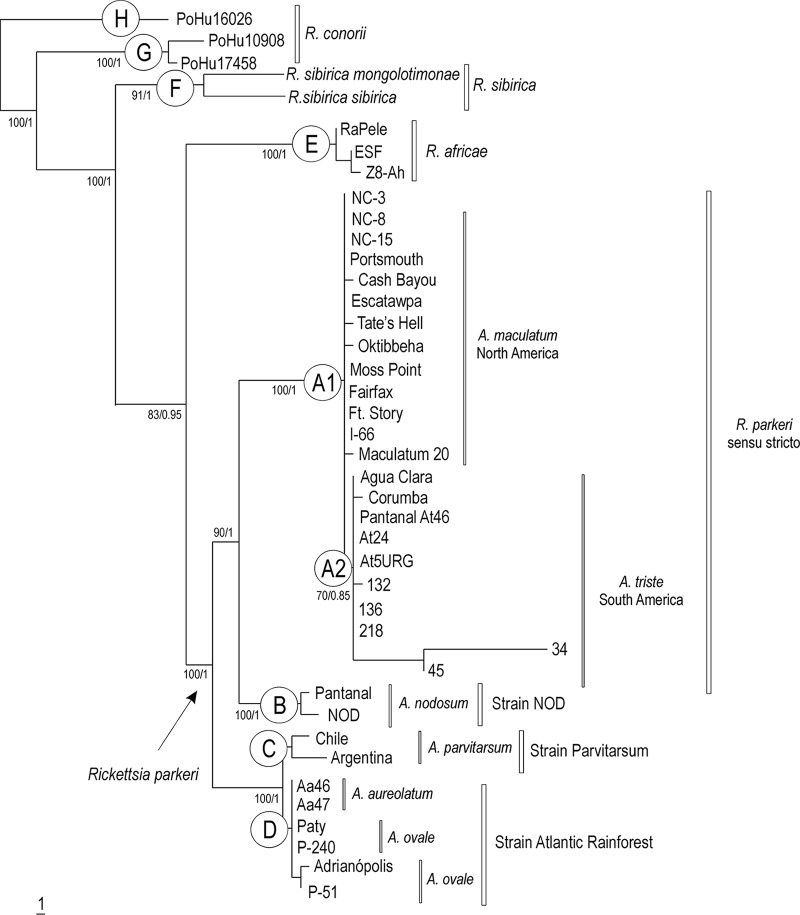
DNA sequences of each of the eight molecular markers were concatenated for each isolate and aligned to be used in the phylogenetic analysis. The final alignment with the 41 rickettsial isolates included 3,603 nucleotides, with 57 informative sites. Under high bootstrap support (MP analysis) or high posterior probabilities (Bayesian [B] analysis), all New World isolates were well separated from the Old World Rickettsia species (R. conorii, R. sibirica, and R. africae) (Fig. 1). The large New World clade was subdivided into the following 4 major clades, all under high bootstrap support or posterior probabilities: the R. parkeri sensu stricto clade (clade A), comprising the type strain Maculatum 20 and all other isolates of R. parkeri from North and South America, associated with ticks of the A. maculatum species complex; the strain NOD clade (clade B), comprising two South American isolates from A. nodosum ticks; the Parvitarsum clade (clade C), comprising two South American isolates from A. parvitarsum ticks; and the strain Atlantic rainforest clade (clade D), comprising six South American isolates from the A. ovale species complex (A. ovale or A. aureolatum). The R. parkeri sensu stricto clade was subdivided clearly into two large clades, one containing all R. parkeri sensu stricto isolates from North America, associated with A. maculatum ticks (clade A1), and one containing all R. parkeri sensu stricto isolates from South America, associated with A. triste (A2). The tree topology shown in Fig. 1 was generally the same for MP and B analyses; the only difference was that the B analysis did not separate North American isolates from the South American isolates of R. parkeri sensu stricto.
FIG 1.
Molecular phylogenetic analysis of New World isolates of Rickettsia parkeri sensu stricto, strains Atlantic rainforest, NOD, and Parvitarsum, in relation to Old World isolates of Rickettsia africae, Rickettsia sibirica, and Rickettsia conorii. A total of 3,603 aligned nucleotide sites of 5 protein-coding genes (gltA, ompA, virB4, dnaA, and dnaK) and 3 intergenic spacers (mppE-pur, rrl-rrf-ITS, rpmE-tRNAfMet) were concatenated and subjected to Bayesian analysis. Numbers at nodes are support values derived from posterior probability. Scale bar, units of expected substitutions per site. Each main clade is indicated by a capital letter (A to H) shown inside a circle.
The overall divergence values of the concatenated sequences were 0.64 to 1.75% between American (clades A to D) and European/Asian (clades F to H) isolates and 0.83 to 1.15% between American and African (clade E) isolates (Table 2). The divergence between European/Asian and African isolates was 0.86 to 1.68%. The divergence values among the American clades (A to D) were generally lower, between 0.19 and 0.93%. Within-clade divergence values were even lower, ranging from 0.0 to 0.27%.
TABLE 2.
Matrix of the divergence of the Rickettsia isolates used in the present studya
| Cladeb | Clade |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | B | C | D | E | F | G | H | |
| A1 | 0.12 | ||||||||
| A2 | 0.19 | 0.21 | |||||||
| B | 0.74 | 0.87 | 0.27 | ||||||
| C | 0.93 | 0.80 | 0.77 | 0.24 | |||||
| D | 0.85 | 0.93 | 0.85 | 0.23 | 0.08 | ||||
| E | 1.02 | 1.15 | 0.95 | 0.83 | 0.85 | 0,03 | |||
| F | 0.83 | 0.97 | 0.80 | 0.64 | 0.66 | 0.86 | 0,13 | ||
| G | 1.62 | 1.74 | 1.55 | 1.33 | 1.35 | 1.61 | 1.37 | 0.10 | |
| H | 1.61 | 1.75 | 1.57 | 1.45 | 1.47 | 1.68 | 1.42 | 1.23 | 0.00 |
Based on an alignment of a concatenated sequence of 3,579 nucleotides (nt), composed of the genes gltA (257 nt), ompA (490 nt), virB4 (684 nt), dnaA (663 nt), and dnaK (615 nt) and the intergenic spacers mppE-pur (197 nt), rrl-rrf-ITS (330 nt), and rpmE-tRNAfMet (343 nt).
Each letter represents a clade in Fig. 1 as follows: A1, Rickettsia parkeri isolates from North America; A2, R. parkeri isolates from South America; B, strain NOD isolates; C, strain Parvitarsum isolates; D, strain Atlantic rainforest isolates; E, R. africae isolates; F, R. sibirica sibirica; G, R. conorii Malish isolates; H, R. conorii Israeli.
DISCUSSION
Since the initial molecular characterization of R. parkeri sensu stricto from A. maculatum ticks and human patients in the United States (4), this Rickettsia species has also been reported in South America, where it infected A. triste (8, 12, 14) and, subsequently, human patients (13, 18). These molecular characterizations were based on the most commonly used molecular markers (portions of the gltA, ompA, and ompB genes), which showed no polymorphism among North and South American isolates. Interestingly, until some decades ago, the taxa A. maculatum and A. triste represented the same tick species (A. maculatum). In a morphological study, Kohls (34) proposed A. triste as a valid species, which had been accepted until the present (35). On the other hand, because of the high morphological similarities between A. maculatum and A. triste, associated with low genetic polymorphism between North American populations of A. maculatum and South American populations of A. triste (20), the possibility of conspecificity of these ticks has not been discarded, and further studies are needed to evaluate this hypothesis (36). Presently, A. maculatum and A. triste, together with A. tigrinum, form the A. maculatum species complex (20), with which R. parkeri sensu stricto has been associated. Our phylogenetic analysis corroborates such an assumption by showing all R. parkeri sensu stricto isolates from A. maculatum and A. triste in a single large clade (clade A). On the other hand, the separation of this clade into two subgroups, clade A1 containing North American isolates and clade A2 with South American isolates, could be a result of the geographical distance of the isolates, the host tick species, or a combination of both. Further studies employing South American isolates of R. parkeri sensu stricto from A. maculatum, as well as from A. tigrinum, would help to elucidate these subgroups.
In the original reports of the strains Atlantic rainforest, NOD, and Parvitarsum in South America, the limited phylogenetic analysis provided enough data to only demonstrate a close relatedness to R. parkeri, R. africae, and R. sibirica (21, 32, 33). Herein, we present a robust phylogenetic analysis with strong statistical support to demonstrate a monophyletic group formed by strains Atlantic rainforest, NOD, and Parvitarsum and isolates of R. parkeri sensu stricto from North and South America. In addition, the genetic divergence values between New World isolates were generally ≤1.00, whereas values between New World isolates and Old World isolates (R. africae, R. sibirica, and R. conorii) were generally ≥1.00 (Table 2). On the basis of this evidence, we propose that Atlantic rainforest, NOD, and Parvitarsum are South American strains of R. parkeri. In fact, Paddock et al. (31) recently provided molecular evidence to classify Rickettsia sp. strain Atlantic rainforest as a distinct strain of R. parkeri. Interestingly, each of these R. parkeri strains seems to be primarily associated with a tick species or a tick species group, namely, R. parkeri sensu stricto with the A. maculatum species group (includes A. triste), R. parkeri strain NOD with A. nodosum, R. parkeri strain Parvitarsum with A. parvitarsum, and R. parkeri strain Atlantic rainforest with the A. ovale species group (includes A. aureolatum). Such rickettsial strain-tick species specificity suggests a coevolution of each tick-strain association.
Our study evaluated multiple isolates of a strain of R. parkeri from North America (R. parkeri sensu stricto) and four distinct strains from South America (R. parkeri sensu stricto, R. parkeri strain Atlantic rainforest, R. parkeri strain NOD, and R. parkeri strain Parvitarsum). During the course of the present study, another strain of R. parkeri was reported from the United States, namely, R. parkeri strain Black Gap, isolated recently from D. parumapertus in the United States, and showed to be nearly identical to R. parkeri strain Atlantic rainforest (31). In addition to these established strains, other unique R. parkeri-like genotypes have been characterized genetically from South American ticks. These include Rickettsia sp. strain Cooperi in Amblyomma dubitatum (37), Rickettsia sp. strain ApPR in Amblyomma parkeri (38), Rickettsia sp. strain PA in Amblyomma naponense (39), all from Brazil, and Rickettsia sp. strain tuberculatum in Amblyomma tuberculatum from the United States (40). Collectively, these data reveal that North American strains of R. parkeri are thus far associated predominantly with 3 species of ticks (A. maculatum, D. parumapertus, and A. tuberculatum), and the South American strains of R. parkeri are associated predominantly with at least 7 species of South American ticks (A. triste, A. ovale, A. nodosum, A. parvitarsum, A. dubitatum, A. parkeri, and A. naponense). It also seems likely that additional strains of R. parkeri will be discovered in the Americas in the years to come.
Nonetheless, the greater diversity of R. parkeri in South America, associated with the genus Amblyomma, suggests that this species radiated first in this continent and thereafter entered into North America. This could have occurred during the great American biotic interchange ca. 3 million years ago, when the formation of the Isthmus of Panama was completed (41). This period coincides with the most likely introduction of the genus Amblyomma into North America (42, 43). Therefore, it is possible that R. parkeri radiated with the genus Amblyomma within South America and thereafter entered with this tick genus into North America, where the bacterium subsequently adapted to other tick genera, such as Dermacentor.
Rickettsia parkeri sensu stricto and R. parkeri strain Atlantic rainforest are emerging agents of tick-borne spotted fever rickettsiosis in the New World, where they cause acute febrile illness characterized by fever, rash, inoculation eschar, and lymphadenopathy, but no deaths so far (6, 13, 18, 44). In the New World, Rocky Mountain spotted fever (also known as Brazilian spotted fever), caused by Rickettsia rickettsii, is the most commonly reported tick-borne spotted fever, which is characterized by more severe symptoms, including high fatality rates in some areas (44). Because the usual serological tests for the diagnosis of spotted fever are not able to distinguish between the infections caused by spotted fever group agents (45), it is possible that many spotted fever cases in the New World could be caused by R. parkeri sensu lato agents. Such an assumption was recently corroborated in the United States, where human cases previously assigned as Rocky Mountain spotted fever were in fact shown to be caused by R. parkeri (46). This scenario becomes even more unresolved if we consider that spotted fever is considered to be highly unreported in Latin America.
MATERIALS AND METHODS
A total of 34 rickettsial isolates, mostly from ticks and a few from humans, were used in this study. The origin of each isolate, as well as the rickettsial collection that provided it for the present study, is described in Table 1. All isolates were grown in Vero cells by using the standard techniques of each laboratory (described in the references cited in Table 1). When >90% of the cells were infected, the monolayer was harvested and subjected to DNA extraction using the DNeasy blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations. In addition, we also processed DNA samples from 5 A. triste ticks from the study by Nava et al. (12), who showed that these 5 tick samples were infected by R. parkeri. For the 39 rickettsia samples (34 isolates and 5 tick samples), the amplification of fragments of five rickettsial genes and three intergenic spacers was attempted with the primer pairs listed in Table 3. DNA fragments amplified by PCR were separated by 1.5% agarose gel electrophoresis, stained with Sybr Safe (Thermo Fisher Scientific, Waltham, MA), and visualized in a photo gel documentation system (AlphaImager HP system, San Jose, CA). Amplicons were purified with ExoSap (USB Corporation, Cleveland, OH) and DNA sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA), in an ABI automated sequencer (model ABI 3500 genetic analyzer; Applied Biosystems/Thermo Fisher Scientific, Foster City, CA) according to the manufacturer's specifications.
TABLE 3.
Primer pairs used for amplification of rickettsial genes or intergenic regions in the present study
| Primer pair | Target | Primer sequence (5′ to 3′) |
Amplicon size (nt)a | Reference | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| 1 | gltA | GCAAGTATCGGTGAGGATGTAAT | GCTTCCTTAAAATTCAATAAATCAGGAT | 401 | 37 |
| 2 | ompA | ATGGCGAATATTTCTCCAAAA | GTTCCGTTAATGGCAGCATCT | 632 | 61 |
| 3 | virB4 | TCTATAGTACATGATTCTGCT | TGATTACCGAGTGTAGTATTATG | 840 | 62 |
| 4 | dnaA | CTTTACAATCATTACGGTG | GCAACTAAGCCCCATCC | 788 | 62 |
| 5 | dnaK | GCATTCTAGTCATACCGCC | CAAAAAATGAAAGAAACTGCTGA | 650 | 62 |
| 6 | mppA-purC | GCAATTATCGGTCCGAATG | TTTCATTTATTTGTCTCAAAATTCA | 160 | 63 |
| 7 | rpmE-tRNAfMet | TTCCGGAAATGTAGTAAATCAATC | TCAGGTTATGAGCCTGACGA | 144 | 63 |
| 8 | rrl-rrf-ITS | GCAACTAAGCCCCATCC | GATAGGTCGGGTGTGGAAG | 350 | 62 |
nt, nucleotides.
DNA sequences of the different target genes or intergenic spacers were edited for the removal of primer sequences by using the SeqMan software (DNAStar, Inc., Madison, WI). The sequences were subjected to multiple alignments by using the program Clustal X (47) by changing the parameters related to the insertion of indels (insertion weight, 1; extension, 1) and then manually adjusted by using GeneDoc v. 2.6.01 (48). The genome sequences of R. africae strain ESF (accession number NC_012633.1) and R. sibirica sibirica strain 246 (accession number AABW01000001.1) were downloaded from the GenBank database. The fragments of the five rickettsial genes and three intergenic spacers listed in Table 3 were saved and included in our alignments, which included a total of 41 rickettsial isolates. Phylogenetic trees were inferred by Bayesian (B) and maximum parsimony (MP) methods. The concatenated alignment of all markers (gltA, ompA, virB4, dnaA, dnaK, mppE-pur, rrl-rrf-ITS, and rpmE-tRNAfMet) was analyzed by B and MP methods. The markers were analyzed individually only by the MP method. MP trees were constructed using the PAUP* v4.0b10 program (49) via a heuristic search with 100 replicates of random additions of the terminals followed by branching (RAS-TBR branch-breaking). Bootstrap support analyses were performed on 100 replicates with the same parameters used in the search. Bayesian analyses were performed in the MrBayes v.3.1.2 program (50); 1,000,000 generations were employed using GTR as a substitution model and four range categories plus an invariant proportion of sites. For the verification of support of branches in the Bayesian analyses, the “posteriori” probability values obtained using the MrBayes program were used. Similarity matrices (based on uncorrected p-distance) were constructed using the Pontos program provided by J. M. Alves at http://sourceforge.net/projects/pontos/.
Accession number(s).
The GenBank accession numbers for the DNA sequences generated in this study for the 39 rickettsial isolates shown in Table 1 are the following: gltA gene, MF737524 to MF737556, MF737558 to MF737562, and MF737564; ompA gene, MF737606 to MF737643; virB4 gene, MF925495 to MF925531, MF925534, and MF925699; dnaA gene, MF737565 to MF737578, MF737580 to MF737602, MF737604, and MF737605; dnaK gene, MF925658 to MF925689, MF925691 to MF925696, and MF925698; mppA-purC intergenic spacer, MF925535 to MF925568, MF925570 to MF925573, and MF925575; rpmE-tRNAfMet intergenic spacer, MF925576 to MF925608, MF925610 to MF925614, and MF925616; and rrl-rrf-ITS intergenic spacer, MF925617 to MF925649, MF925651 to MF925655, and MF925657.
Supplementary Material
ACKNOWLEDGMENTS
We thank David H. Walker and Patricia A. Valdes for providing DNA of R. africae strain Z8-Ah.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (grant 2011/51979-1 to F.A.N.-B.) and by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior CAPES/PROEX 1841/2016.
The findings and conclusions are those of the authors and do not necessarily reflect the views of the U.S. Department of Health and Human Services.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02872-17.
REFERENCES
- 1.Lackman DB, Parker RR, Gerloff RK. 1949. Serological characteristics of a pathogenic rickettsia occurring in Amblyomma maculatum. Public Health Rep 64:1342–1349. doi: 10.2307/4587134. [DOI] [PubMed] [Google Scholar]
- 2.Parker RR, Kohls GM, Cox GW, Davis GE. 1939. Observations on an infectious agent from Amblyomma maculatum. Public Health Rep 54:1482–1484. doi: 10.2307/4582985. [DOI] [Google Scholar]
- 3.Lackman DB, Bell EJ, Stoenner HG, Pickens EG. 1965. The Rocky Mountain spotted fever group of rickettsias. Health Lab Sci 2:135–141. [PubMed] [Google Scholar]
- 4.Paddock CD, Summer JW, Comer JA, Zaki SR, Goldsmith CS, Goddard J, Mclellan SLF, Tamminga CL, Ohl CA. 2004. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin Infect Dis 38:805–811. doi: 10.1086/381894. [DOI] [PubMed] [Google Scholar]
- 5.Paddock CD, Goddard J. 2015. The evolving medical and veterinary importance of the Gulf Coast tick (Acari: Ixodidae). J Med Entomol 52:230–252. doi: 10.1093/jme/tju022. [DOI] [PubMed] [Google Scholar]
- 6.Straily A, Feldpausch A, Ulbrich C, Schell K, Casillas S, Zaki SR, Denison AM, Condit M, Gabel J, Paddock CD. 2016. Notes from the Field: Rickettsia parkeri rickettsiosis—Georgia, 2012–2014. MMWR Morb Mortal Wkly Rep 65:718–719. doi: 10.15585/mmwr.mm6528a3. [DOI] [PubMed] [Google Scholar]
- 7.Herrick KL, Pena SA, Yaglom HD, Layton BJ, Moors A, Loftis AD, Condit ME, Singleton J, Kato CY, Denison AM, Ng D, Mertins JW, Paddock CD. 2016. Rickettsia parkeri rickettsiosis, Arizona, USA. Emerg Infect Dis 22:780–785. doi: 10.3201/eid2205.151824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venzal JM, Portillo A, Estrada Penã A, Castro O, Cabrera PA, Oteo JA. 2004. Rickettsia parkeri in Amblyomma triste from Uruguay. Emerg Infect Dis 10:1493–1495. doi: 10.3201/eid1008.030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conti Díaz IA. 2001. Rickettsiosis por Rickettsia conorii (fiebre botonosa del Mediterráneo o fiebre de Marsella). Estado actual en Uruguay. Rev Med Urug (Montev) 17:119–124. (In Spanish.) [Google Scholar]
- 10.Conti Díaz IA. 2003. Rickettsiosis caused by Rickettsia conorii in Uruguay. Ann N Y Acad Sci 990:264–266. doi: 10.1111/j.1749-6632.2003.tb07375.x. [DOI] [PubMed] [Google Scholar]
- 11.Conti-Díaz IA, Moraes-Filho J, Pacheco RC, Labruna MB. 2009. Serological evidence of Rickettsia parkeri as the etiological agent of rickettsiosis in Uruguay. Rev Inst Med Trop Sao Paulo 51:337–339. doi: 10.1590/S0036-46652009000600005. [DOI] [PubMed] [Google Scholar]
- 12.Nava S, Elshenawy Y, Eremeeva ME, Sumner JW, Mastropaolo M, Paddock CD. 2008. rickettsia parkeri in Argentina. Emerg Infect Dis 14:1894–1897. doi: 10.3201/eid1412.080860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romer Y, Seijo AC, Crudo F, Nicholson WL, Varela-Stokes A, Lash RR, Paddock CD. 2011. Rickettsia parkeri rickettsiosis, Argentina. Emerg Infect Dis 17:1169–1173. doi: 10.3201/eid1707.101857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silveira I, Pacheco RC, Szabó MPJ, Ramos HGC, Labruna MB. 2007. First report of Rickettsia parkeri in Brazil. Emerg Infect Dis 13:1111–1113. doi: 10.3201/eid1307.061397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores-Mendoza C, Florin D, Felices V, Pozo EJ, Graf PC, Burrus RG, Richards AL. 2013. Detection of Rickettsia parkeri from within Piura, Peru, and the first reported presence of Candidatus Rickettsia andeanae in the tick Rhipicephalus sanguineus. Vector Borne Zoonotic Dis 13:505–508. doi: 10.1089/vbz.2012.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lado P, Castro O, Labruna MB, Venzal JM. 2014. First molecular detection of Rickettsia parkeri in Amblyomma tigrinum and Amblyomma dubitatum ticks from Uruguay. Ticks Tick Borne Dis 5:660–662. doi: 10.1016/j.ttbdis.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Tomassone L, Conte V, Parrilla G, De Meneghi D. 2010. Rickettsia infection in dogs and Rickettsia parkeri in Amblyomma tigrinum ticks, Cochabamba Department, Bolivia. Vector Borne Zoonotic Dis 10:953–958. doi: 10.1089/vbz.2009.0126. [DOI] [PubMed] [Google Scholar]
- 18.Romer Y, Nava S, Govedic F, Cicuttin G, Denison AM, Singleton J, Kelly AJ, Kato CY, Paddock CD. 2014. Rickettsia parkeri rickettsiosis in different ecological regions of Argentina and its association with Amblyomma tigrinum as a potential vector. Am J Trop Med Hyg 91:1156–1160. doi: 10.4269/ajtmh.14-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weck B, Dall'Agnol B, Souza U, Webster A, Stenzel B, Klafke G, Martins JR, Reck J. 2016. Spotted fever group Rickettsia in the Pampa biome, Brazil, 2015–2016. Emerg Infect Dis 22:2014–2016. doi: 10.3201/eid2211.160859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrada-Peña A, Venzal JM, Mangold AJ, Cafrune MM, Guglielmone AA. 2005. The Amblyomma maculatum Koch, 1844 (Acari: Ixodidae: Amblyomminae) tick group: diagnostic characters, description of the larva of A. parvitarsum Neumann, 1901, 16S rDNA sequences, distribution and hosts. Syst Parasitol 60:99–112. doi: 10.1007/s11230-004-1382-9. [DOI] [PubMed] [Google Scholar]
- 21.Spolidorio MG, Labruna MB, Mantovani E, Brandao PE, Richtzenhain LJ, Yoshinari NH. 2010. Novel spotted fever group rickettsiosis, Brazil. Emerg Infect Dis 16:521–523. doi: 10.3201/eid1603.091338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva N, Eremeeva ME, Rozental T, Ribeiro GS, Paddock CD, Ramos EAG, Favacho ARM, Reis MG, Dasch GA, de Lemos ERS, Ko AI. 2011. Eschar-associated spotted fever rickettsiosis, Bahia, Brazil. Emerg Infect Dis 17:275–278. doi: 10.3201/eid1702.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krawczak FS, Muñoz-Leal S, Guztzazky AC, Oliveira SV, Santos FC, Angerami RN, Moraes-Filho J, de Souza JC Jr, Labruna MB. 2016. Rickettsia sp. strain Atlantic rainforest infection in a patient from a spotted fever-endemic area in southern Brazil. Am J Trop Med Hyg 95:551–553. doi: 10.4269/ajtmh.16-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabó MP, Nieri-Bastos FA, Spolidorio MG, Martins TF, Barbieri AM, Labruna MB. 2013. In vitro isolation from Amblyomma ovale (Acari: Ixodidae) and ecological aspects of the Atlantic rainforest Rickettsia, the causative agent of a novel spotted fever rickettsiosis in Brazil. Parasitology 140:719–728. doi: 10.1017/S0031182012002065. [DOI] [PubMed] [Google Scholar]
- 25.Nieri-Bastos FA, Horta MC, Barros-Battesti DM, Moraes-Filho J, Ramirez DG, Martins TF, Labruna MB. 2016. Isolation of the pathogen Rickettsia sp. strain Atlantic rainforest from its presumed tick vector, Amblyomma ovale (Acari: Ixodidae), from two areas of Brazil. J Med Entomol 53:977–981. doi: 10.1093/jme/tjw062. [DOI] [PubMed] [Google Scholar]
- 26.Barbieri AR, Filho JM, Nieri-Bastos FA, Souza JC Jr, Szabó MP, Labruna MB. 2014. Epidemiology of Rickettsia sp. strain Atlantic rainforest in a spotted fever-endemic area of southern Brazil. Ticks Tick Borne Dis 5:848–853. doi: 10.1016/j.ttbdis.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Guglielmone AA, Estrada-Peña A, Mangold AJ, Barros-Battesti DM, Labruna MB, Martins JR, Venzal JM, Arzua M, Keirans JE. 2003. Amblyomma aureolatum (Pallas, 1772) and Amblyomma ovale Koch, 1844 (Acari: Ixodidae): hosts, distribution and 16S rDNA sequences. Vet Parasitol 113:273–288. doi: 10.1016/S0304-4017(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 28.Krawczak FS, Agostinho WC, Polo G, Moraes-Filho J, Labruna MB. 2016. Comparative evaluation of Amblyomma ovale ticks infected and noninfected by Rickettsia sp. strain Atlantic rainforest, the agent of an emerging rickettsiosis in Brazil. Ticks Tick Borne Dis 7:502–507. doi: 10.1016/j.ttbdis.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Londoño AF, Díaz FJ, Valbuena G, Gazi M, Labruna MB, Hidalgo M, Mattar S, Contreras V, Rodas JD. 2014. Infection of Amblyomma ovale by Rickettsia sp. strain Atlantic rainforest, Colombia. Ticks Tick Borne Dis 5:672–675. doi: 10.1016/j.ttbdis.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 30.Lopes MG, May J Jr, Foster RJ, Harmsen BJ, Sanchez E, Martins TF, Quigley H, Marcili A, Labruna MB. 2016. Ticks and rickettsiae from wildlife in Belize, Central America. Parasit Vectors 9:62. doi: 10.1186/s13071-016-1348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paddock CD, Allerdice MEJ, Karpathy SE, Nicholson WL, Levin ML, Smith TC, Becker T, Delph RJ, Knight RN, Ritter JM, Sanders JH, Goddard J. 2017. Unique strain of Rickettsia parkeri associated with the hard tick Dermacentor parumapertus Neumann in the western United States. Appl Environ Microbiol 83:e03463-16. doi: 10.1128/AEM.03463-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogrzewalska M, Pacheco RC, Uezu A, Richtzenhain LJ, Ferreira F, Labruna MB. 2009. Rickettsial infection in Amblyomma nodosum ticks (Acari: Ixodidae) from Brazil. Ann Trop Med Parasitol 103:413–425. doi: 10.1179/136485909X451744. [DOI] [PubMed] [Google Scholar]
- 33.Ogrzewalska M, Nieri-Bastos FA, Marcili A, Nava S, González-Acuña D, Muñoz-Leal S, Ruiz-Arrondo I, Venzal JM, Mangold A, Labruna MB. 2016. A novel spotted fever group Rickettsia infecting Amblyomma parvitarsum (Acari: Ixodidae) in highlands of Argentina and Chile. Ticks Tick Borne Dis 7:439–442. doi: 10.1016/j.ttbdis.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Kohls GM. 1956. Concerning the identity of Amblyomma maculatum, A. tigrinum, A. triste and A. ovatum of Koch, 1844. Proc Entomol Soc Wash 58:143–147. [Google Scholar]
- 35.Guglielmone AA, Robbins RG, Apanaskevich DA, Petney TN, Estrada-Peña A, Horak IG. 2014. The hard ticks of the world (Acari: Ixodida: Ixodidae). Springer, Dordrecht, Netherlands. [Google Scholar]
- 36.Nava S, Venzal JM, González-Acuña DG, Martins TF, Guglielmone AA. 2017. Ticks of the southern cone of America: diagnosis, distribution, and hosts with taxonomy, ecology and sanitary importance, 1st ed Elsevier, London, United Kingdom. [Google Scholar]
- 37.Labruna MB, Whitworth T, Horta MC, Bouyer DH, McBride JW, Pinter A, Popov V, Gennari SM, Walker DH. 2004. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J Clin Microbiol 42:90–98. doi: 10.1128/JCM.42.1.90-98.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pacheco RC, Arzua M, Nieri-Bastos FA, Moraes-Filho J, Marcili A, Richtzenhain LJ, Barros-Battesti DM, Labruna MB. 2012. Rickettsial infection in ticks (Acari: Ixodidae) collected on birds in southern Brazil. J Med Entomol 49:710–716. doi: 10.1603/ME11217. [DOI] [PubMed] [Google Scholar]
- 39.Soares HS, Barbieri ARM, Martins TF, Minervino AHH, de Lima JTR, Marcili A, Gennari SM, Labruna MB. 2015. Ticks and rickettsial infection in the wildlife of two regions of the Brazilian Amazon. Exp Appl Acarol 65:125–140. doi: 10.1007/s10493-014-9851-6. [DOI] [PubMed] [Google Scholar]
- 40.Zemtsova GE, Gleim E, Yabsley MJ, Conner LM, Mann T, Brown MD, Wendland L, Levin ML. 2012. Detection of a novel spotted fever group Rickettsia in the gopher tortoise tick. J Med Entomol 49:783–786. doi: 10.1603/ME11264. [DOI] [PubMed] [Google Scholar]
- 41.Smith BT, Klicka J. 2010. The profound influence of the Late Pliocene Panamanian uplift on the exchange, diversification, and distribution of New World birds. Ecography 33:333–342. doi: 10.1111/j.1600-0587.2009.06335.x. [DOI] [Google Scholar]
- 42.Balashov YS. 1994. Importance of continental drift in the distribution and evolution of ixodid ticks. Entomol Rev 73:42–50. [Google Scholar]
- 43.Beati L, Nava S, Burkman EJ, Barros-Battesti DM, Labruna MB, Guglielmone AA, Cáceres AG, Guzmán-Cornejo CM, León R, Durden LA, Faccini JL. 2013. Amblyomma cajennense (Fabricius, 1787) (Acari: Ixodidae), the Cayenne tick: phylogeography and evidence for allopatric speciation. BMC Evol Biol 13:267. doi: 10.1186/1471-2148-13-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paddock CD, Lane RS, Staples JE, Labruna MB. 2016. Changing paradigms for tick-borne diseases in the Americas, p 221–258. In Global health impacts of vector-borne diseases: workshop summary. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 45.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, Abdad MY, Stenos J, Bitam I, Fournier PE, Raoult D. 2013. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev 26:657–702. doi: 10.1128/CMR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raoult D, Paddock CD. 2005. Rickettsia parkeri infection and other spotted fevers in the United States. N Engl J Med 353:626–627. doi: 10.1056/NEJM200508113530617. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholas KB, Nicholas HB Jr, Deerfield DW II. 1997. GeneDoc: analysis and visualization of genetic variation. Embnet.news 4:14. [Google Scholar]
- 49.Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4b10. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 50.Ronquist F, Huelsenbeck JP. 2003. Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 51.Paddock CD, Fournier PE, Sumner JW, Goddard J, Elshenawy Y, Metcalfe MG, Loftis AD, Varela-Stokes A. 2010. Isolation of Rickettsia parkeri and identification of a novel spotted fever group Rickettsia sp. from Gulf Coast ticks (Amblyomma maculatum) in the United States. Appl Environ Microbiol 76:2689–2696. doi: 10.1128/AEM.02737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varela-Stokes AS, Paddock CD, Engber B, Toliver M. 2011. Rickettsia parkeri in Amblyomma maculatum ticks, North Carolina, USA, 2009–2010. Emerg Infect Dis 17:2350–2353. doi: 10.3201/eid1712.110789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitman TJ, Richards AL, Paddock CD, Tamminga CL, Sniezek PJ, Jiang J, Byers DK, Sanders JW. 2007. Rickettsia parkeri infection after tick bite, Virginia. Emerg Infect Dis 13:334–336. doi: 10.3201/eid1302.061295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fornadel CM, Zhang X, Smith JD, Paddock CD, Arias JR, Norris DE. 2011. High rates of Rickettsia parkeri infection in Gulf Coast ticks (Amblyomma maculatum) and identification of “Candidatus Rickettsia andeanae” from Fairfax County, Virginia. Vector Borne Zoonotic Dis 11:1535–1539. doi: 10.1089/vbz.2011.0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nieri-Bastos FA, Szabó MPJ, Pacheco RC, Soares JF, Soares HS, Moraes-Filho J, Dias RA, Labruna MB. 2013. Comparative evaluation of infected and noninfected Amblyomma triste ticks with Rickettsia parkeri, the agent of an emerging rickettsiosis in the New World. Biomed Res Int 2013:402737. doi: 10.1155/2013/402737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melo ALT, Alves AS, Nieri-Bastos FA, Martins TF, Witter R, Pacheco TA, Soares HS, Marcili A, Chitarra CS, Dutra V, Nakazato L, Pacheco RC, Labruna MB, Aguiar DM. 2015. Rickettsia parkeri infecting free-living Amblyomma triste ticks in the Brazilian Pantanal. Ticks Tick Borne Dis 6:237–241. doi: 10.1016/j.ttbdis.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 57.Pacheco RC, Venzal JM, Richtzenhain LJ, Labruna MB. 2006. Rickettsia parkeri in Uruguay. Emerg Infect Dis 12:1804–1805. doi: 10.3201/eid1211.060577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelly PJ, Beati L, Matthewman LA, Mason PR, Dasch GA, Raoult D. 1994. A new pathogenic spotted fever group rickettsia from Africa. J Trop Med Hyg 97:129–137. [PubMed] [Google Scholar]
- 59.De Sousa R, França A, Dória Nòbrega S, Belo A, Amaro M, Abreu T, Poças J, Proença P, Vaz J, Torgal J, Bacellar F, Ismail N, Walker DH. 2008. Host- and microbe-related risk factors for and pathophysiology of fatal Rickettsia conorii infection in Portuguese patients. J Infect Dis 198:576–585. doi: 10.1086/590211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Sousa R, Barata C, Vitorino L, Santos-Silva M, Carrapato C, Torgal J, Walker D, Bacellar F. 2006. Rickettsia sibirica isolation from a patient and detection in ticks, Portugal. Emerg Infect Dis 12:1103–1108. doi: 10.3201/eid1207.051494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roux V, Fournier PE, Raoult D. 1996. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J Clin Microbiol 34:2058–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vitorino L, Chelo IM, Bacellar F, Zé-Zé L. 2007. Rickettsiae phylogeny: a multigenic approach. Microbiology 153:160–168. doi: 10.1099/mic.0.2006/001149-0. [DOI] [PubMed] [Google Scholar]
- 63.Fournier PE, Zhu Y, Ogata H, Raoult D. 2004. Use of highly variable intergenic spacer sequences for multispacer typing of Rickettsia conorii strains. J Clin Microbiol 42:5757–5766. doi: 10.1128/JCM.42.12.5757-5766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.