ABSTRACT
Streptococcus suis, a global zoonosis of pigs, shows regional differences in the prevalence of human-associated disease for Asian and non-Asian countries. The isolation rates and diversities of S. suis on tonsils of healthy slaughter pigs in China and the United Kingdom were studied for effects of geography, temperature, pig age, and farm type. Isolates underwent analysis of molecular serotype and multilocus sequence type and virulence-associated genotyping. Although we found no significant difference in positive isolation rates between Chinese and UK farms, the prevalences of serotypes previously associated with human disease were significantly greater in the Chinese collection (P = 0.003). A significant effect of temperature was found on the positive isolation rate of the Chinese samples and the prevalence of human disease-associated serotypes in the UK S. suis population (China, P = 0.004; United Kingdom, P = 0.024) and on the prevalence of isolates carrying key virulence genes in China (P = 0.044). Finally, we found marked diversity among S. suis isolates, with statistically significant temperature effects on detection of multiple strain types within individual pigs. This study highlighted the high carriage prevalence and diversity of S. suis among clinically healthy pig herds of China and the United Kingdom. The significant effect of temperature on prevalence of isolation, human disease-associated serotypes, and diversity carried by individual pigs may shed new light on geographic variations in human S. suis-associated disease.
IMPORTANCE Streptococcus suis is a global zoonotic pathogen and also a normal colonizer mainly carried on the tonsil of pigs. Thus, it is important to study the effect of environmental and management-associated factors on the S. suis populations in clinically healthy pigs. In this research, we investigated the similarities and differences between the S. suis populations obtained from different pig ages, seasons, and farm management systems and discovered the relationship between high climatic temperature and the prevalence of S. suis.
KEYWORDS: Streptococcus suis, clinically healthy pig herds, environmental factors, management-associated factors
INTRODUCTION
Streptococcus suis, a globally important pig pathogen that infects pigs and humans (1), is a normal colonizer of the upper respiratory tracts (URT) of healthy pigs (2) and is a major worldwide driver of antibiotic administration to pigs. Colonization of pigs by S. suis is widespread globally, but disease-associated strains are found only rarely in the URT (3). The high level of genomic diversity between isolates of S. suis, as well as evidence for high levels of recombination, indicates the potential for emergence of additional disease-associated strains (3) and the importance of gaining a deeper understanding of S. suis population diversity in the URT of healthy pigs.
Classical typing methods identified 35 serotypes of S. suis with a large number of nonserotypeable isolates, but serotypes 32 and 34 were excluded after analysis of the 16S rRNA gene sequence showed that they should be reclassified as S. orisratti (4). It was recently proposed that serotypes 20, 22, 26, and 33 should also be reclassified as another species based on the phylogeny of one or two genes (5), but whole-genome analysis of a larger collection of divergent isolates of S. suis found them to be more closely associated with S. suis than with any other species (6, 7). Of the 33 serotypes, several are overrepresented among the isolates from clinical samples. Serotypes 1, 2, 7, 9, and 14 were the most prevalent disease-associated serotypes in Europe, Southeast Asia, and South America, while serotypes 3 and 8 were more frequently isolated from cases in North America (8). However, human cases of disease are dominated by serotype 2, followed by serotype 14, with serotypes 4, 5, 16, 21, 24, and 31 occasionally reported (9–13). The introduction of molecular typing by PCR, based on polymorphisms in the cps loci (14), has assisted in assigning molecular serotype to most, but not all, previously nonserotypeable isolates commonly found in healthy pigs. A new approach, known as NCLs (novel cps loci), was introduced to address the classification of remaining nonserotypeable S. suis based on eight novel cps loci (15).
Multilocus sequence typing (MLST) (16), based on polymorphisms in seven housekeeping gene fragments to identify discrete sequence types (STs), not only showed serotype to be a poor indicator of genetic relatedness but also identified significant geographical differences in ST distribution. For example, ST7 was only found in China, where it was associated with a severe outbreak of pig and human disease in Sichuan Province in 2005, while ST1 was predominant in meningitis and septicemia among humans and pigs in Asia, Europe, and South America (8, 17). To date, 956 STs have been described (see PubMLST [https://pubmlst.org/ssuis/]), the majority of these STs belonging to disease-associated strains. However, the MLST method is based on a small fraction of the genome and, although useful as an epidemiological tool, it has been proved ineffective in the discrimination of disease-associated from non-disease-associated isolates.
Reports of the first cases of human disease dated back to the 1950s in Europe, where disease was historically linked to occupational exposure, but recent surveillance shows that most human cases currently occur in Asia (18). In China, S. suis has been implicated in summertime outbreaks of toxic shock-like syndrome (19, 20) and was identified as the third most common cause of adult bacterial meningitis in Hong Kong (21). Meningitis and septicemia are widely linked to S. suis in other Asian countries, including Thailand and especially Vietnam, where it was the most frequent reported cause of adult bacterial meningitis (22). The widespread occurrence in Asia of zoonotic cases of S. suis might be explained by the high density of pigs in the region, traditional slaughtering practices without preventive measures, and the consumption of uncooked or lightly cooked pig products (23). While these factors have been shown to be relevant, no large-scale genomic comparative studies have yet been undertaken on the diversity and prevalence of carriage of S. suis in pigs entering the pig meat supply chain in Asian versus European countries.
This study set out to apply emergent molecular epidemiological methods to gain insights into the factors affecting S. suis carriage in pigs entering the human food chain. A longitudinal study was performed to investigate the genomic characteristics of the S. suis populations in clinically healthy pig populations in pig meat supply chains in China and the United Kingdom. We set out to describe relationships between strain prevalence and diversity with pig age, climatic temperature, and farm management system.
RESULTS
Factors associated with S. suis positive isolation rate at farm level. (i) Overall and farm-level isolation rates.
The Chinese collection included 223 isolates of S. suis, confirmed by whole-genome sequence (WGS), obtained from 137/500 (27.4%) tonsil swabs from clinical healthy pigs. The positive isolation rate of each farm at the 5-week-old sample point ranged from 8.00 to 62.96%, and at 20 weeks the range was 4.00 to 52.00%. For the UK collection, 127 isolates, confirmed by WGS, were isolated from 89/250 (35.6%) tonsil swabs. The positive isolation rate ranged from 4.00 to 92.00% at the 5-week-old sampling occasion and from 4.00 to 40.00% at 20 weeks (Table 1). There was no significant difference between the positive isolation rate of the Chinese samples and that of the UK samples (two-tailed P = 0.174 [Student t test]).
TABLE 1.
Distribution of S. suis-positive isolation rates, mean temperatures, and farm types in China and the United Kingdoma
| Country and farm | Farm type | 5-wk-old pigs |
20-wk-old pigs |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MT (°C) | Size | N (F) | PP | PIR (%) | MT (°C) | Size | N (F) | PP | PIR (%) | ||
| China | |||||||||||
| C1 | Intensive | 29.5 | 23 | 10 (9) | 6 | 26.09 | 29 | 25 | 26 (21) | 11 | 44.00 |
| C2 | Intensive | 32 | 27 | 33 (30) | 17 | 62.96 | 19.5 | 25 | 1 (1) | 1 | 4.00 |
| C3 | Intensive | 29.5 | 25 | 28 (26) | 15 | 60.00 | 17.5 | 25 | 2 (2) | 2 | 8.00 |
| C4 | Intensive | 15.5 | 25 | 12 (12) | 9 | 36.00 | 4.8 | 25 | 2 (2) | 2 | 8.00 |
| C5 | Intensive | 15.5 | 25 | 12 (12) | 10 | 40.00 | 4.8 | 25 | 12 (12) | 8 | 32.00 |
| C6 | Small | 28.5 | 24 | 21 (19) | 12 | 50.00 | 29 | 25 | 20 (19) | 13 | 52.00 |
| C7 | Small | 29 | 24 | 11 (11) | 8 | 33.33 | 28.5 | 25 | 6 (6) | 4 | 16.00 |
| C8 | Small | 30 | 27 | 13 (13) | 8 | 29.63 | 17.5 | 25 | 7 (7) | 4 | 16.00 |
| C9 | Small | 14 | 25 | 2 (2) | 2 | 8.00 | 14 | 25 | 1 (1) | 1 | 4.00 |
| C10 | Small | 8.5 | 25 | 3 (3) | 2 | 12.00 | 14 | 25 | 1 (1) | 1 | 4.00 |
| United Kingdom | |||||||||||
| U1 | Intensive | 14 | 25 | 17 (17) | 11 | 44.00 | 15 | 25 | 14 (14) | 10 | 40.00 |
| U2 | Intensive | 19 | 25 | 42 (40) | 23 | 92.00 | 12.5 | 25 | 1 (1) | 1 | 4.00 |
| U3 | Intensive | 16.5 | 25 | 23 (23) | 17 | 68.00 | 4 | 25 | 9 (9) | 8 | 32.00 |
| U4 | Intensive | 11.5 | 25 | 8 (8) | 7 | 28.00 | 7.5 | 25 | 5 (5) | 4 | 16.00 |
| U5 | Intensive | 7 | 25 | 1 (1) | 1 | 4.00 | 7 | 25 | 7 (7) | 7 | 28.00 |
MT, mean temperature (the average of the highest and lowest air temperatures of the sampling day); size, sample size (the number of pigs sampled each time); N, the number of S. suis isolates; F, the number of isolates in the filtered subset for direct comparison of China and UK data; PP, positive pigs (the number of S. suis-positive pigs); PIR, positive isolation rate (the proportion of S. suis-positive pigs).
(ii) Effect of temperature, age, and farm type on positive isolation rate at farm level.
The 20 Chinese sampling occasions and the 10 UK sampling occasions were, respectively, divided into two groups across the mean values of the corresponding set of daily mean air temperature values of the sampling times.
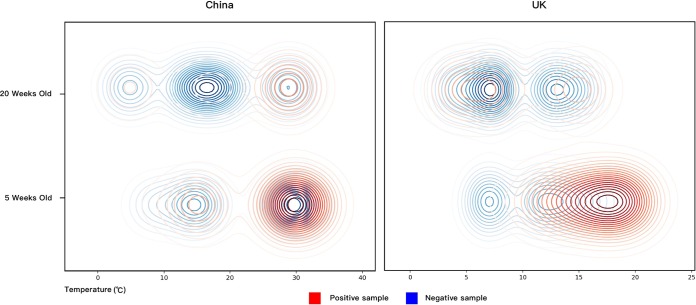
For the Chinese sampling occasions, a three-way analysis of variance (ANOVA) was used to evaluate the impact of temperature, farm type, and age, which showed that temperature was the only significant factor affecting a positive isolation rate at farm level. The positive isolation rates for pig farms sampled on high-temperature occasions were significantly greater than for those sampled on low-temperature occasions (temperature, P = 0.004; age, P = 0.203; farm type, P = 0.064) (see Table 3). For the UK sampling occasions, since the equal variance test for two-way ANOVA was not passed, t tests and paired t tests were used to evaluate the effect of temperature and age separately. Although there was no detectable significant effect of temperature or age (temperature, two-tailed P = 0.111, t test; age, two-tailed P = 0.285, paired t test), the distribution of the UK samples showed a similar pattern to that of the Chinese samples, where the frequency of positive samples was higher for sampling occasions of 5-week-old pigs at high air temperatures (Fig. 1).
TABLE 3.
Summary of statistical analysis of the influence of environmental and management-associated factors on the distribution and composition of S. suis populationsa
| Analysis | Data set | Temp | Age | Farm type | Country |
|---|---|---|---|---|---|
| Positive isolation rate at farm level | China | * | – | – | / |
| UK | – | – | / | / | |
| Filtered subsets | / | / | / | – | |
| Prevalence of pigs with multiple STs per swab | China | * | – | – | / |
| UK | * | – | / | / | |
| Filtered subsets | / | / | / | – | |
| Prevalence of HDAS isolates | China | – | – | – | / |
| UK | * | – | / | / | |
| Filtered subsets | / | / | / | * | |
| Prevalence of PVGT isolates | China | * | – | – | / |
| UK | – | – | / | / | |
| Filtered subsets | / | / | / | – |
HDAS, human disease-associated serotype; PVGT, positive virulence genotype (the virulence genes including epf, mrp, and/or sly); ST, sequence type; *, statistically significant (P < 0.05); −, not significant; /, not involved in the analysis.
FIG 1.
Distribution of S. suis-positive and -negative samples across the sampling occasions accounting for different pig ages and air temperatures. Kernel density estimation was used to describe the bivariate distribution of 500 Chinese and 250 UK samples across the air temperatures of the sampling times and the ages of the sampled pig herds.
When the Chinese and the UK sampling occasions were combined and two-way ANOVA was used to evaluate the effect of temperature and age, there was no significant interaction between temperature and age, and temperature showed a significant effect (P = 0.001), whereas age also had the noteworthy effect that 5-week-old pigs may carry more S. suis (P = 0.051).
Typing characteristics of the S. suis populations. (i) Molecular serotyping.
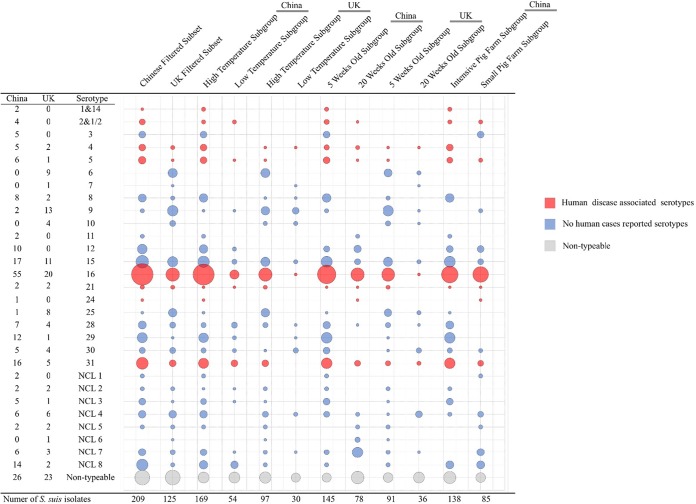
In total, 71.75% (160/223) of the Chinese S. suis isolates could be allocated to a molecular serotype across 18 serotypes, which included all 8 serotypes that had human infection cases reported (serotypes 2, 4, 5, 14, 16, 21, 24, and 31). A total of 68.50% (87/127) of the UK isolates were serotypeable covering 15 serotypes and 5 human disease-associated serotypes (HDAS; serotypes 4, 5, 16, 21, and 31). Serotype 16 was the most dominant isolates in both collections (China, n = 55; United Kingdom, n = 20), followed by serotypes 15 (n = 17) and 31 (n = 16) in the Chinese collection and serotypes 9 (n = 13) and 15 (n = 11) in UK collection. Isolates that could not be allocated a molecular serotype by this method were analyzed according to the eight novel categories identified in the NCL scheme. Both collections showed seven NCL types, while NCL type 6 and NCL type 1 could not be found in the Chinese and the UK collections, respectively (Fig. 2).
FIG 2.
Molecular serotyping composition of different S. suis subgroups. The absence or presence and the sizes of the bubbles represents the number of isolates of each serotype in each population. Red indicates serotypes that have previously been obtained from human cases of disease. The Chinese filtered subset and the Chinese high-temperature subgroup had all eight human disease-associated serotypes. Serotype 2 and 1/2 isolates were identified in all of the Chinese subgroups but not in the UK collation. Serotype 16 isolates could be identified in all subgroups.
(ii) Multilocus sequence typing.
For the Chinese collection, a total of 207 of the original 223 S. suis isolates could be classified into 93 different kinds of STs. The STs with 6 or 7 identical alleles were grouped as a clonal complexes, while the STs that belonged to no clonal complex were named as singletons. The 93 STs of Chinese isolates were finally grouped as 15 clonal complexes and 60 singletons. Of the 93 STs, only 11 STs were reported previously. The two serotype 1 and 14 isolates were typed as ST11; for the four serotype 2 and 1/2 isolates, one was typed as ST1, and the other three were typed as ST7.
Of the original 127 UK S. suis isolates, 118 were classified into 70 different kinds of STs, clustered as 53 singletons and 7 clonal complexes, only one of which had been previously reported (ST29). There were 30 isolates that could not be typed by the MLST system due to the absence of one or more alleles. There were no identical STs found in both the Chinese and UK collections (Fig. 3) (see Table S1 in the supplemental material).
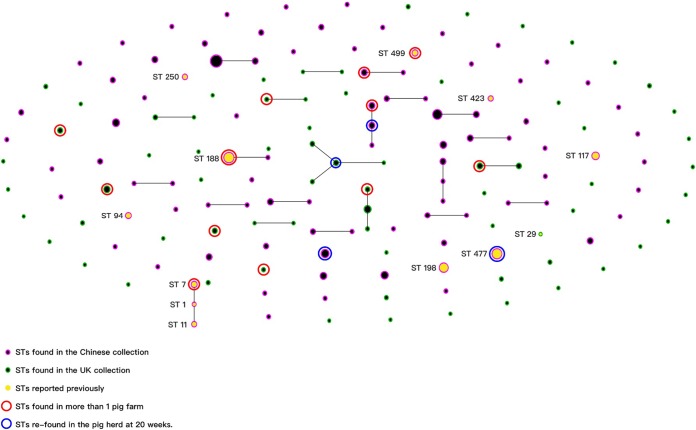
FIG 3.
Organization of STs within the S. suis population. To show the relationship between the STs of the whole S. suis population in this study, the 93 STs of the Chinese S. suis collection and the 70 STs of the UK S. suis collection were clustered using eBURST (http://eburst.mlst.net/). Each dot in the figure stands for an ST; STs with 6 or 7 identical alleles were treated as clonal complexes (CC) and are connected with lines. The sizes of the dots represent the numbers of isolates. All previously reported 12 STs are labeled. There were 7 UK STs (all new STs) and 6 Chinese STs (4 reported STs and 2 new STs) identified at more than one pig farm. Four STs (three from China and one from the United Kingdom) could be identified from both the 5-week-old and the corresponding 20-week-old sampling occasions.
(iii) Genotypes of mrp, epf, and sly.
Genotype was allocated to 223 Chinese isolates and, in total, eight virulence genotypes (VGTs) were found: Δepf Δmrp Δsly (n = 174), Δepf mrpNA1 Δsly (n = 10), Δepf mrpNA1 sly+ (n = 12), Δepf mrpNA2 sly+ (n = 12), Δepf mrpEU sly+ (n = 4); epf+ mrpNA1 sly+ (n = 1), epf+ mrpNA2 sly+ (n = 5), and epf+ mrpEU sly+ (n = 4) (where NA1 and NA2 represent the two North American subtypes and EU represents the European subtype). All 49 positive VGT (PVGT) isolates (representing 21.9% of the collection) possessed the mrp gene. Furthermore, 10/223 (4.38%) isolates in the Chinese collection were both PVGT and belonged to serotypes previously reported in human disease: serotypes 1 and 14 (2/2 in the collection were PVGT), serotypes 2 and 1/2 (4/4), serotype 4 (3/5), and serotype 21 (1/2). One nontypeable isolate possessed all three studied virulence genes. There were 6 of the total 20 sampling occasions (5 weeks old = 1, 20 weeks old = 5) that had no PVGT isolate. The genotypes Δepf mrpNA2 sly+ and Δepf mrpNA1 sly+ were the most prevalent PVGTs that could be detected in all subgroups.
The UK collection (n = 127) possessed five different VGTs: Δepf Δmrp Δsly (n = 109), Δepf mrpNA1 Δsly (n = 1), Δepf mrpNA1 sly+ (n = 7), Δepf mrpNA2 sly+ (n = 9), and epf+ mrpNA1 sly+ (n = 1). The only isolate positive for mrp, epf, and sly was nonserotypeable. As with the Chinese collection, there were 18 PVGT isolates (representing 14.1% of the collection), the mrp gene was detected in all of the virulence genes harboring PVGT isolates, and the Δepf mrpNA2 sly+ and Δepf mrpNA1 sly+ genotypes were the most prevalent PVGTs that could be detected in all subgroups (Table 2). No isolate in the UK collection was both PVGT and HDAS. Three of the total ten sampling occasions (5 weeks old = 1, 20 weeks old = 2) had no PVGT isolate.
TABLE 2.
Genomic characteristics of different S. suis populationsa
| Isolate | No. of isolates | Simpson′s diversity index |
Proportion of HDAS (%) | Virulence genotype pattern (%) (epf mrp sly) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ds | CI− | CI+ | − − − | − mrpNA2 sly | − mrpNA1 − | − mrpNA1 sly | − mrpEU sly | epf mrpNA2 sly | epf mrpNA1 sly | epf mrpEU sly | |||
| All S. suis isolates | |||||||||||||
| China_HTS | 168 | 0.971 | 0.962 | 0.981 | 43.45 | 75.00 | 6.55 | 5.95 | 6.55 | 2.38 | 1.19 | 0.60 | 1.79 |
| China_LTS | 55 | 0.976 | 0.960 | 0.991 | 32.73 | 89.09 | 1.82 | 0 | 1.82 | 0 | 5.45 | 0 | 1.82 |
| UK_HTS | 97 | 0.965 | 0.947 | 0.982 | 28.86 | 85.57 | 8.25 | 0 | 5.15 | 0 | 0 | 1.03 | 0 |
| UK_LTS | 30 | 0.966 | 0.929 | 1.000 | 6.67 | 86.67 | 3.33 | 3.33 | 6.67 | 0 | 0 | 0 | 0 |
| China_5 WOS | 145 | 0.969 | 0.959 | 0.979 | 42.76 | 75.17 | 7.59 | 5.52 | 6.90 | 0 | 2.07 | 0.69 | 2.07 |
| China_20 WOS | 78 | 0.972 | 0.957 | 0.987 | 37.18 | 84.62 | 1.28 | 2.56 | 2.56 | 5.13 | 2.56 | 0 | 1.28 |
| UK_5 WOS | 91 | 0.960 | 0.940 | 0.980 | 28.57 | 84.62 | 8.79 | 0 | 5.49 | 0 | 0 | 1.098 | 0 |
| UK_20 WOS | 36 | 0.987 | 0.975 | 0.999 | 11.11 | 88.89 | 2.78 | 2.78 | 5.56 | 0 | 0 | 0 | 0 |
| China_IPFS | 138 | 0.972 | 0.964 | 0.981 | 39.86 | 76.81 | 7.25 | 3.62 | 7.25 | 0 | 1.45 | 0.72 | 2.90 |
| China_SPFS | 85 | 0.941 | 0.908 | 0.973 | 42.35 | 81.18 | 2.35 | 5.88 | 7.06 | 0 | 3.53 | 0 | 0 |
| China_all | 223 | 0.979 | 0.973 | 0.985 | 40.81 | 78.48 | 5.38 | 4.48 | 5.38 | 1.79 | 2.24 | 0.45 | 1.79 |
| UK_all | 127 | 0.972 | 0.959 | 0.984 | 23.62 | 85.83 | 7.09 | 0.79 | 5.51 | 0 | 0 | 0.79 | 0 |
| Filtered S. suis isolates | |||||||||||||
| China_filtered | 209 | 0.980 | 0.974 | 0.986 | 39.71 | 78.47 | 5.26 | 4.78 | 5.47 | 1.44 | 2.39 | 0.48 | 1.44 |
| UK_filtered | 125 | 0.972 | 0.960 | 0.985 | 23.20 | 85.60 | 7.20 | 0.80 | 5.60 | 0 | 0 | 0.80 | 0 |
HDAS, human disease-associated serotype; HTS, high-temperature subgroup; LTS, low-temperature subgroup; 5 WOS, 5-week-old subgroup; 20 WOS, 20-week-old subgroup; IPFS, intensive pig farm subgroup; SPFS, small pig farm subgroup; Ds, Simpson′s diversity index; CI+, confidence interval upper limit (95%); CI−, confidence interval lower limit (95%).
Factors associated with pig-level diversity of S. suis.
It was not rare for more than one S. suis isolate to be obtained from the same tonsil scrape, each with different genomic characteristics. In summary, 28.47% (39/137) carried more than one S. suis ST, 27.01% (37/137) of the positive Chinese samples carried more than one S. suis ST, and 13.87% (19/137) carried isolates with multiple virulence gene profiles. For the UK samples, 25.84% (23/89) carried more than one S. suis ST, 22.47% (20/89) of positive tonsil scrapes carried more than one S. suis ST, and 10.11% (9/89) carried isolates with multiple virulence gene profiles.
Individual isolates were also identified based on their STs. The proportion of pigs carrying S. suis isolates with multiple STs at each sampling occasion was used to evaluate the influence of temperature, farm type, and age on the diversity of S. suis within sampled pigs. For the Chinese samples, three-way ANOVA results showed that there was a significant interaction between temperature, age, and farm type (P = 0.043). Since the further three-way interaction term analysis found no significant interaction between temperature and age at both intensive and small traditional type farms (intensive level, P = 0.062; small level, P = 0.198), simple main effect tests were performed at intensive and small farms, respectively. The results showed that, at both types of farm, temperature was a significant factor, whereas age was not a significant factor (temperature, intensive [P = 0.01] and small [P = 0.02]; age, intensive [P = 0.945] and small [P = 0.454]). Since the UK data set failed to pass the equal variance test for two-way ANOVA, a t test and a paired t test were used instead to evaluate temperature and age factors, respectively. The UK samples from high-temperature sampling occasions had significantly higher pig-level diversity (more STs per tonsil scrape) than those from low-temperature sampling occasions (two-tailed P = 0.0296 [t test]), whereas no significant difference was found between samples from 5-week-old and 20-week-old pigs (two-tailed P = 0.401 [paired t test]). To compare the pig-level diversity of S. suis between Chinese and UK S. suis-positive pigs, a filtering process was undertaken to maximize the direct comparability of the Chinese and UK collections, reducing the former collection from 223 to 209 isolates and the latter from 127 to 125 isolates. The result showed that the pig-level diversity of S. suis isolates was not significantly different for the Chinese and UK collections (P = 1.000 [rank sum test]) (Table 3).
Factors associated with HDAS prevalence, positive virulence genotype prevalence, and the diversity of S. suis at population level. (i) High-temperature subgroup versus the low-temperature subgroup.
In summary, higher temperatures were associated with an increased proportion of human disease-associated serotypes (HDAS), which reached statistical significance in the UK subgroups, but not in the Chinese subgroups. HDAS represented 43.45% (73/168) of the Chinese high-temperature subgroup (with all eight HDAS represented), while 32.73% (18/55) of the Chinese low-temperature subgroup isolates were HDAS (serotypes 2, 5, 16, 21, and 31; P = 0.213 [chi-square]). The UK high-temperature subgroup had 28.86% (28/97) HDAS isolates (serotype 4, 5, 16, 21, and 31), while the low-temperature subgroup had 6.67% (2/30) HDAS isolates (serotypes 4 and 16) (P = 0.024 [chi-square]).
Higher temperatures were also associated with increased prevalence of PVGT (i.e., isolates carrying at least one virulence-associated gene), reaching statistical significance for the Chinese high-temperature subgroup, at 25.00% (42/168) (PVGT), compared to that of the Chinese low-temperature subgroup (10.91% [6/55]; P = 0.044, chi-square). For the UK collection, the high- and the low-temperature subgroups included 14/97 isolates and 4/30 PVGT isolates, respectively, with no significant difference (P = 0.882 [chi-square]) (Table 3). The diversity indices for the high- and low-temperature subgroups based on MLST analyses were not significantly different in either China or the United Kingdom (Table 2).
(ii) Five-week subgroup versus 20-week subgroup.
The prevalence of HDAS was not significantly different for the 5-week-old versus the 20-week-old pig subgroups in China or the United Kingdom. Briefly, there was a 42.76% (62/145) prevalence of HDAS in the Chinese 5-week-old subgroup and a 41.02% (32/78) prevalence of HDAS in the 20-week-old subgroup. The proportions of HDAS in the UK 5-week-old and 20-week-old pig subgroups were 28.57% (26/91) and 11.11% (4/36), respectively (China, P = 0.914; United Kingdom, P = 0.063 [chi-square]).
Similarly, there were no significant differences in the prevalence of PVGT isolates between age subgroups in either country. For the Chinese collection, 24.83% (36/145) of the isolates from the 5-week-old subgroup carried virulence gene(s), while 15.38% (12/78) of the isolates from the 20-week-old subgroup carried virulence gene(s). The UK 5-week-old subgroup had 15.38% (14/91) virulence gene-positive isolates, while the 20-week-old subgroup had 11.11% (4/36) virulence gene-positive isolates. The differences between the 5-week-old subgroups and 20-week-old subgroups were not significant (China, P = 0.143; United Kingdom, P = 0.743 [chi-square]) (Table 3). The diversity indices for the 5- and 20-week-old subgroups, based on STs, were not significantly different in China or in the United Kingdom (Table 2).
(iii) Intensive-farm subgroup versus small pig farm subgroup.
There were no significant differences in the prevalence of HDAS between the Chinese intensive pig farm subgroup 39.86% (55/138) and the small pig farm subgroup 42.35% (36/85) (P = 0.819 [chi-square]), nor were there any significant differences between these two subgroups in terms of the prevalence of PVGT isolates. In the intensive pig farm subgroup, 23.19% (32/138) of isolates carried virulence gene(s), and in the small pig farm subgroup the prevalence was 18.82% (16/85). The differences between the subgroups were not significant (P = 0.547 [chi-square]) (Table 3). Finally, significant differences were not found between the diversity indices of these two subgroups, based on the STs (Table 2).
(iv) Chinese subset versus UK subset.
HDAS were found at a significantly higher prevalence (39.71%, 83/209) in the Chinese filtered subset than in the UK filtered subset (23.20%, 29/125) (P = 0.003 [chi-square]). However, there was no significant difference in the prevalence of PVGT isolates or in the diversity index based on STs between the two filtered subsets (Tables 2 and 3).
DISCUSSION
Increased awareness of S. suis as a zoonotic pathogen has directed attention to the role of pigs as healthy carriers of S. suis, enabling transmission to humans by exposure to unprocessed pig meat or pork products and also through close contact with pigs (18). The strikingly high incidence of human cases of S. suis infection in some Asian countries, such as Vietnam, versus non-Asian countries was linked to culturally based practices, including the consumption of lightly cooked pig meat products (24), but, so far, there has been much less focus on other potential predisposing differences in prevalence or the diversity of carriage of S. suis among healthy slaughter pigs. China and the United Kingdom each have a large pig industry but with different farming systems and different climates. Unlike the United Kingdom, China has reported occasional large-scale outbreaks of human disease (20). In the present study, we provided a comparative characterization of the prevalence, population structure, and diversity of S. suis among clinically healthy pig herds entering the pig meat supply chain in Asian and non-Asian countries. Direct comparison of the Chinese and UK collections was hampered by subtle but potentially important differences in the methods used for isolate collection—notably up to six colonies were selected per swab in China, whereas only three were selected in the United Kingdom. This was addressed by applying filters to the two country level collections to result in a reduced but more comparable subset of isolates.
This 1-year longitudinal study found that S. suis is widely carried in clinically healthy pig herds of China and the United Kingdom, with no significant difference in positive isolation rate for S. suis among the 10 Chinese pig farms and the 5 UK pig farms represented here. It should be noted that the use of API Strep 20 kit as a selection step for suspected S. suis isolates prior to their confirmation by whole-genome sequencing data might have resulted in the exclusion of true S. suis isolates from the study (25), with a systematic underestimation of true prevalence across the collections. The Chinese collection had 18 different serotypes based on molecular typing, 9 of which carried one or more of the virulence genes epf, mrp, and sly. The UK collection contained 15 different serotypes, only 3 of which contained epf, mrp, and/or sly genes (Table 4). Serotype 16 was the most prevalent serotype in both countries, followed by serotypes 15 and 31 in China and serotypes 9 and 15 in the United Kingdom, respectively. Since serotypes 16 and 31 have been reported in human infections (9, 10), we used a filtered subset to directly compare the prevalence of HDAS in the Chinese and UK collections and found that the Chinese collection had more HDAS serotypes represented and also a significantly higher proportion of HDAS overall compared to the United Kingdom (China, 83/209 [39.71%] isolates, including serotypes 1 and 14, 2 and 1/2, 4, 5, 16, 24, 21, and 31; United Kingdom, 29/125 [23.20%] isolates, including serotypes 4, 5, 16, 21, and 31; P = 0.003 [chi-square]). These findings indicate that healthy pig populations in the United Kingdom and China are important reservoirs of zoonotic S. suis and also that geographic differences in the prevalence of HDAS isolates of S. suis, and therefore the risk for zoonotic transmission, may exist.
TABLE 4.
Distribution of virulence factor genotype patterns described for each serotypea
| Serotype(s) or NCL type | Country | VG pattern (epf mrp sly) | No. of isolates |
|---|---|---|---|
| Serotypes | |||
| Serotypes 1 and 14 | China | epf mrpEU sly | 2 |
| Serotypes 2 and 1/2 | China | epf mrpEU sly | 1 |
| epf mrpNA2 sly | 3 | ||
| Serotype 3 | China | − mrpNA1 − | 5 |
| Serotype 4 | China | − mrpNA1 sly | 3 |
| UK | − − − | 4 | |
| Serotype 5 | China | − − − | 6 |
| UK | − − − | 1 | |
| Serotype 6 | UK | − − − | 9 |
| Serotype 7 | UK | − mrpNA1 − | 1 |
| Serotype 8 | China | − mrpNA2 sly | 8 |
| UK | − − − | 2 | |
| Serotype 9 | China | − − − | 2 |
| UK | − − − | 13 | |
| Serotype 10 | UK | − − − | 4 |
| Serotype 11 | China | − mrpNA1 − | 2 |
| Serotype 12 | China | − mrpNA1 sly | 1 |
| − − − | 9 | ||
| Serotype 15 | China | − − − | 1 |
| − mrpNA1 − | 3 | ||
| − mrpNA1 sly | 7 | ||
| − mrpNA2 sly | 4 | ||
| epf mrpNA1 sly | 1 | ||
| epf mrpNA2 sly | 1 | ||
| UK | − mrpNA1 sly | 7 | |
| − mrpNA2 sly | 4 | ||
| Serotype 16 | China | − − − | 55 |
| UK | − − − | 20 | |
| Serotype 21 | China | − − − | 1 |
| epf mrpNA2 sly | 1 | ||
| UK | − − − | 2 | |
| Serotype 24 | China | − − − | 1 |
| Serotype 25 | China | − − − | 1 |
| UK | − − − | 8 | |
| Serotype 28 | China | − − − | 7 |
| UK | − mrpNA2 sly | 4 | |
| Serotype 29 | China | − − − | 12 |
| UK | − − − | 1 | |
| Serotype 30 | China | − − − | 5 |
| UK | − − − | 4 | |
| Serotype 31 | China | − − − | 16 |
| UK | − − − | 5 | |
| NCL types | |||
| NCL1 | China | − − − | 2 |
| NCL2 | China | − − − | 2 |
| UK | − − − | 2 | |
| NCL3 | China | − − − | 5 |
| UK | − − − | 1 | |
| NCL4 | China | − − − | 6 |
| UK | − − − | 6 | |
| NCL5 | China | − − − | 2 |
| UK | − − − | 2 | |
| NCL6 | UK | − − − | 1 |
| NCL7 | China | − − − | 6 |
| UK | − − − | 3 | |
| NCL8 | China | − − − | 14 |
| UK | − − − | 2 |
NCL, novel capsule locus; VG, virulence gene; −, none/not applicable.
A key finding of this study was the significant association between air temperature not only with a positive isolation rate at the farm level but also with an increased prevalence of HDAS isolates and of isolates carrying virulence-associated genes, a finding that sheds light on previous observations of increased levels of S. suis in pig meat in hot weather (26) and potential associations with increased incidence of human disease in during the summer (21). Observations on the prevalence of isolates bearing the virulence-associated genes sly, epf, and mrp should be tempered by the caveat that associations between the presence of these genes and virulence have only been studied for serotype 2 isolates (27). Sampling occasions on relatively hot days had significantly higher positive isolation rates than on cold days; furthermore, the proportion of isolates carrying virulence genes was significantly higher in the high-temperature subgroup in China. Meanwhile, increased temperature was associated with an increased prevalence of HDAS isolates in the UK collection. These results align with observations that S. suis infection in humans occurs more frequently during the warmer months of the year but also emphasize the potential importance of enhanced carriage and shedding of zoonotic S. suis from healthy pigs, as well as the potential contamination of pig meat, during warmer months. The biological processes underlying this observed effect of temperature on the prevalence of disease-associated S. suis are worthy of deeper investigation. The relationship is likely to be complex, since an opposite relationship between temperature and transmission of the human pathogen Streptococcus pneumoniae was recently described, with transmission and the prevalence of carriage being enhanced during cooler and drier months across the studied populations (28). That study tracked regional monthly minimum temperatures rather than local mean temperatures on the specific day of sampling, as in the present study, and did not include an analysis of the diversity of carriage at the individual host or population level. Recent comparative genomic studies of disease-associated and non-disease-associated S. suis found a significantly reduced genome size among disease-associated isolates (3). It might be hypothesized that disease-associated isolates have comparatively reduced competitive fitness in the upper respiratory tract and or environment, with survival and successful transmission of this globally prevalent bacterium being supported by higher ambient temperatures. Nevertheless, although a statistical association cannot be interpreted as causality, these findings indicate the potential for further exploration of the relationship between ambient temperature and the infection dynamics of S. suis.
The effects of the age of pigs and the farm type were also evaluated as factors affecting a positive S. suis isolation rate. No significant difference was found between intensive and smaller pig farms in China, but both types were operated in continuous flow systems. A larger and more detailed risk factor analysis might reveal underlying management effects. Although not statistically significant, markedly more S. suis isolates were obtained from 5-week-old pigs than from 20-week-old pigs in both countries (Table 2). Aligning this finding with the reported observation that clinical S. suis disease of pigs is most prevalent in the postweaning period (29), when pigs are commonly mixed and maternally derived passive antibody titers are declining, whereas S. suis-associated disease is less prevalent in older pigs approaching slaughter age, suggests that future and larger studies focused on the carriage of disease-associated strains might reveal subtle but significant age associations with positive isolation rates.
The extraordinary diversity of S. suis has been described elsewhere (3). Our findings confirmed that it is not rare for an individual pig to carry multiple S. suis isolates (strains) as determined from different multilocus sequence types, serotypes, or virulence genotypes. The higher temperature sampling occasions were associated with a statistically increased number of isolates recovered per pig, based on MLST, highlighting the likelihood that any temperature effect on the diversity of S. suis populations is active at the individual pig level and not only at the pig population level.
The isolates of S. suis from clinical healthy pigs in this study also showed a high diversity at pig population level, again based on MLST (Fig. 3). In total, a total of 163 STs were identified from the China and UK collections in this study, only 12 of which had been previously reported. In our study, the STs found among serotypes with strong potential of virulence (serotype 2 and 1/2 and serotypes 1 and 14) found in the Chinese collection were ST1, ST7, and ST11, similar to other published reports (8, 16, 30). This finding suggests that the S. suis isolates from healthy pigs are more diverse than those from diseased pigs. A similar finding was mentioned in a previous study in which 115 STs were identified among 179 S. suis isolates from throat swabs obtained from healthy pigs (31).
The low overlap of serotypes and STs from pigs sampled on the same farm at 5 and 20 weeks (Fig. 3) was unexpected and might reflect high rates of exchange and infection dynamics between sampled and nonsampled pigs and the environment, or it might be a consequence of the low sensitivity of the sampling approach taken: it was practical to pick and characterize only a small number of isolates from each tonsil swab culture. Future studies exploiting direct metagenomic sequencing methods might enable greater depth of characterization of the highly diverse populations of S. suis in healthy and diseased pig populations. Nevertheless, further analysis of the genome sequences and background data of the isolates from these new collections promises to shed further light on the genomic diversity and evolutionary characteristics of this important pathogen.
Taken together, this study highlights the high diversity and carriage prevalence of S. suis among clinically healthy pig herds in China and United Kingdom. The significant effect of temperature on the prevalence of isolation, the prevalence of human disease-associated serotypes and virulence genotypes, and diversity at the individual pig level may shed new light on geographic variations in human S. suis-associated disease.
MATERIALS AND METHODS
Sample collection and S. suis isolation. (i) Sampling and S. suis isolation in China.
Five intensive pig farms (500 to 3,000 sows, herds 1 to 5) and five traditional small pig farms (70 to 300 sows, herds 6 to 10) were sampled to study the prevalence of S. suis in different farm types. The intensive pig farms (>500 sows) followed standardized management with modern feeding systems, while the traditional small pig farms (<300 sows) used variable management and feeding methods. All 10 farms held slaughter pigs from farrowing until slaughter at around 5 to 6 months of age on a continuous flow basis. To study any effect of pig age on S. suis carriage or diversity, tonsil scrapes were collected twice from each farm (at 5 weeks of age and again at 20 weeks of age), sampling from between 23 and 27 individuals randomly selected from the same group within the herd. Finally, these 20 sampling visits were scheduled across different seasons from March 2013 to March 2014, enabling each sample set to be classified into different groups according to ambient air temperature, farm type, and pig age. Tonsil scrape material was transferred to swabs and then cultured on Columbia sheep blood agar at 37°C for 24 h aerobically. Six colonies with alpha-hemolytic activity and compatible colony morphology were then subcultured. Isolates were biochemically profiled using an API 20 Strep kit (bioMérieux); those with positive results were stored as S. suis.
(ii) Sampling and S. suis isolation in the United Kingdom.
Five intensive farms were sampled, each holding pigs from weaning at 4 weeks of age until slaughter at 20 to 25 weeks of age on an all-in all-out basis. In accordance with the common sampling protocol applied in China, swabs from tonsil scrapes were collected at 5 and 20 weeks of age from 25 randomly selected pigs from the same group within the herd. Swabs were cultured on Columbia sheep blood agar under aerobic conditions at 37°C for 24 h. A maximum of three alpha-hemolytic colonies per plate, with compatible S. suis morphologies, were subcultured and profiled using an API 20 Strep kit; those with positive results were stored as S. suis.
(iii) Subset for direct comparison of Chinese and UK isolates.
In order to directly compare the prevalence and diversity of S. suis between the United Kingdom and China, a subset of each collection was generated by filtering according to the following rules. (i) Since six Chinese colonies were picked per swab for identification compared to three in the United Kingdom, if more than three S. suis isolates were obtained from a single swab, only the first three isolates were kept to limit the S. suis isolates obtained from a swab from either country to not more than three. (ii) For this maximum of three isolates selected per pig, if any of these isolates shared the same API profile, then only one representative of a given API profile per pig was kept. The filtered Chinese subset included 209 isolates, whereas the UK subset included 125 isolates.
Data grouping.
The sampling points under different conditions were grouped into three pairs of subgroups (age, farm type, and temperature subgroups for both the United Kingdom and China). Based on the ages of the sampled pigs, the 20 sampling points of China and the 10 sampling points of the United Kingdom were divided into two pairs of age subgroups: the 5-week-old subgroup and the 20-week-old subgroup, respectively. The farm type subgroups were relevant only to the Chinese collection and included 10 sampling points of the five intensive pig farms (intensive pig farm subgroup) and 10 sampling points of small pig farms (small pig farm subgroup). As a stable and unambiguous attribution, air temperature was used in this study to represent the seasonal variation of the S. suis-positive isolation rate or diversity. The average value of the daily maximum and the minimum temperature was used to represent the air temperature of the sampling day. For the three-way ANOVA, the different temperature values of the Chinese and the UK sampling occasions were divided into binary groups by the mean values, respectively (China, 20.8°C; United Kingdom, 11.9°C). The high-temperature subgroups included sampling points where the temperature was above the mean value, and the low-temperature subgroups represented sampling points that were equal to or below the mean value. The binary temperature grouping strategy was also used when evaluating the diversity difference.
Sequencing and assembly.
For Chinese isolates, genomic DNA was prepared from isolates grown overnight at 37°C in tryptone soy broth (BD Biosciences) plus 10% bovine serum, using bacterial DNA kits (Omega Bio-Tek). Genomic DNA was prepared from UK isolates grown overnight at 37°C in Todd-Hewitt broth plus 0.2% yeast (Oxoid, Ltd.) using a MasterPure Gram-positive DNA isolation kit (Epicentre). All genomic DNA samples were qualified with an optical density at 260 nm (OD260)/OD280 ratio between 1.8 and 2 (NanoDrop; Thermo Scientific). Genomic DNA (typically 500 ng) was used to prepare multiplexed libraries (32, 33) for sequencing on Illumina HiSeq 2000 instruments operated according to the manufacturer's instructions with 100 cycle paired-end runs.
For the S. suis genomes of both countries, a Perl program (Fastq_screen [http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen]) was used to map the raw reads to the published genomes of S. suis. Samples with high levels of unmapped data (>50%) were confirmed by BLAST analysis against the NCBI genome database; those found to be contaminated with genomes from other species were excluded from further analysis. Genome assemblies of China and UK isolates were generated using SPAdes 3.5.0 and Velvet 1.2.08 (with Velvet Optimizer 2.2.5), respectively. Assemblies with an N50 less than 15,000 were discarded.
Molecular serotyping.
A BLAST database was built, based on a published multiple PCR method (14), including all unique cps genes for S. suis serotype identification. Cutoff values were used to determine gene presence or absence (a sequence homology of >95% with an alignment length >95% of the target gene). The cps genes of serotypes 2 and 1/2 and serotypes 1 and 14 are too similar to distinguish at the draft genomic level (14); thus, serotypes 2 and 1/2 were grouped together, as were serotypes 1 and 14. The nonserotypeable isolates were further classified into 8 different groups according to eight novel cps loci (NCL) (15). The isolates that could not be typed by both methods were recorded as “nontypeable.”
Multilocus sequence typing.
A database including all published alleles of seven housekeeping gene fragments was built to run the BLAST process (16). BLAST results yielding 100% homology were treated as the same alleles, whereas results with a sequence homology of >80% and an alignment length of >80% were considered new alleles. Isolates yielding results with a lower similarity were not assigned a sequence type (ST) and were treated as MLST-nontypeable isolates, since the corresponding housekeeping genes were considered to be absent.
Genotyping of mrp, sly, and epf gene.
The full lengths of mrp, sly, and epf genes were extracted from the draft genome sequences. Based on the three classic virulence-associated genes, the genotype of the three virulence genes, namely, the virulence genotype (VGT), of each isolate was detected. Sequences with a homology of >80% and an alignment length of >80% of the corresponding reference gene were considered alleles. Isolates in which none of the three virulence genes were detected were termed negative VGTs (NVGTs); otherwise, the isolates were termed positive VGTs (PVGTs).
Statistical analysis.
Statistical analyses were performed using SigmaPlot software (version 12.0). A P value of ≤0.05 (5) was considered significant. ANOVA, a t test, or a paired t test was used to evaluate the influence of different factors on the distribution of S. suis populations, while a chi-square test was used to evaluate the differences between the compositions of different S. suis populations. For data that failed to pass the normality test, the Mann-Whitney rank sum test, Wilcoxon signed-rank test, and ANOVA on ranks were used instead of the t test, paired t test, and ANOVA, respectively.
Analysis of diversity.
Diversity was analyzed at multiple levels; namely, individual pig level, group level, and country level. At pig level, diversity was assessed for the China and UK collections separately by comparing the number of different STs obtained from each tonsil scrape. Simpson's diversity index (Ds) values, again based on STs, were used to evaluate the diversity of the S. suis populations at the subgroup and country level. The STs grouped into a clonal complex were calculated as the same type. The MLST-nontypeable isolates with different ST genes patterns were also identified as different types. Due to differences in the isolation protocols in China and the United Kingdom, the diversity between subgroups within countries was calculated first, and then diversity at the country level was compared directly using the filtered subsets produced as described above. The Simpson's diversity indices of different S. suis subgroups were calculated using the equation defined by Hunter and Gaston (34):
| (1) |
Ds represents the probability of two independent isolates being placed into different types. N is the total number of the population. s is the total number of the types, while nj is number of the individuals belonging to the jth type. The variance for Ds (σ2) was estimated by using the following equation:
| (2) |
The confidence interval (CI) of the Ds was used to evaluated statistical significance when comparing the diversities of different populations. The difference between two Ds values without overlapped CI was treated as significant. According to the Chebyshev's theorem, for normal distributions, about 95% of the results will fall between +2 and −2 standard deviations from the mean. Thus, the proposed equation of the CI is calculated as follows:
| (3) |
Accession number(s). The genes (GenBank accession numbers) examined in this study include epf (AY341262.1), mrpEU (X64450.1), mrpNA2 (FJ685609.1), and sly (Z36907.1). The mrpNA1 reference gene was extracted from Canadian isolate 89/1591 (AAFA00000000) (35). The GenBank accession numbers of the 223 Chinese isolates are as follows: POID00000000 to POIZ00000000, POJA00000000 to POJZ00000000, POKA00000000 to POKZ00000000, POLA00000000 to POLZ00000000, POMA00000000 to POMZ00000000, PONA00000000 to PONZ00000000, POOA00000000 to POOZ00000000, POPA00000000 to POPZ00000000, and POQA00000000 to POQR00000000 (see Table S1 in the supplemental material).
The Sequence Read Archive (SRA) accession numbers of the 127 UK isolates are as follows: ERS454741, ERS454745 to ERS454747, ERS454749 to ERS454753, ERS454755 to ERS454762, ERS454764 to ERS454783, ERS454785 to ERS454800, ERS454802, ERS454803, ERS454805 to ERS454807, ERS454809, ERS454811 to ERS454817, ERS454819 to ERS454823, ERS454827 to ERS454830, ERS454832, ERS454835 to ERS454837, ERS454839 to ERS454841, ERS454843 to ERS454858, ERS454860 to ERS454865, ERS454867 to ERS454876, ERS454878 to ERS454881, ERS454882, ERS454884, ERS454887, ERS454889, ERS454890, and ERS454892 to ERS454895 (see Table S1).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the International S&T Cooperation Program of China (ISTCP grant 2013DFG32360 for R.Z.), the Biotechnology and Biological Sciences Research Council (BBSRC grant BB/L003902/1 for A.W.T.), National Key R&D Program of China (2017YFD0500201 for R.Z.), and the Hubei Province Natural Science Foundation for Innovative Research Groups (2016CFA015 for R.Z.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02590-17.
REFERENCES
- 1.Feng YJ, Zhang HM, Wu ZW, Wang SH, Cao M, Hu D, Wang CJ. 2014. Streptococcus suis infection. Virulence 5:477–497. doi: 10.4161/viru.28595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottschalk M, Xu JG, Calzas C, Segura M. 2010. Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol 5:371–391. doi: 10.2217/fmb.10.2. [DOI] [PubMed] [Google Scholar]
- 3.Weinert LA, Chaudhuri RR, Wang JH, Peters SE, Corander J, Jombart T, Baig A, Howell KJ, Vehkala M, Valimaki N, Harris D, Chieu TT, Chau NV, Campbell J, Schultsz C, Parkhill J, Bentley SD, Langford PR, Rycroft AN, Wren BW, Farrar J, Baker S, Hoa NT, Holden MTG, Tucker AW, Maskell DJ. 2015. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat Commun 6:6740. doi: 10.1038/ncomms7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill JE, Gottshalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. 2005. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs are Streptococcus orisratti. Vet Microbiol 107:63–69. doi: 10.1016/j.vetmic.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Ishida S, Tien LHT, Osawa R, Tohya M, Nomoto R, Kawamura Y, Takahashi T, Kikuchi N, Kikuchi K, Sekizaki T. 2014. Development of an appropriate PCR system for the reclassification of Streptococcus suis. J Microbiol Methods 107:66–70. doi: 10.1016/j.mimet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Baig A, Weinert LA, Peters SE, Howell KJ, Chaudhuri RR, Wang J, Holden MT, Parkhill J, Langford PR, Rycroft AN, Wren BW, Tucker AW, Maskell DJ. 2015. Whole genome investigation of a divergent clade of the pathogen Streptococcus suis. Front. Microbiol 6:1191. doi: 10.3389/fmicb.2015.01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okura M, Osaki M, Nomoto R, Arai S, Osawa R, Sekizaki T, Takamatsu D. 2016. Current taxonomical situation of Streptococcus suis. Pathogens 24:E45. doi: 10.3390/pathogens5030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang JH, Shang KX, Kashif J, Wang LP. 2015. Genetic diversity of Streptococcus suis isolated from three pig farms of China obtained by acquiring antibiotic resistance genes. J Sci Food Agr 95:1454–1460. doi: 10.1002/jsfa.6841. [DOI] [PubMed] [Google Scholar]
- 9.Huong VTL, Ha N, Huy NT, Horby P, Nghia HDT, Thiem VD, Zhu XT, Hoa NT, Hien TT, Zamora J, Schultsz C, Wertheim HFL, Hirayama K. 2014. Epidemiology, clinical manifestations, and outcomes of Streptococcus suis infection in humans. Emerg Infect Dis 20:1105–1114. doi: 10.3201/eid2011.140915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callejo R, Prieto M, Salamone F, Auger JP, Goyette-Desjardins G, Gottschalk M. 2014. Atypical Streptococcus suis in man, Argentina, 2013. Emerg Infect Dis 20:500–502. doi: 10.3201/eid2003.131148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callejo R, Zheng H, Du P, Prieto M, Xu J, Zielinski G, Auger JP, Gottschalk M. 2016. Streptococcus suis serotype 2 strains isolated in Argentina (South America) are different from those recovered in North America and present a higher risk for humans. JMM Case Rep 3:e005066. doi: 10.1099/jmmcr.0.005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. 2014. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect 3:e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatrongjit R, Kerdsin A, Gottshalk M, Takeuchi D, Hamada S, Oishi K, Akeda Y. 2015. First human case report of sepsis due to infection with Streptococcus suis serotype 31 in Thailand. BMC Infect Dis 15:392. doi: 10.1186/s12879-015-1136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okura M, Lachance C, Osaki M, Sekizaki T, Maruyama F, Nozawa T, Nakagawa I, Hamada S, Rossignol C, Gottschalk M, Takamatsu D. 2014. Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J Clin Microbiol 52:1714–1719. doi: 10.1128/JCM.03411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H, Ji SB, Liu ZJ, Lan RT, Huang Y, Bai XM, Gottschalk M, Xu JG. 2015. Eight novel capsular polysaccharide synthesis gene loci identified in nontypeable Streptococcus suis isolates. Appl Environ Microbiol 81:4111–4119. doi: 10.1128/AEM.00315-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King SJ, Leigh JA, Heath PJ, Luque I, Tarradas C, Dowson CG, Whatmore AM. 2002. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol 40:3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segura M, Fittipaldi N, Calzas C, Gottschalk M. 2017. Critical Streptococcus suis virulence factors: are they all really critical? Trends Microbiol 25:585–599. doi: 10.1016/j.tim.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Goyette-Desjardins G, Auger JP, Xu JG, Segura M, Gottschalk M. 2014. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect 3:e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang J, Wang C, Feng Y, Yang W, Song H, Chen Z, Yu H, Pan X, Zhou X, Wang H, Wu B, Wang H, Zhao H, Lin Y, Yue J, Wu Z, He X, Gao F, Khan AH, Wang J, Zhao GP, Wang Y, Wang X, Chen Z, Gao GF. 2006. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med 3:e151. doi: 10.1371/journal.pmed.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, Wang S, Liu L, Zu R, Luo L, Xiang N, Liu H, Liu X, Shu Y, Lee SS, Chuang SK, Wang Y, Xu J, Yang W. 2006. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis 12:914–920. doi: 10.3201/eid1206.051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma E, Chung PH, So T, Wong L, Choi KM, Cheung DT, Kam KM, Chuang SK, Tsang T. 2008. Streptococcus suis infection in Hong Kong: an emerging infectious disease? Epidemiol Infect 136:1691–1697. doi: 10.1017/S0950268808000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wertheim HF, Nguyen HN, Taylor W, Lien TT, Ngo HT, Nguyen TQ, Nguyen BN, Nguyen HH, Nguyen HM, Nguyen CT, Dao TT, Nguyen TV, Fox A, Farrar J, Schultsz C, Nguyen HD, Nguyen KV, Horby P. 2009. Streptococcus suis, an important cause of adult bacterial meningitis in northern Vietnam. PLoS One 4:e5973. doi: 10.1371/journal.pone.0005973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wertheim HF, Nghia HD, Taylor W, Schultsz C. 2009. Streptococcus suis: an emerging human pathogen. Clin Infect Dis 48:617–625. doi: 10.1086/596763. [DOI] [PubMed] [Google Scholar]
- 24.Nghia HD, Tu le TP, Wolbers M, Thai CQ, Hoang NV, Nga TV, Thao le TP, Phu NH, Chau TT, Sinh DX, Diep TS, Hang HT, Truong H, Campbell J, Chau NV, Chinh NT, Dung NV, Hoa NT, Spratt BG, Hien TT, Farrar J, Schultsz C. 2011. Risk factors of Streptococcus suis infection in Vietnam: a case-control study. PLoS One 6:e17604. doi: 10.1371/journal.pone.0017604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. 1991. Isolation and characterization of Streptococcus suis capsular types 9-22. J Vet Diagn Invest 3:60–65. doi: 10.1177/104063879100300113. [DOI] [PubMed] [Google Scholar]
- 26.Cheung PY, Lo KL, Cheung TT, Yeung WH, Leung PH, Kam KM. 2008. Streptococcus suis in retail markets: How prevalent is it in raw pork? Int J Food Microbiol 127:316–320. doi: 10.1016/j.ijfoodmicro.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Staats JJ, Plattner BL, Stewart GC, Changappa MM. 1999. Presence of the Streptococcus suis suilysin gene and expression of MRP and EF correlates with high virulence in Streptococcus suis type 2 isolates. Vet Microbiol 70:201–211. doi: 10.1016/S0378-1135(99)00147-9. [DOI] [PubMed] [Google Scholar]
- 28.Numminen E, Chewapreecha C, Turner C, Goldblatt D, Nosten F, Bentley SD, Turner P, Corander J. 2015. Climate induces seasonality in pneumococcal transmission. Sci Rep 5:11344. doi: 10.1038/srep11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottschalk M. 2012. Streptococcosis, p 841–853. In Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW (ed), Diseases of swine, 10th ed John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 30.Wang KC, Zhang W, Li XC, Lu CP, Chen JM, Fan WX, Huang BX. 2013. Characterization of Streptococcus suis isolates from slaughter swine. Curr Microbiol 66:344–349. doi: 10.1007/s00284-012-0275-4. [DOI] [PubMed] [Google Scholar]
- 31.Zheng H, Ji SB, Lan RT, Liu ZJ, Bai XM, Zhang W, Gottschalk M, Xu JG. 2014. Population analysis of Streptococcus suis isolates from slaughtered swine by use of minimum core genome sequence typing. J Clin Microbiol 52:3568–3672. doi: 10.1128/JCM.00536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. 2008. A large genome center's improvements to the Illumina sequencing system. Nat Methods 5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quail MA, Otto TD, Gu Y, Harris SR, Skelly TF, McQuillan JA, Swerdlow HP, Oyola SO. 2011. Optimal enzymes for amplifying sequencing libraries. Nat Methods 9:10–11. doi: 10.1038/nmeth.1814. [DOI] [PubMed] [Google Scholar]
- 34.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fittipaldi N, Fuller TE, Teel JF, Wilson TL, Wolfram TJ, Lowery DE, Gottschalk M. 2009. Serotype distribution and production of muramidase-released protein, extracellular factor, and suilysin by field strains of streptococcus suis isolated in the united states. Vet Microbiol 139:310–317. doi: 10.1016/j.vetmic.2009.06.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.