ABSTRACT
The master regulator CcpA (catabolite control protein A) manages a large and complex regulatory network that is essential for cellular physiology and metabolism in Gram-positive bacteria. Although CcpA can affect the expression of target genes by binding to a cis-acting catabolite-responsive element (cre), whether and how the expression of CcpA is regulated remain poorly explored. Here, we report a novel dual-cre motif that is employed by the CcpA in Clostridium acetobutylicum, a typical solventogenic Clostridium species, for autoregulation. Two cre sites are involved in CcpA autoregulation, and they reside in the promoter and coding regions of CcpA. In this dual-cre motif, creP, in the promoter region, positively regulates ccpA transcription, whereas creORF, in the coding region, negatively regulates this transcription, thus enabling two-way autoregulation of CcpA. Although CcpA bound creP more strongly than creORF in vitro, the in vivo assay showed that creORF-based repression dominates CcpA autoregulation during the entire fermentation. Finally, a synonymous mutation of creORF was made within the coding region, achieving an increased intracellular CcpA expression and improved cellular performance. This study provides new insights into the regulatory role of CcpA in C. acetobutylicum and, moreover, contributes a new engineering strategy for this industrial strain.
IMPORTANCE CcpA is known to be a key transcription factor in Gram-positive bacteria. However, it is still unclear whether and how the intracellular CcpA level is regulated, which may be essential for maintaining normal cell physiology and metabolism. We discovered here that CcpA employs a dual-cre motif to autoregulate, enabling dynamic control of its own expression level during the entire fermentation process. This finding answers the questions above and fills a void in our understanding of the regulatory network of CcpA. Interference in CcpA autoregulation leads to improved cellular performance, providing a new useful strategy in genetic engineering of C. acetobutylicum. Since CcpA is widespread in Gram-positive bacteria, including pathogens, this dual-cre-based CcpA autoregulation would be valuable for increasing our understanding of CcpA-based global regulation in bacteria.
KEYWORDS: CcpA, Clostridium acetobutylicum, dual-cre motif, two-way autoregulation
INTRODUCTION
Transcriptional autoregulation is an essential feature for some key regulatory proteins in bacteria, enabling precise feedback control of their expressional levels in response to extra- or intracellular signals (1–3). Autoregulation can be positive or negative, and both play important roles in genetic networks. For example, in the most-studied organism, Escherichia coli, a variety of transcription factors (TFs) that are involved in substrate utilization (2, 3), DNA replication (1), polysaccharide capsule formation (4), and two-component systems (5, 6) are capable of dynamically controlling their own transcription. Autoregulated TFs have also been discovered in several other microorganisms, including pathogens (7–9). However, the vast majority of these autoregulated TFs are pathway specific and rarely act globally.
Catabolite control protein A (CcpA) is an essential global regulator involved in mediating many important cellular processes in Gram-positive bacteria, including pathogens (10–12). CcpA targets genes by recognizing a 14- to 16-nucleotide (nt) cre (catabolite-responsive element) site in the coding or promoter regions (13). In previous work, we identified the CcpA protein in Clostridium acetobutylicum, a typical solventogenic Clostridium species that has been widely used in industrial acetone-butanol-ethanol (ABE) production (14). During the following analysis, many potential CcpA-binding cre sites were uncovered, and a cre was found inside the coding region for CcpA (15). We recently scanned the surrounding regions of CcpA more carefully, and in addition to the above-mentioned cre site inside the CcpA-coding region, a noncanonical CcpA-binding site may be present in the promoter region. If this is true, this new CcpA-binding site, combined with the cre inside the CcpA-coding region, will yield a novel dual-cre motif that enables the autoregulation of CcpA in C. acetobutylicum. In Bacillus subtilis, CcpA seems to be constitutively expressed (16); however, it is still unclear whether and how CcpA expression is regulated in other Gram-positive bacteria, although in Lactobacillus pentosus and Staphylococcus xylosus, potential single cre sites have been detected in the ccpA promoter region (17, 18). Understanding the dual-cre form employed by the C. acetobutylicum CcpA may assist in answering these regulation questions.
In this work, we performed a detailed investigation into how CcpA employs a novel dual-cre motif to achieve its autoregulation in C. acetobutylicum. The results from in vitro and in vivo experiments showed that the cre sites in the promoter and the coding regions activate and repress, respectively, CcpA expression. These two cre sites bind CcpA with different affinities, suggesting a potential leveraging of these two sites in CcpA autoregulation. Based on the understanding of dual-cre-based CcpA autoregulation in C. acetobutylicum, we modulated this autoregulation process by mutating the cre site in the coding region, releasing the self-repression effect on CcpA expression, and this novel engineering strategy led to an encouraging improved performance of C. acetobutylicum in growth and product synthesis.
RESULTS
Identification of the cre site inside the CcpA-coding region in C. acetobutylicum.
In our previous work, the pleiotropic regulatory functions of CcpA were uncovered in C. acetobutylicum, yielding a large number of genes that CcpA targets as well as a consensus motif of CcpA-binding cre sites (15). A possible cre sequence (AAGAAAAAGATTACAT) was found in the CcpA-coding region, but confirmation of its function awaits experimental verification. We name this putative cre site “creORF.” If creORF is really a CcpA-binding site, CcpA in C. acetobutylicum can regulate itself at the transcriptional level. Such an autoregulation mechanism, to our knowledge, has not been reported in CcpA, the global regulator in Gram-positive bacteria.
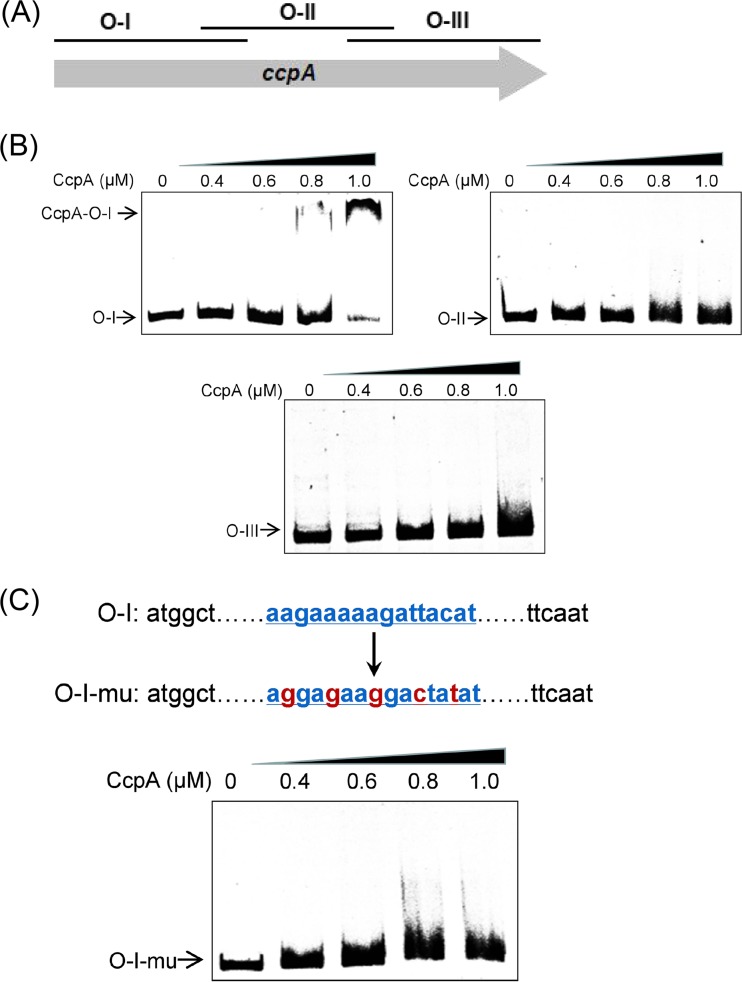
To verify this hypothesis, we first examined the binding of CcpA to creORF by using electrophoretic mobility shift assays (EMSAs). The whole sequence of the CcpA-coding region (1,005 bp) was divided into three fragments that overlapped each other by 50 bp (Fig. 1A); these fragments were used as probes in the EMSAs. As expected, a substantial DNA binding shift was observed in the case in which the first 400-bp segment (O-I) (which contains just the creORF sequence) was used (Fig. 1B), whereas no obvious DNA binding shift was detected for the other DNA probes, i.e., O-II and O-III (Fig. 1B). When creORF was mutated, this 400-bp DNA fragment was not bound by CcpA (Fig. 1C), further demonstrating the binding between CcpA and creORF. Together, these results verified that creORF is a major binding site of CcpA, although other binding sites with very low affinity to CcpA in its coding region may exist.
FIG 1.
Identification of the CcpA-binding site inside the ccpA coding region. (A) The sequence of the whole ccpA coding region was divided into three fragments, O-I (400 bp), O-II (400 bp), and O-III (305 bp). (B) EMSAs for investigating the in vitro CcpA binding to O-I, O-II, and O-III. (C) Mutation of the potential CcpA-binding site (AAGAAAAAGATTACAT) inside the O-I fragment and EMSAs for investigating the in vitro CcpA binding to fragment O-I-mu, which contained a mutated creORF site. The final concentration of Cy5-labeled DNA probe used was 0.04 pM, and 0 to 1.0 μM CcpA was used.
Identification of a noncanonical cre site in the CcpA promoter region.
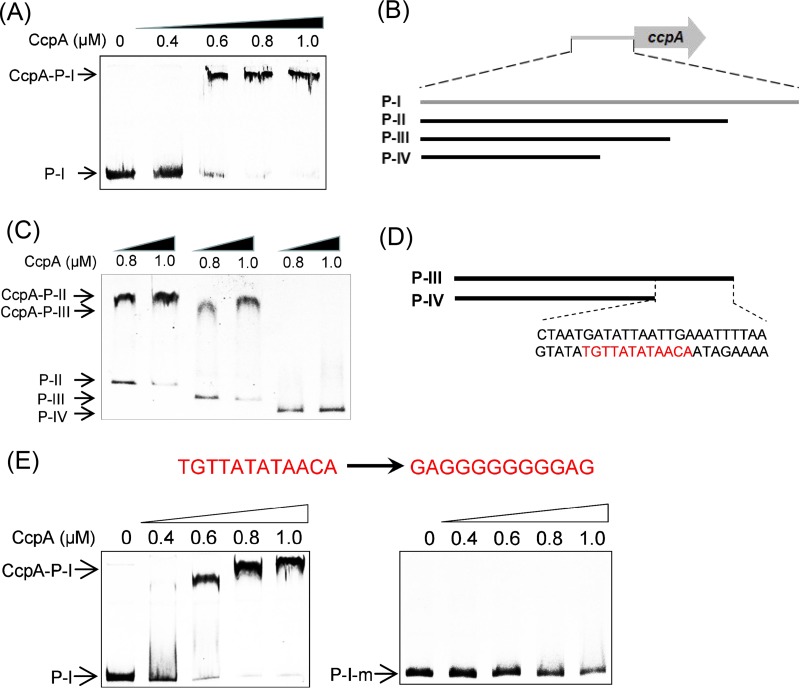
Since creORF is the cre site in the CcpA-coding region, we sought potential cre sites in the promoter region of ccpA despite negative results from previous genome screening based on the typical cre motif (15). EMSAs using the whole promoter region (P-I) sequence as a probe were performed to find these sites. An obvious DNA binding shift was observed (Fig. 2A), indicating the existence of potential noncanonical CcpA-binding sites in the promoter region. To localize these noncanonical cre sites, the 244-bp promoter region of ccpA was gradually truncated, yielding the fragments P-I (244 bp), P-II (194 bp), P-III (154 bp), and P-IV (104 bp) as probes for EMSAs (Fig. 2B). Figure 2C shows that a significantly shifted band was observed in assays with the P-II and P-III probes, while the band was almost completely eliminated when the P-IV probe was used, thereby indicating that a potential CcpA-binding site resides in the truncated fragment from the P-III region. Since this truncated fragment is only 50 bp long, we visually scanned this region and found a 12-bp palindromic sequence (TGTTATATAACA), which we named “creP” (Fig. 2D). The creP site is quite different from the typical cre site as well as the newly identified, flexible crevar architecture in C. acetobutylicum (15, 19). To further examine whether creP is a CcpA-binding site, it was mutated, which resulted in a derived P-I fragment (P-I-mu) that was used as the probe in EMSAs (Fig. 2E). As expected, the mutation of creP almost completely abolished the band shift of CcpA-P-I (Fig. 2E), thereby verifying the role of creP.
FIG 2.
Identification of the noncanonical CcpA-binding site inside the promoter region of ccpA. (A) EMSAs for investigating the in vitro CcpA binding to the promoter region (P-I). (B) Truncation of P-I. Fragments of 50, 100 and 150 bp in length were cut off from P-I, yielding the three shortened fragments P-II, P-III, and P-IV. (C) EMSAs for investigating the in vitro CcpA-binding to P-II, P-III, and P-IV. (D) Detection of a potential CcpA-binding site in the truncated fragment from P-III via visual analysis. The putative binding site (TGTTATATAACA) is highlighted in red. (E) EMSAs for investigating the in vitro CcpA binding to the probe P-I and its derivative P-I-m, in which TGTTATATAACA was mutated to GAGGGGGGGGAG. The final concentration of Cy5-labeled DNA probe used was 0.04 pM, and 0 to 1.0 μM CcpA was used.
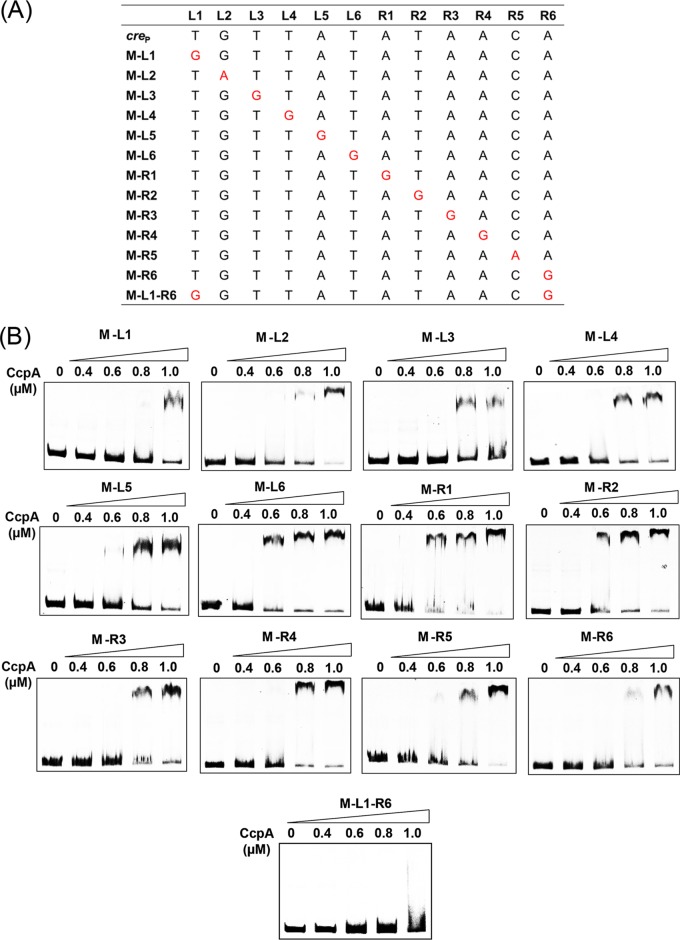
Given that creP is a noncanonical CcpA-binding site, we next sought to determine the contribution of each nucleotide in creP to CcpA binding. Each nucleotide in creP was mutated individually, and the resulting 12 derivative probes were used in EMSAs (Fig. 3A). The EMSA results showed that compared to the binding between CcpA and the original probe, P-I (Fig. 2E), the CcpA-P-I binding of all creP mutants was weakened, and while the magnitude of weakening differed among mutation sites, it was particularly obvious for the two outermost nucleotides (M-L1 and M-R6) (Fig. 3B). Simultaneous mutations at both L1 and R6 were also performed (Fig. 3A), and the resulting probe (M-L1-R6) was used to examine its binding to CcpA. As expected, this double-nucleotide mutation completely eliminated the CcpA-creP binding (Fig. 3B), further indicating the importance of the nucleotides at L1 and R6 site for creP. Therefore, it can be concluded that each nucleotide within creP contributes to CcpA-creP binding; moreover, this binding is influenced more by the outer nucleotides than by the inner nucleotides.
FIG 3.
Characterization of the noncanonical CcpA-binding site creP. (A) Single and double point mutations of creP. The mutation sites are marked in red. (B) EMSAs of CcpA binding to the probes containing the mutated creP sites. The final concentration of Cy5-labeled DNA probe used was 0.04 pM, and 0 to 1.0 μM CcpA was used.
Characterization of dual-cre-based CcpA autoregulation.
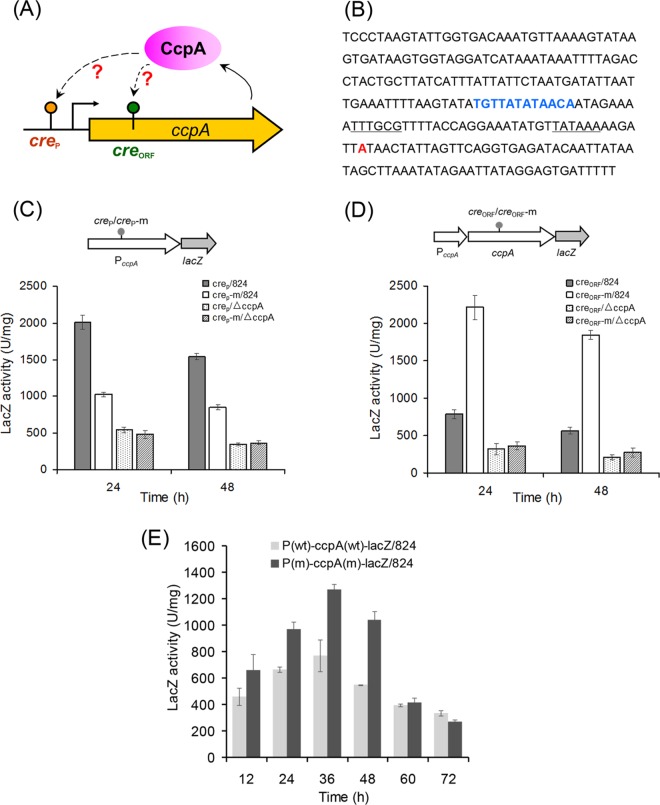
These results strongly suggested that CcpA employs a novel dual-cre (creP and creORF) motif to regulate CcpA expression (Fig. 4A). This autoregulation model was then investigated in vivo. The promoter fragment and the coding region of CcpA, containing mutated creP and creORF sites, respectively, were obtained by using overlapping PCR. These two mutated DNA fragments (creP-m and creORF-m) and the corresponding original sequences (creP and creORF) were then separately linked to the reporter gene lacZ, yielding a series of expression plasmids for in vivo β-galactosidase activity assays.
FIG 4.
Characterization of the roles of the dual cre sites in CcpA autoregulation. (A) The proposed dual-cre form for CcpA autoregulation. (B) Characteristics of the promoter region of ccpA. The transcriptional start site A is highlighted in red. The −10 and −35 regions are underlined. The CcpA-binding site creP is highlighted in blue. (C) Role of the creP site in controlling CcpA expression. creP, original promoter sequence of ccpA; creP-m, derived promoter sequence of ccpA that was mutated in the creP site; 824, C. acetobutylicum wild-type strain; ΔccpA, C. acetobutylicum strain with ccpA inactivation. lacZ was used here as the reporter gene, and the gray circle represents the original and mutated CcpA-binding sites, creP and creP-m. PccpA, promoter of the ccpA gene. (D) Role of the creORF site in controlling CcpA expression. creORF, ccpA coding region; creORF-m, derived ccpA coding region that was mutated in the creORF site. The gray circle represents the original and mutated CcpA-binding sites, creORF and creORF-m. The ccpA and lacZ genes were coexpressed to form a fusion protein. (E) Comparison of the influences of the creP and creORF sites on CcpA expression. P(wt) and P(m), wild-type and creP mutant promoter region sequences of ccpA, respectively. ccpA(wt) and ccpA(m), wild-type and creORF mutant ccpA genes, respectively. All data from the in vivo assay represent averages from two independent samples.
A 5′ rapid amplification of cDNA ends (5′-RACE) experiment showed that creP is located upstream of the −35 region of the ccpA promoter (Fig. 4B). The mutation of creP (by replacing TGTTATATAACA with GGGGATATAACA) resulted in an approximately 50% decrease in LacZ activity from that of the original construct in wild-type C. acetobutylicum, whereas these types of mutations did not significantly change LacZ activity in ccpA-inactivated C. acetobutylicum (Fig. 4C). This result further verified CcpA-creP binding activity in vivo and also suggested that the binding of CcpA to creP can promote the expression of CcpA. A following question is whether the CcpA-creP binding has a phenotypic effect on C. acetobutylicum. Thus, the plasmid-carried ccpA gene, under the control of its native or creP-mutated (by replacing TGTTATATAACA with GGGGATATAACA) promoter, was introduced into the above-described ccpA-inactivated C. acetobutylicum mutant for expression. The resulting strains, 824m-P-ccpA and 824m-Pm-ccpA, were compared to see the phenotypic changes. As shown in Fig. S1 in the supplemental material, the mutation of the creP site caused an impaired cellular performance in fermentation using glucose as the sole carbon source. Therefore, it seems that the transcriptional autoactivation of CcpA by CcpA-creP binding plays an important role in the metabolic regulation of C. acetobutylicum.
For the creORF site, its mutation (by replacing AAGAAAAAGATTACAT with AGGAGAAGGACTATAT) yielded LacZ activity that was approximately 3-fold greater than that of the original sequence in the wild-type C. acetobutylicum (Fig. 4D); no significant change resulted from the mutation of creORF in ccpA-inactivated C. acetobutylicum (Fig. 4D). These results also validated in vivo CcpA-creORF binding, but in contrast to the case for creP, the CcpA-creORF binding represses ccpA expression (Fig. 4D).
Next, both creP and creORF were mutated to examine whether they caused changes in ccpA expression in vivo. As shown in Fig. 4E, the double mutations led to increased LacZ activities before 48 h; in contrast, LacZ activity in the mutated and the original constructs did not differ after 60 h. This result indicates that creORF-based repression plays a more dominant role than creP-based activation in CcpA autoregulation during the early and middle cultivation phases; in the late phase, these two-way autoregulations based on creP and creORF tend to balance each other.
Since CcpA can bind to both creP and creORF, a question that arises is whether the CcpA binding affinities for these two sites differ. To explore this possibility, competitive EMSAs were performed in which four DNA fragments, namely, Cy5-labeled DNA fragments that contained either creP or creORF (Cy5-Df1 and Cy5-Df2) and nonlabeled DNA fragments that contained either creP or creORF (Df1 and Df2), were used as probes. The results show that CcpA-creORF binding was almost completely abolished in the presence of 25- to 100-fold-higher concentrations of Df1 (Fig. 5A); 25- to 100-fold-higher concentrations of nonlabeled DNA fragment Df2 (the specific competitor) were used as the positive control for the competitive EMSAs (Fig. 5A). In contrast, CcpA-creP binding was less affected by 25- to 100-fold-higher concentrations of Df2 (Fig. 5B); the 25- to 100-fold-higher concentrations of nonlabeled Df1 were adopted here as the positive control in the competitive EMSAs (Fig. 5B). Together, these results suggested that CcpA has a higher binding affinity to creP than to creORF.
FIG 5.
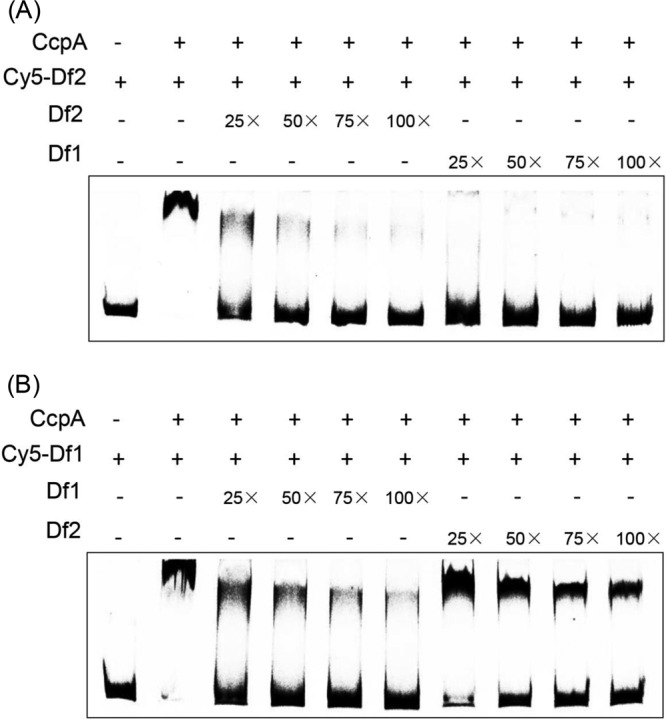
Competitive EMSAs for calculation of binding affinities of the CcpA protein with the creP and creORF sites. (A) Competitive effect of creP on CcpA-creORF binding. (B) Competitive effect of creORF on CcpA-creP binding. The initial concentration of Cy5-Df1 and Cy5-Df2 DNA was 0.04 pM, and 1.0 μM CcpA was used.
Interference with CcpA autoregulation improves cellular performance.
CcpA is able to control its own expression, but it is unknown what phenotypic changes will occur if ccpA expression is free from this autoregulation. As a preliminary test, we overexpressed the ccpA gene under the control of a widely used constitutive Pthl promoter (the promoter of gene CAC2873) in C. acetobutylicum, which is not affected by CcpA (unpublished data). Interestingly, this design led to significantly improved cellular performance using glucose as the sole carbon source, i.e., higher growth rate and glucose consumption, greater acid reassimilation, and greater solvent formation (Fig. 6). After 72 h of fermentation, the ccpA-overexpressing strain 824-ccpA produced 21.98 g/liter of total solvents (5.71 g/liter acetone, 2.88 g/liter ethanol, and 13.39 g/liter butanol), approximately 15% higher than that (19.11 g/liter) of the control strain (824-P). Next, a quantitative reverse transcription-PCR (qRT-PCR) experiment was performed to examine the difference in ccpA transcription levels between the wild-type and ccpA-overexpressing strains, and as expected, the results confirmed successful plasmid-borne ccpA overexpression that led to a significant increase in ccpA transcription (Table 1). These results demonstrate that enhanced CcpA expression can positively change the phenotype of C. acetobutylicum.
FIG 6.
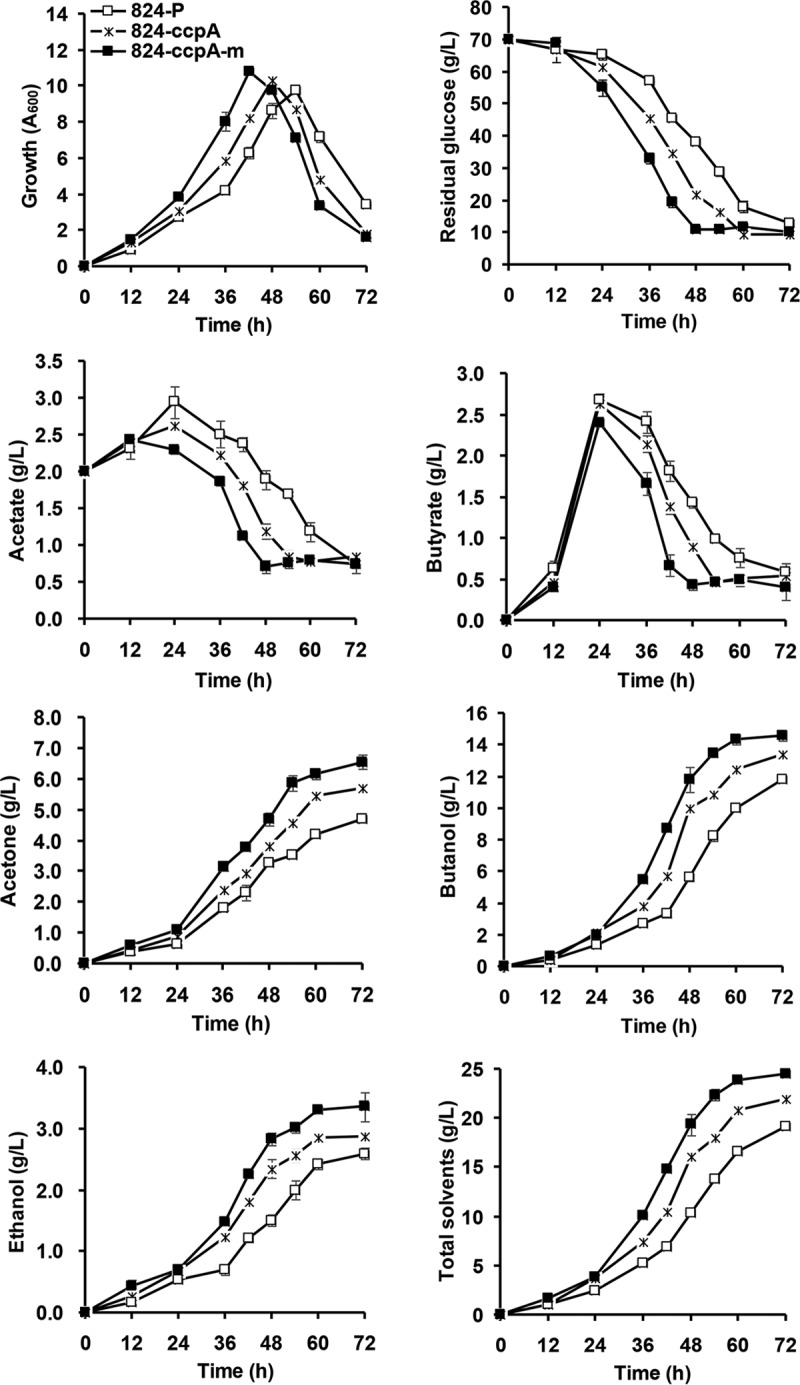
Fermentation profiles of the ccpA-overexpressing strains and the wild-type strain using glucose as the sole carbon resource. 824-P, wild-type C. acetobutylicum strain harboring the control plasmid pIMP1-Pthl. 824-ccpA, C. acetobutylicum strain harboring plasmid pIMP1-Pthl-ccpA, which carries a wild-type ccpA gene for overexpression. 824-ccpA-m, C. acetobutylicum strain harboring plasmid pIMP1-Pthl-ccpAmut, which carries the creORF mutant ccpA gene for overexpression. Total solvents represent the sum of acetone, butanol, and ethanol. The mean from three independent biological replicates and the standard deviation are shown.
TABLE 1.
Transcriptional changes of the ccpA gene after ccpA overexpression in C. acetobutylicum
| Straina | Fold change in ccpA transcription (mean ± SD) after the following fermentation time (h): |
||
|---|---|---|---|
| 24 | 48 | 72 | |
| 824-P | 1 | 1.28 ± 0.26 | 0.53 ± 0.04 |
| 824-CcpA | 3,879.7 ± 188.1 | 2,767.7 ± 398.2 | 1,029.2 ± 198.6 |
824-P, wild-type C. acetobutylicum strain harboring the control plasmid pIMP1-Pthl; 824-CcpA, the engineered C. acetobutylicum strain harboring the ccpA-overexpressing plasmid pIMP1-Pthl-ccpA (Table 2).
However, the CcpA-binding creORF site in the coding region still existed in the above-described ccpA-overexpressing construct and could somewhat repress ccpA expression. To assess this problem, a synonymous mutation was made in creORF, namely, replacing AAGAAAAAGATTACAT with AGGAGAAGGACTATAT. The ccpA gene harboring the mutated creORF sequence was then introduced into the wild-type strain for overexpression, yielding a new variant, 824-ccpA-m. In the fermentation using glucose as the sole carbon source, the growth, sugar consumption rate, and solvent synthesis of the 824-ccpA-m strain were, as expected, even better than those of 824-ccpA (the first ccpA-overexpressing strain mentioned above), producing 24.48 g/liter total solvents (6.55 g/liter acetone, 3.36 g/liter ethanol, and 14.57 g/liter butanol), which is 28% higher than the level for strain 824-P (Fig. 6). This result indicates that the synonymous creORF mutation might relieve the binding of CcpA to this site, thus eliminating CcpA self-repression and releasing more CcpA protein to exert its regulatory function.
The utilizations of xylose and a glucose-xylose mixture by the three strains described above were also compared. In the fermentation using xylose as the sole carbon source, both 824-ccpA and 824-ccpA-m behaved worse than the control strain 824-P, showing slower growth, sugar consumption, and formation of solvents (see Fig. S2 in the supplemental material). These phenotypic changes should be attributed to the enhanced carbon catabolite repression (CCR) that is caused by intracellular overexpression of ccpA. Hence, when using the mixture of glucose and xylose, compared to 824-P, both 824-ccpA and 824-ccpA-m showed slightly decreased xylose utilization, although they exhibited faster growth and glucose consumption (see Fig. S3 in the supplemental material). This is consistent with the phenotypic changes of 824-ccpA and 824-ccpA-m in total solvent production compared to 824-P, namely, a higher production rate but slightly lower final titer (Fig. S3).
DISCUSSION
As an important global regulator in Gram-positive bacteria, CcpA directly or indirectly regulates many genes. Thus, the CcpA-mediated regulatory system is always associated with many important cellular phenotypes (20–23). In light of the data here, we propose an autoregulation mechanism of CcpA based on a novel dual-cre motif in C. acetobutylicum, a typical species of solventogenic clostridia. This model includes two CcpA-binding sites: CcpA-creP binding, which occurs within the promoter region, promotes ccpA transcription, whereas CcpA-creORF binding, which occurs within the coding region, leads to transcriptional repression. Importantly, we verified that CcpA autoregulation is essential to physiological and metabolic processes of C. acetobutylicum, indicating its significance in the regulatory network.
The binding of CcpA to cre sites in gene coding regions or the regions downstream of promoters generally leads to transcriptional repression, whereas binding in the regions upstream of promoters activates transcription (24). The self-promotion of CcpA expression via the creP site should therefore be due to the relative position of creP in the promoter region, i.e., the area upstream of the −35 region. This positioning allows CcpA to bind at this location and then recruit RNA polymerase to activate and enhance the transcription of ccpA (25, 26). Here, the 12-bp creP (TGTTATATAACA) site shares low homology with the typical cre motif as well as the newly reported flexible crevar architecture in C. acetobutylicum (15, 19). This explains why the creP site was missed in the previous screening, which used the typical cre motif (15).
Although both creP and creORF were bound by CcpA, the former exhibited higher in vitro binding affinity to CcpA (Fig. 5). The higher binding affinity may establish an advantage for creP over creORF in binding to CcpA when the intracellular CcpA level is still low during the early stages of the cell life cycle, thereby enabling rapid initiation of CcpA expression as well as the whole CcpA regulatory network. However, although creP bound more strongly than creORF to CcpA, creP-based CcpA self-promotion was unexpectedly weaker in intensity than creORF-based self-repression during most of the culturing time (Fig. 4E). This result indicates that CcpA autoregulation in C. acetobutylicum is more strongly influenced by the “roadblock” effect in the coding region that results from CcpA binding to the creORF site.
Most TFs identified in prokaryotic microorganisms employ one binding site to achieve their autoregulation; even in the few autoregulation cases involving multiple binding sites, these cis elements nearly always occur in the promoter region (5, 7, 27–29). Specifically considering the global regulator CcpA, only Lactobacillus pentosus and Staphylococcus xylosus putative cre sites were predicted to be present in the promoter region of ccpA (17, 18), indicating a potential autoregulation mechanism that has yet to be experimentally verified. Thus, the dual-cre motif discovered in this study revealed the first appearance of a novel two-way autoregulation mode for CcpA in Gram-positive bacteria. Since the two cre sites (creP and creORF) in this model bound CcpA with different strength, it can be speculated that such a dual-cre model enables more flexible regulation than the single-cre form.
It should be noted that creORF is actually a dual-use codon (i.e., not only a regulatory site but also a region encoding amino acids). Such dual-use codons occur frequently in many microorganisms but are rarely involved in autoregulation. We are still unclear whether creORF-like TF-binding sites within exons have greater or special significance in autoregulation compared to typical binding sites in promoter regions, and to find an answer requires extensive exploration by studying more examples.
In the case of CcpA-dependent carbon catabolite activation (CCA) in Gram-positive bacteria, our understanding is limited to its role in activating gene transcription, which is often caused by the interaction of the complex of P-Ser-HPr/P-Ser-Crh and CcpA with RNA polymerase (30). For example, CcpA can directly or indirectly activate the transcription of several essential genes involved in central carbon catabolism (31–33) and amino acid synthesis pathways (34, 35) that are crucial for maintaining cellular viability. However, in this study, enhanced CcpA expression levels could lead to improved cell performance in fermenting glucose. This improvement may be attributed to the reinforcement of CCA by overexpression of the ccpA gene, leading to the transcriptional upregulation of some relevant genes. However, the enhanced CcpA level caused by ccpA overexpression also had negative effects, namely, strengthening of its repression of the metabolic pathways of some nonpreferable sugars (e.g., xylose here) and then inhibiting the use of these sugars. To further address this problem, identification and mutation of CcpA-binding sites surrounding the essential genes in the pathways will be a useful strategy (15). In conclusion, the results here indicate that the CcpA regulatory network in C. acetobutylicum, as well as other Gram-positive bacteria, may be optimized further.
Here, we have discovered a novel CcpA autoregulation mechanism in C. acetobutylicum, which is based on a previously unreported dual-cre motif. The two cre sites in this motif reside in the promoter and coding regions for CcpA and have two opposite effects on CcpA autoregulation. This two-way autoregulation mechanism may enable CcpA to more effectively control and modulate the downstream regulatory network and may simultaneously provide new insights into regulation patterns of global TFs. Finally, considering the phenotypic changes caused by changes in the intracellular CcpA level, modification of the CcpA autoregulation model may be valuable in future metabolic engineering applications.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The strains and plasmids used in this work are listed in Table 2. E. coli strains DH5α and ER2275 were grown in Luria-Bertani (LB) medium or on LB agar. E. coli strain Rosetta(DE3) was used as the host for protein purification. LB medium was supplemented with appropriate concentrations of antibiotics (100 μg/ml ampicillin and 100 μg/ml spectinomycin) when needed. The wild-type C. acetobutylicum strain ATCC 824 was maintained in 75% glycerol and stored at −70°C. The C. acetobutylicum ccpA-disrupted mutant was constructed previously by using the ClosTron system (14). The in vivo methylation of plasmids was performed in E. coli strain ER2275, and they were then transformed into C. acetobutylicum strains through electroporation. CGM medium (36) and P2 medium (37) were used for inoculum preparation and fermentation of C. acetobutylicum strains, respectively. Erythromycin (10 μg/ml) was added to P2 medium when needed.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Bacterial strains | ||
| C. acetobutylicum | ||
| 824 | ATCC 824, wild-type strain | ATCC |
| ΔccpA | ccpA::intron | 14 |
| 824-P | ATCC 824 carrying the pIMP1-Pthl plasmid | This study |
| 824-ccpA | ATCC 824 carrying the pIMP1-Pthl-ccpA plasmid | This study |
| 824-ccpA-m | ATCC 824 carrying the pIMP1-Pthl-ccpAmut plasmid | This study |
| 824 m-P-ccpA | ΔccpA carrying the pIMP1-PccpA-ccpA plasmid | This study |
| 824 m-Pm-ccpA | ΔccpA carrying the pIMP1-PccpAmut-ccpA plasmid | This study |
| creP/824 | ATCC 824 carrying the pIMP1-PccpA-lacZ plasmid | This study |
| creP-m/824 | ATCC 824 carrying the pIMP1-PccpAmut-lacZ plasmid | This study |
| creORF/824 | ATCC 824 carrying the pIMP1-PccpA-ccpA-lacZ plasmid | This study |
| creORF-m/824 | ATCC 824 carrying the pIMP1-PccpA-ccpAmut-lacZ plasmid | This study |
| creORF/ΔccpA | ΔccpA carrying the pIMP1-PccpA-ccpA-lacZ plasmid | This study |
| creORF-m/ΔccpA | ΔccpA carrying the pIMP1-PccpA-ccpAmut-lacZ plasmid | This study |
| E. coli | ||
| Top10 | General cloning host strain | Invitrogen |
| ER2275 | Strain used to methylate the expression vector | New England Biolabs |
| Rosetta(DE3) | Strain used for protein overexpression | Novagen |
| Plasmids | ||
| pIMP1-Pthl | Vector with thl promoter | This study |
| pIMP1-Pthl-ccpA | ccpA overexpression vector derived from pIMP1-Pthl | This study |
| pIMP1-Pthl-ccpAmut | ccpAmut overexpression vector derived from pIMP1-Pthl | This study |
| pIMP1-PccpA-ccpA | ccpA overexpression vector derived from pIMP1-PccpA | This study |
| pIMP1-PccpAmut-ccpA | ccpA overexpression vector derived from pIMP1-PccpAmut | This study |
| pIMP1-PccpA-lacZ | lacZ reporter gene driven by ccpA promoter | This study |
| pIMP1-PccpAmut-lacZ | lacZ reporter gene driven by mutant ccpA promoter | This study |
| pIMP1-PccpA-ccpA-lacZ | Fusion of a segment of the ccpA gene and lacZ reporter driven by the ccpA promoter | This study |
| pIMP1-PccpA-ccpAmut-lacZ | Fusion of a segment of the ccpAmut gene and lacZ reporter driven by the ccpA promoter | This study |
| pIMP1-PccpAmut-ccpAmut-lacZ | Fusion of a segment of the ccpAmut gene and lacZ reporter driven by the ccpAmut promoter | This study |
Plasmid construction.
The plasmids and primers used in this work are listed in Table 2 and in Table S1 in the supplemental material, respectively. For ccpA overexpression, the promoter Pthl was first PCR amplified from the plasmid pIMP1-Pthl by using primers Pthl-PstI-for/Pthl-ccpA-rev and then linked to the ccpA gene via overlapping PCR by using primers ccpA-Pthl-for/ccpA-EcoRI-rev. The resulting PCR fragment was digested with PstI/EcoRI and then inserted into plasmid pIMP1, which was digested with the same restriction enzymes, yielding plasmid pIMP1-Pthl-ccpA. Plasmid pIMP1-PccpAmut-ccpA, containing creP-m, was constructed by using primers PccpA-PstI-for/PccpAmut-rev and PccpAmut-for/ccpA-EcoRI-rev, two fragments were linked via overlapping PCR, and then it was digested with PstI/EcoRI and inserted into plasmid pIMP1.
The construction of plasmid pIMP1-PccpA-ccpA-lacZ was performed as follows. A DNA fragment containing the promoter region and part of the coding region (from nucleotide −244 to +372) of the ccpA gene was PCR amplified from the genome of C. acetobutylicum ATCC 824 by using primers PccpA-PstI-for/OccpA-lacZ-rev and then linked to the lacZ reporter gene via overlapping PCR by using primers lacZ-OccpA-for/lacZ-SmaI-rev. The resulting DNA fragment was digested with PstI/SmaI and then inserted into plasmid pIMP1, which was digested with the same restriction enzymes, yielding plasmid pIMP1-PccpA-ccpA-lacZ.
The construction of the other derivative plasmid (pIMP1-PccpAmut-ccpAmut-lacZ) was similar to the protocol described above except that the primers (PccpAmut-for/PccpAmut-rev and ccpAmut-for/ccpAmut-rev) used for overlapping PCR contained mutated creP and creORF sites.
β-Galactosidase assay.
The plasmids carrying the lacZ gene were transformed into the C. acetobutylicum wild-type strain and derivative mutants. Cells were grown anaerobically in P2 medium (containing 70 g/liter glucose) at 37°C for 24 h or 48 h, reaching an optical density at 600 nm (OD600) of 3 or 7, respectively. Pellets of 5 ml of grown cells were collected by centrifugation at 5,000 × g for 10 min at 4°C and then resuspended in 300 μl B-PER reagent (Thermo Scientific Pierce) and vortexed for 1 min. The cell lysate was heat treated at 60°C for 30 min, and the precipitate was removed by centrifugation at 12,000 × g for 30 min. The supernatant was used for LacZ (β-galactosidase) activity assay according to a protocol reported previously (38).
Protein overexpression and purification.
To purify the CcpA protein, the ccpA gene (CAC3037) of C. acetobutylicum ATCC 824 was PCR amplified and then inserted into plasmid pET-28a (Invitrogen). The resulting plasmid, pET-28a-ccpAcac, was transformed into the E. coli Rosetta(DE3) strain for protein expression. The subsequent purification procedure was the same as previously described (15).
EMSA.
The probes used for electrophoretic mobility shift assay (EMSA) were generated by two-step PCR amplification. First, the unlabeled DNA fragments were PCR amplified from the C. acetobutylicum genome using specific primer pairs containing a universal sequence (5′-AGCCAGTGGCGATAAG-3′) at the 5′ terminus. Next, the labeled DNA fragments were obtained by PCR using a 5′-Cy5-labeled universal primer, 5′-AGCCAGTGGCGATAAG-3′, and then recovered by agarose gel electrophoresis as probes for EMSAs. The EMSAs were performed as previously described (15). The voltage used in EMSA was 110 V.
Real-time qRT-PCR.
The C. acetobutylicum cells were grown in P2 medium using d-glucose (70 g/liter) as the sole carbon source. Samples were harvested at three time points (24, 48, and 72 h). Total RNA was extracted with a kit (catalog number cw0581; CWbio) according to the manufacturer's instructions. Contaminating DNA was eliminated by DNase I (TaKaRa) digestion, which was verified by PCR analysis using the RNA as the template. The concentration of RNA was determined by using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). cDNA was obtained by reverse transcription using the PrimeScript RT reagent kit (TaKaRa). Real-time quantitative reverse transcription-PCRs (qRT-PCRs) were preformed in a MyiQ2 real-time PCR detection system (Bio-Rad). The reaction mixtures (20 μl) contained 1× iQ SYBR green Supermix (Bio-Rad), 0.5 μM each primer, and the diluted cDNA template (0.625 ng/μl), and the reaction conditions were as follows: an initial denaturation at 95°C for 10 min, 40 cycles of 15 s at 95°C, and a final incubation at 60°C for 1 min. The primers used for qRT-PCR are listed in Table S1. The CAC2679 gene was used as the internal control.
5′-RACE.
The transcriptional start site of the ccpA gene was determined by the 5′ rapid amplification of cDNA ends (5′-RACE) method. C. acetobutylicum was cultivated in P2 medium using d-glucose (70 g/liter) as the carbon source, and the samples were harvested at 24 and 48 h. Total RNA was isolated by using a TRIzol (Invitrogen) extraction kit. The 5′-RACE experiment was performed using a 5′-full RACE kit with TAP (catalog number 6107; TaKaRa). Primers ccpA-outer-Dw/ccpA-inner-Dw were used for PCR by the cDNA fragment of ccpA as the template. Finally, the PCR products were cloned into T-vector for sequencing.
Fermentation.
Inoculum preparation and fermentations of C. acetobutylicum were performed anaerobically in CGM and P2 media, respectively, to which erythromycin (10 μg/ml) was added when needed. The detailed manipulations were similar to those previously described (39). In brief, 150 μl frozen stock of C. acetobutylicum was transferred into 5 ml liquid CGM medium and then incubated anaerobically at 37°C. When the OD600 of grown cells reached 0.8 to 1.0, 2.5 ml of the grown cells was inoculated into 50 ml liquid P2 medium for fermentation. In the fermentations, d-glucose (70 g/liter), d-xylose (60 g/liter), or a mixture of d-glucose and d-xylose (40 g/liter glucose and 20 g/liter xylose) was used as the carbon source.
Analytical methods.
Cell growth was monitored based on the OD600 of the culture using a spectrophotometer (DU730; Beckman Coulter). For analysis of fermentation products (acetone, acetic acid, butyric acid, butanol, and ethanol) and sugars (glucose and xylose), samples were taken at appropriate time intervals and then centrifuged at 7,000 × g for 10 min at 4°C. The supernatant was then analyzed. The concentrations of fermentation products were determined by gas chromatography (7890A; Agilent, Wilmington, DE, USA) equipped with a capillary column (Alltech EC-Wax) and a flame ionization detector (Agilent). The glucose and xylose concentrations were measured with a high-pressure liquid chromatography system (model 1200; Agilent) equipped with a Sugar-PakTMI column (Waters Corp., MA, USA) and a refractive index detector (Agilent). The analyses of fermentation products and sugars were carried out as described previously (14).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 31630003, 31700030, 31570043, and 31670043), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (membership 2012213), and the Science and Technology Commission of Shanghai Municipality (grant 17JC1404800).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00114-18.
REFERENCES
- 1.Saggioro C, Olliver A, Sclavi B. 2013. Temperature-dependence of the DnaA-DNA interaction and its effect on the autoregulation of dnaA expression. Biochem J 449:333–341. doi: 10.1042/BJ20120876. [DOI] [PubMed] [Google Scholar]
- 2.Samarasinghe S, El-Robh MS, Grainger DC, Zhang W, Soultanas P, Busby SJ. 2008. Autoregulation of the Escherichia coli melR promoter: repression involves four molecules of MelR. Nucleic Acids Res 36:2667–2676. doi: 10.1093/nar/gkn119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semsey S, Krishna S, Erdossy J, Horvath P, Orosz L, Sneppen K, Adhya S. 2009. Dominant negative autoregulation limits steady-state repression levels in gene networks. J Bacteriol 191:4487–4491. doi: 10.1128/JB.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbett D, Bennett HJ, Askar H, Green J, Roberts IS. 2007. SlyA and H-NS regulate transcription of the Escherichia coli K5 capsule gene cluster, and expression of slyA in Escherichia coli is temperature-dependent, positively autoregulated, and independent of H-NS. J Biol Chem 282:33326–33335. doi: 10.1074/jbc.M703465200. [DOI] [PubMed] [Google Scholar]
- 5.Clarke MB, Sperandio V. 2005. Transcriptional autoregulation by quorum sensing Escherichia coli regulators B and C (QseBC) in enterohaemorrhagic E. coli (EHEC). Mol Microbiol 58:441–455. doi: 10.1111/j.1365-2958.2005.04819.x. [DOI] [PubMed] [Google Scholar]
- 6.Miyashiro T, Goulian M. 2008. High stimulus unmasks positive feedback in an autoregulated bacterial signaling circuit. Proc Natl Acad Sci U S A 105:17457–17462. doi: 10.1073/pnas.0807278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson J, McKenzie JL, Cursons R, Cook GM, Arcus VL. 2009. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J Mol Biol 390:353–367. doi: 10.1016/j.jmb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalo-Asensio J, Soto CY, Arbues A, Sancho J, del Carmen Menendez M, Garcia MJ, Gicquel B, Martin C. 2008. The Mycobacterium tuberculosis phoPR operon is positively autoregulated in the virulent strain H37Rv. J Bacteriol 190:7068–7078. doi: 10.1128/JB.00712-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams CL, Cotter PA. 2007. Autoregulation is essential for precise temporal and steady-state regulation by the Bordetella BvgAS phosphorelay. J Bacteriol 189:1974–1982. doi: 10.1128/JB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang C, Bongiorni C, Perego M. 2011. Glucose-dependent activation of Bacillus anthracis toxin gene expression and virulence requires the carbon catabolite protein CcpA. J Bacteriol 193:52–62. doi: 10.1128/JB.01656-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shelburne SA III, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A 105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer R, Baliga NS, Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol 187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher MA, Sprehe M, Bartholomae M, Hillen W, Brennan RG. 2011. Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators. Nucleic Acids Res 39:2931–2942. doi: 10.1093/nar/gkq1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren C, Gu Y, Hu S, Wu Y, Wang P, Yang Y, Yang C, Yang S, Jiang W. 2010. Identification and inactivation of pleiotropic regulator CcpA to eliminate glucose repression of xylose utilization in Clostridium acetobutylicum. Metab Eng 12:446–454. doi: 10.1016/j.ymben.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Ren C, Gu Y, Wu Y, Zhang W, Yang C, Yang S, Jiang W. 2012. Pleiotropic functions of catabolite control protein CcpA in Butanol-producing Clostridium acetobutylicum. BMC Genomics 13:349. doi: 10.1186/1471-2164-13-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miwa Y, Saikawa M, Fujita Y. 1994. Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology 140:2567–2575. doi: 10.1099/00221287-140-10-2567. [DOI] [PubMed] [Google Scholar]
- 17.Egeter O, Bruckner R. 1996. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol 21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- 18.Mahr K, Hillen W, Titgemeyer F. 2000. Carbon catabolite repression in Lactobacillus pentosus: analysis of the ccpA region. Appl Environ Microbiol 66:277–283. doi: 10.1128/AEM.66.1.277-283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Zhang L, Huang H, Yang C, Yang S, Gu Y, Jiang W. 2017. A flexible binding site architecture provides new insights into CcpA global regulation in Gram-positive bacteria. mBio 8:e02004-16. doi: 10.1128/mBio.02004-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varga J, Stirewalt VL, Melville SB. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J Bacteriol 186:5221–5229. doi: 10.1128/JB.186.16.5221-5229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varga JJ, Therit B, Melville SB. 2008. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the Gram-positive anaerobic pathogen Clostridium perfringens. Infect Immun 76:4944–4951. doi: 10.1128/IAI.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendez M, Huang IH, Ohtani K, Grau R, Shimizu T, Sarker MR. 2008. Carbon catabolite repression of type IV pilus-dependent gliding motility in the anaerobic pathogen Clostridium perfringens. J Bacteriol 190:48–60. doi: 10.1128/JB.01407-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol 79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 24.Marciniak BC, Pabijaniak M, de Jong A, Duhring R, Seidel G, Hillen W, Kuipers OP. 2012. High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis. BMC Genomics 13:401. doi: 10.1186/1471-2164-13-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shivers RP, Dineen SS, Sonenshein AL. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol Microbiol 62:811–822. doi: 10.1111/j.1365-2958.2006.05410.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Yang YK, Chambliss GH. 2005. Evidence that Bacillus catabolite control protein CcpA interacts with RNA polymerase to inhibit transcription. Mol Microbiol 56:155–162. doi: 10.1111/j.1365-2958.2005.04496.x. [DOI] [PubMed] [Google Scholar]
- 27.Cramer A, Eikmanns BJ. 2007. RamA, the transcriptional regulator of acetate metabolism in Corynebacterium glutamicum, is subject to negative autoregulation. J Mol Microbiol Biotechnol 12:51–59. doi: 10.1159/000096459. [DOI] [PubMed] [Google Scholar]
- 28.Bagchi G, Chauhan S, Sharma D, Tyagi JS. 2005. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology 151:4045–4053. doi: 10.1099/mic.0.28333-0. [DOI] [PubMed] [Google Scholar]
- 29.Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. 2004. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol 53:871–888. doi: 10.1111/j.1365-2958.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 30.Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem 73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 31.Tobisch S, Zuhlke D, Bernhardt J, Stulke J, Hecker M. 1999. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J Bacteriol 181:6996–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fillinger S, Boschi-Muller S, Azza S, Dervyn E, Branlant G, Aymerich S. 2000. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J Biol Chem 275:14031–14037. doi: 10.1074/jbc.275.19.14031. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig H, Rebhan N, Blencke HM, Merzbacher M, Stulke J. 2002. Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation. Mol Microbiol 45:543–553. doi: 10.1046/j.1365-2958.2002.03034.x. [DOI] [PubMed] [Google Scholar]
- 34.Ludwig H, Meinken C, Matin A, Stulke J. 2002. Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J Bacteriol 184:5174–5178. doi: 10.1128/JB.184.18.5174-5178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, Fujita Y. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol Microbiol 56:1560–1573. doi: 10.1111/j.1365-2958.2005.04635.x. [DOI] [PubMed] [Google Scholar]
- 36.Wiesenborn DP, Rudolph FB, Papoutsakis ET. 1988. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl Environ Microbiol 54:2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baer SH, Blaschek HP, Smith TL. 1987. Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl Environ Microbiol 53:2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tummala SB, Welker NE, Papoutsakis ET. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 65:3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, Yang Y, Ren C, Yang C, Yang S, Gu Y, Jiang W. 2015. Molecular modulation of pleiotropic regulator CcpA for glucose and xylose coutilization by solvent-producing Clostridium acetobutylicum. Metab Eng 28:169–179. doi: 10.1016/j.ymben.2015.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.