ABSTRACT
Fresh vegetables harbor diverse bacterial populations on their surfaces. However, information on this microbiota is limited to a few types of fresh vegetables, and little is known about how it varies with geography and host condition. Here, we analyzed bacterial communities on the floret surfaces of 66 field-grown broccoli collected from 22 farms in four farming regions of Jeju Island, South Korea, using 454 pyrosequencing of 16S rRNA amplicons, and we determined their relationships to farming region and host-associated factors. Geographic variations in bacterial community composition and diversity were observed among farming regions, which partly reflected their relative humidity and insolation. The most abundant phyla were Proteobacteria, followed by Actinobacteria, Firmicutes, and Bacteroidetes; core operational taxonomic units (OTUs) assigned to Pseudomonas, Acinetobacter, Oxalobacteraceae, Comamonadaceae, and Enterobacteriaceae contributed to the community differences. Bacterial community composition differed between immature and mature samples, with mature samples harboring higher bacterial diversity. In comparison with communities on other types of fresh vegetables and fruits, bacterial communities on broccoli florets were unique but more similar to those of ground vegetables than to those of tree fruits/vegetables. This study presents novel data on the variability of floret-associated bacterial populations of field-grown broccoli relative to environmental and host-associated factors.
IMPORTANCE Fresh vegetables harbor diverse and complex bacterial populations on their surfaces. These indigenous bacteria may play a role in human and crop health; however, the diversity and variability of bacterial communities on fresh vegetables require further study. A popular crop of leafy vegetables, broccoli, is of great agricultural and industrial importance. This study provides a detailed description of the bacterial community composition and diversity on the surfaces of broccoli florets. The variability of bacterial communities is associated with the geographic location of farming sites and is affected by host growth and health. The bacterial communities specific to broccoli were identified and showed greater similarity to those found on ground vegetables than to those found on tree fruits/vegetables. This study presents novel data on the impact of environmental and host-associated conditions on the variability of floret-associated bacterial populations present on field-grown broccoli.
KEYWORDS: broccoli, microbial communities, microbial ecology, phyllosphere-inhabiting microbes
INTRODUCTION
Fresh vegetables harbor bacterial populations on produce surfaces (1). Human consumers are directly exposed to these bacteria, with potential health consequences. Fresh vegetables typically act as a vehicle for foodborne pathogens, and their consumption is linked to the occurrence of foodborne illness (2). Fresh vegetables can be a main source of microbes colonizing indoor environments (3). Exposure to these bacteria can induce allergic responses in the respiratory tract (4) and could affect commensal microbiota in the gastrointestinal tract (5, 6). Many studies of bacteria associated with fresh vegetables have focused on potential pathogens.
The surface of fresh vegetables is an environment that supports a variety of microbes (7). Culture-independent studies based on high-throughput rRNA gene sequencing data show that foliar microbiota are more complex and diverse than previously thought (8). Distinct produce type-specific bacterial populations have been identified (9), the community composition of which is influenced by farming and environmental conditions (9–13). However, the microbial ecology on preharvest fresh vegetables is still poorly understood, and these issues have been addressed with few types of broad-leafed vegetables (10, 11, 13, 14). Since bacteria residing on plant surfaces play a positive or negative role in host physiology (15), a better understanding of their ecology may help improve the safety and quality of fresh vegetables. For example, cointeractions between plant leaves and resident bacteria confer resistance to human and plant pathogens (16, 17), while exposure to resident bacteria triggers the spoilage of fresh vegetables (18). In this context, a detailed characterization of the bacterial communities within the foliar microbiota of fresh vegetables can make an important contribution to the field of phyllosphere microbiology.
Since fresh leafy vegetables contain health-promoting phytochemicals (19), their consumption has increased in recent years. The commercial market for fresh vegetables is faced with the challenges posed by crop diseases (20, 21) and foodborne illness (22, 23). Therefore, economically important vegetables, particularly leafy greens, are important subjects for research in the field of phyllosphere microbiology (8). Broccoli, a main crop of the Brassicaceae family, is of great agricultural and industrial importance. For example, the annual value of broccoli production in the United States is estimated at over $1 billion (24). In contrast with other leafy vegetables, the flowers of broccoli formed by plenty of short narrow leaves are usually consumed. Despite this, little is known about the diversity of bacteria on broccoli florets and how this varies relative to environmental and host-associated conditions.
Jeju Island in South Korea has fertile volcanic soil that is beneficial for agricultural productivity. The island accounts for 84.9% of the broccoli-producing area and 79.9% of the market share of broccoli in South Korea (25). Broccoli farming in Jeju Island is usually conducted in the same season every year (26), which excludes the influence of seasonality on the development of bacterial communities. Here, we describe the composition and diversity of floret-associated bacterial communities on field-grown broccoli and assess community variations with respect to farming region, host growth, and meteorological conditions. We collected 66 broccoli samples from 22 commercial farms located in four different geographic regions of Jeju Island (Fig. 1a; see also Fig. S1 in the supplemental material) and analyzed 16S rRNA gene sequence data from 454 pyrosequencing. We also examined the effect of host health on the development of bacterial communities on broccoli using samples from a region in which black rot and downy mildew occurred before sample collection. The results will improve our understanding of the ecology of phyllosphere microbiota on fresh vegetables.
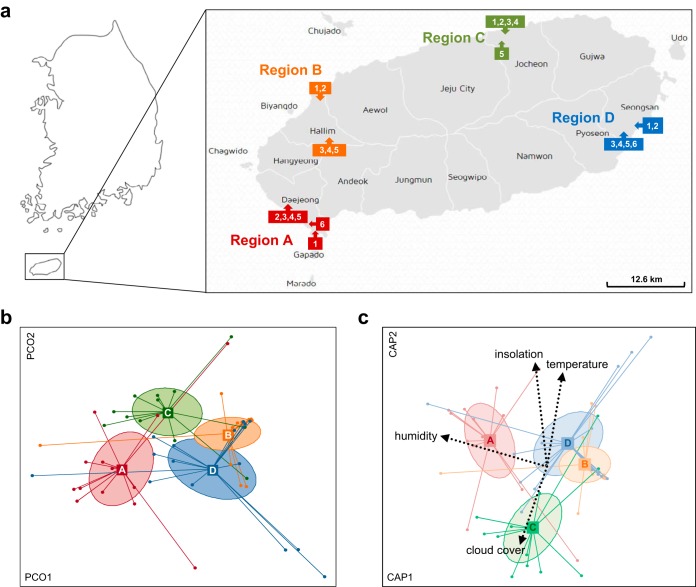
FIG 1.
Regional variations in bacterial communities and their correlation with meteorological variables. (a) The sampling sites in four regions where field-grown broccoli were sampled for this study are indicated on the map of Jeju Island (downloaded from http://www.visitjeju.net/en/index.jto). (b) Bacterial community compositions within the four regions were compared using a UniFrac distance-based principal-coordinate analysis, with 95% confidence ellipses. Statistical significance was evaluated using Adonis, with 999 permutations. (c) Meteorological variables correlated with the constrained ordination of distance-based redundancy analysis specific for farming regions. Only variables showing statistical significance are shown (insolation, P = 0.003; relative humidity, P = 0.003; cloud cover, P = 0.034; and temperature, P = 0.012). Statistical significance was evaluated using envfit, with 999 permutations. The ellipses represent 95% confidence inertia ellipses.
RESULTS
Farming region-dependent variation in bacterial communities.
Using 454 pyrosequencing of 16S rRNA gene amplicons, we obtained a mean ± standard deviation (SD) of 1,319 ± 869 high-quality sequences per sample after quality filtering and removal of chloroplast- and mitochondrion-derived sequences (Table S1). We clustered sequences into OTUs approximating the species level and compared bacterial communities using weighted UniFrac distance-based principal-coordinate analysis (PCoA). The rarefaction curves for bacterial operational taxonomic units (OTUs) in the four regions were close to reaching a plateau, except for region A (Fig. S2a), indicating that sequencing detected the majority of species in the bacterial communities on broccoli florets. We found that bacterial communities were separated significantly by region (Adonis, P = 0.001) (Fig. 1b). We determined variations in bacterial community structure by comparing the indices for species richness (Observed_OTUs) and biodiversity (PD_Whole_Tree). Region A showed the highest values for species richness and biodiversity; no significant differences were found among the other regions (Fig. S2b). Next, we divided the regions into nine subregions based on the geographical proximity of farms. The rarefaction curves for bacterial OTUs in the nine subregions were also close to reaching a plateau, except for subregions A2 and A3 (Fig. S2c). For nine subregions, bacterial communities were clustered by subregion (P = 0.001; region, r2 = 0.2762; subregion, r2 = 0.4137), but regional differences were still exhibited dominantly (Fig. S2d). Also, similar patterns in species richness and biodiversity were seen with respect to subregional comparisons, although region C showed higher values for species richness and biodiversity than regions B and D (Fig. S2e). These results suggest that bacterial community structure and composition on broccoli floret surfaces may be shaped on a large regional scale than on a short spatial scale.
To determine which environmental variables were linked to regional variation of bacterial communities, we correlated five meteorological parameters (mean values for weekly temperature, relative humidity, precipitation, insolation, and cloud cover at the sampling regions) with the ordination constrained to region-specific axes and distance-based redundancy analysis (db-RDA) (Table S2). Regional variation in bacterial communities was correlated significantly with all meteorological parameters except for precipitation (Fig. 1c). Insolation (r2 = 0.1923, P = 0.003) and relative humidity (r2 = 0.1867, P = 0.003) contributed most to regional differences in bacterial communities. Insolation specifically contributed to the separation of region C from the others, while relative humidity contributed to the separation of region A. Cloud cover, which correlated negatively with insolation, contributed to the separation of region C from the others (r2 = 0.1180, P = 0.034). Conversely, temperature (r2 = 0.1599, P = 0.012) contributed to the separation of regions B and D from the others. The results indicated that regional differences in bacterial communities may be attributed to weather conditions in farming regions, particularly insolation and relative humidity.
Identification of core members of bacterial communities.
Sixteen phyla were detected on broccoli florets, but the majority of them accounted for less than 1% total abundance. Four phyla were abundant and commonly observed across the samples (Fig. S3a). Proteobacteria (89.1% ± 11.8%) was the most abundant phylum, followed by Actinobacteria (4.2 ± 5.8%), Firmicutes (3.6% ± 5.5%), and Bacteroidetes (2.5% ± 4.6%). Among Proteobacteria, Gammaproteobacteria (67.5% ± 27.7%), Betaproteobacteria (15.4% ± 17.5%), and Alphaproteobacteria (6.0% ± 7.8%) were the most abundant classes. The relative abundances of four phyla varied among farming regions. Within the Proteobacteria phylum, the abundance of Gammaproteobacteria was negatively correlated with the abundances of Betaproteobacteria and Alphaproteobacteria. A high abundance of Gammaproteobacteria and low numbers of Betaproteobacteria and Alphaproteobacteria were observed in regions B and D, while a low abundance of Gammaproteobacteria with a high abundance of Betaproteobacteria and Alphaproteobacteria were observed in regions A and C (one-way analysis of variance [ANOVA], P < 0.05) (Fig. S3b). Where Gammaproteobacteria were rare, the Actinobacteria and Firmicutes phyla were present in large numbers (P < 0.05) (Fig. S3b). The colonization of Gammaproteobacteria may therefore be a key determinant of regional variation in bacterial communities.
To determine the core members of the bacterial communities on broccoli florets, we identified shared OTUs in the bacterial communities among the four regions. A total of 36 OTU were detected as core members for the bacterial communities on broccoli florets (Fig. 2a). Of those, five OTU (OTU543864, OTU698795, OTU778478, OTU838837, and OTU944197) were prevalent in >50% of the total broccoli samples, which were assigned to the genera Acinetobacter and Pseudomonas and unclassified genera of Oxalobacteraceae, Comamonadaceae, and Enterobacteriaceae. Further, OTUs were selected that had >1% abundance and were also prevalent across at least half of the samples within each region. At the family level, OTUs meeting these criteria were assigned to the Pseudomonadaceae, Enterobacteriaceae, Moraxellaceae, Oxalobacteraceae, and Comamonadaceae families. At the genus level, OTUs assigned to the genus Pseudomonas were the most common in broccoli samples, followed by OTUs assigned to the genus Acinetobacter and unclassified genera of the families Enterobacteriaceae, Oxalobacteraceae, and Comamonadaceae (Fig. 2b). The OTUs of the Enterobacteriaceae, Oxalobacteraceae, and Comamonadaceae families were not classified into any known genus. This is evidence of the existence of novel bacterial species specific to the phyllosphere of fresh produce, including broccoli.
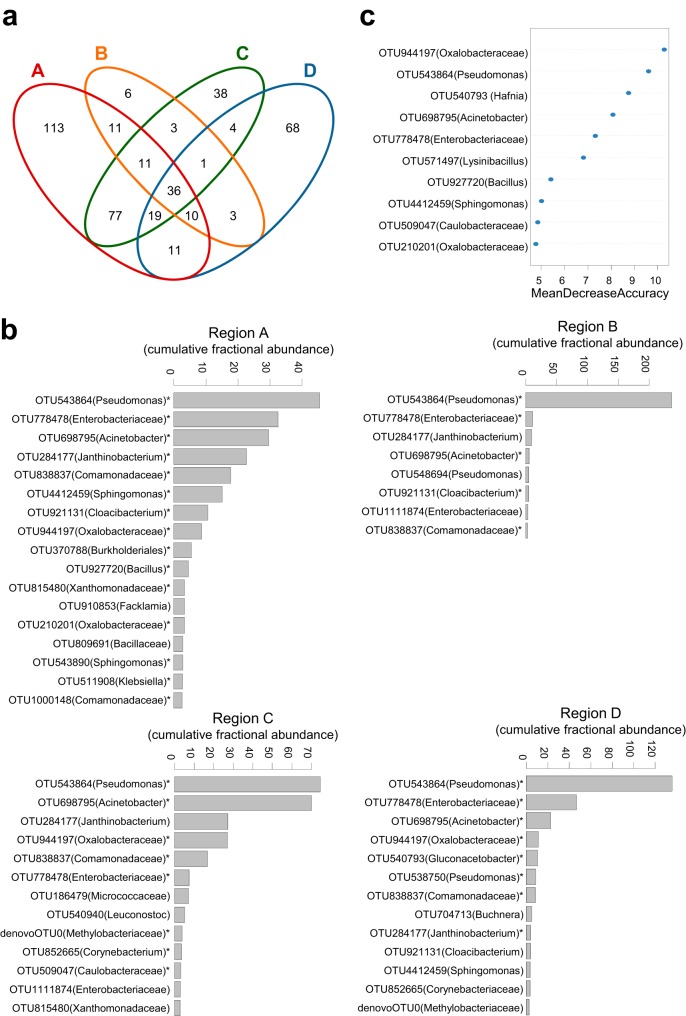
FIG 2.
Core members of the bacterial communities. (a) Venn diagram showing shared OTUs in the four farming regions (A to D). (b) Rank abundance plots showing dominant OTUs (average relative abundance, ≥1%) in the four farming regions. Asterisks indicate the OTUs found in at least ≥50% of the samples within each region. (c) OTUs that discriminate farming regions were determined using random forest analysis. A higher mean decrease in accuracy indicates that an OTU makes a greater contribution to the accuracy of the random forest classification.
To test the contribution of the core genera to differences in community composition, a random forest classifier model was created with 411 OTU, selecting the most discriminant 10 OTU for each observed source of variation. The genera of the core OTUs and the genera Hafnia, Lysinibacillus, and Bacillus made the largest contribution to the observed regional differences (Fig. 2c), indicating that the relative abundances of the core genera are a key determinant of microbial community structure that we detected in the four regions. Together, the phyllosphere microbiota of field-grown broccoli is dominated by few bacterial groups with large minor bacterial groups.
Effects of host growth on bacterial communities.
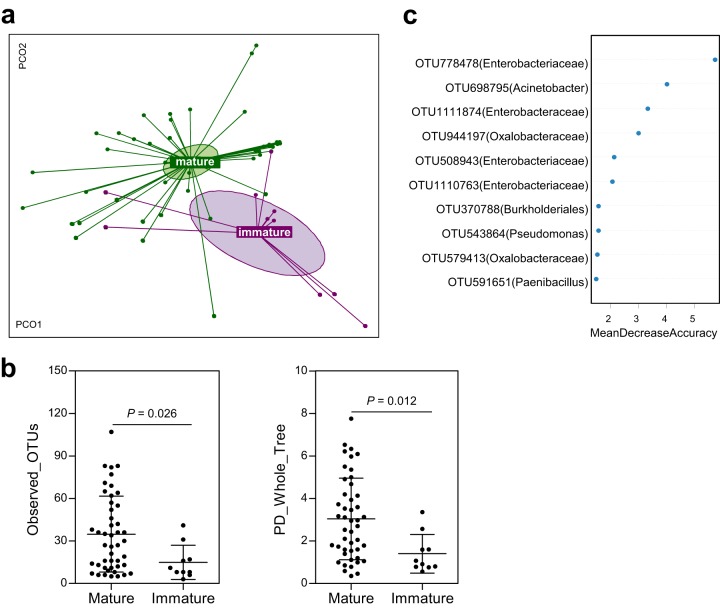
To test the effects of host growth on bacterial community composition on broccoli floret surfaces, we compared immature broccoli sampled in the growing season (8 to 9 weeks after seeding, 80 to 120 g) with mature broccoli sampled at the time of harvest (15 to 16 weeks after seeding, 250 to 400 g) from each region (Fig. S1b). Bacterial community composition differed significantly with host growth (Adonis, P = 0.006) (Fig. 3a). Species richness and biodiversity of bacterial communities were significantly lower in immature samples than in mature samples (Fig. 3b). Considering that temporal variations in sample collection exist, different weather conditions at the time of sampling for immature and mature plants might confound host growth-related differences in bacterial communities. By correlating five meteorological parameters with the ordination constrained to host growth-specific axes of db-RDA, we found that only temperature (r2 = 0.4676, P = 0.001) accounted for the difference in bacterial communities between growth stages. Plant maturation, therefore, influenced the formation of bacterial populations on broccoli florets, although it may not be the sole determinant.
FIG 3.
Changes in bacterial communities according to host growth. Bacterial community composition (a) and diversity (b) were compared according to host growth using UniFrac distance-based principal-coordinate analysis, with 95% confidence ellipses. Statistical significance was evaluated using Adonis, with 999 permutations and two-tailed unpaired Student's t test. (c) OTUs that discriminate host growth were determined using random forest analysis. A higher mean decrease in accuracy indicates that an OTU makes a greater contribution to the accuracy of the random forest classification.
Phylum-level variation was associated with host growth. A high abundance of Gammaproteobacteria with low abundance of Actinobacteria was observed in immature compared with mature broccoli plants (Student's t test, P < 0.05) (Fig. S3c), indicating that Gammaproteobacteria-mediated variation in bacterial communities may be a feature associated with broccoli host physiology. To identify the contribution of the core genera to host growth-specific differences in community composition, a random forest classifier model was conducted. Variations in the abundances of the core OTUs and the occurrence of minor Enterobacteriaceae OTUs made the largest contribution to the host growth differences (Fig. 3c), implying that different bacterial populations may be colonized on broccoli florets according to host growth stage.
Depletion of bacterial populations on unhealthy broccoli florets.
Broccoli plants grown in region C were physically damaged by black rot and downy mildew in late 2014 before sampling. Indeed, the weight of the broccoli floret in region C (approximately 250 g) was much less than that in regions A, B, and D (approximately 400 g). We investigated differences in the abundance, composition, and diversity of bacterial communities on broccoli florets on the basis of host health status, although regional variation was not excluded due to a lack of undamaged broccoli samples from region C. Differences in bacterial community compositions between healthy and unhealthy broccoli florets were determined on the basis of weighted UniFrac distance (P = 0.035) (Fig. S4a) but not unweighted UniFrac distance (P = 0.141) (Fig. S4b); also, there were no significant differences in species richness and biodiversity (Fig. S4c). Thus, this may be the result of regional variations in bacterial communities.
Next, we determined the numbers of viable bacteria on healthy and unhealthy broccoli florets using cell cultivation methods. As undamaged healthy samples from region C were not supported in this study, we additionally collected broccoli samples from local retail stores as alternative controls to minimize regional variations in the abundance of viable bacteria. The abundance of viable bacteria in unhealthy damaged broccoli (1.69 ± 0.87 log CFU · g of tissue−1) was significantly lower than that in healthy broccoli (4.43 ± 1.91 log CFU · g of tissue−1) and retail broccoli (4.73 ± 1.63 log CFU · g of tissue−1) (one-way ANOVA, P < 0.05) (Fig. 4a). Many studies of phyllosphere microbiota report that bacterial communities on the surfaces of fresh vegetables consist mostly of members of the Enterobacteriaceae family (8). In this regard, the numbers of viable Enterobacteriaceae were compared using three selective media specific for Enterobacteriaceae and coliform bacteria. The abundance of viable Enterobacteriaceae in the damaged broccoli (0.09 ± 0.37 log CFU · g of tissue−1) was significantly lower than in the healthy (1.88 ± 1.31 log CFU · g of tissue−1) and retail (2.11 ± 1.04 log CFU · g of tissue−1) broccoli (P < 0.05) (Fig. 4b). The same patterns were observed in coliform bacteria (P < 0.05) (Fig. 4c and d). The results suggest that colonization of broccoli floret surfaces may be influenced by the health state of the host plant, although the culture-based method only reflects approximately 1% to 10% of actual bacteria (27). However, no candidate bacteria that cause black rot were detected in the culture analysis. Taken together, the data suggest that disease-induced physical damage to broccoli may have a larger effect on the entire load of resident bacteria rather than on community composition.
FIG 4.
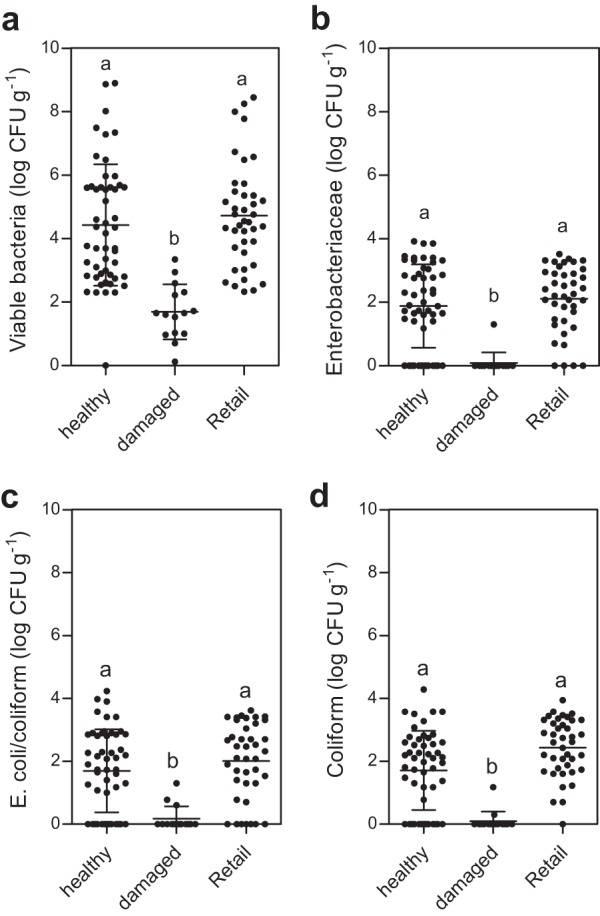
Loss of viable bacteria on the florets of damaged broccoli. Viable bacteria (a), Enterobacteriaceae (b), E. coli/coliform (c), and coliform bacteria (d) were estimated using cell culture and compared by host health. All data are mean ± SD. Statistical significance was determined by one-way ANOVA and Tukey post hoc test.
Global comparisons of bacterial communities on the surfaces of fresh produce.
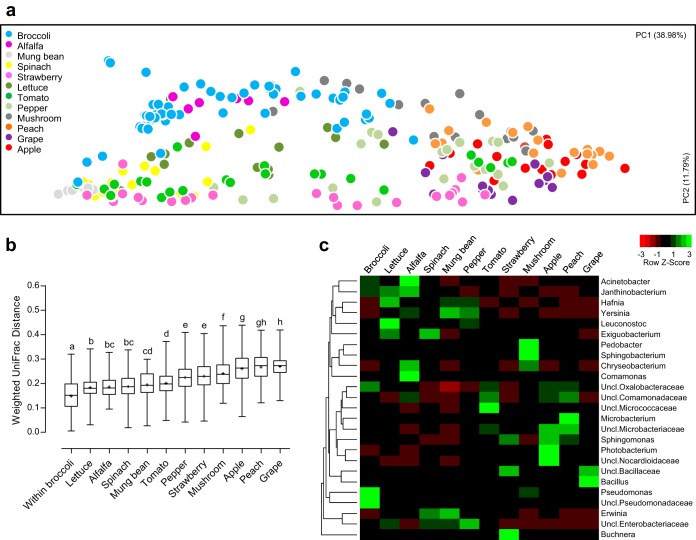
We next compared bacterial populations on broccoli with those on other fresh produce. To identify the taxa responsible for differences, we compared the bacterial communities on 11 fruits and vegetables described by Leff and Fierer (9). The bacterial community on broccoli florets clustered separately from that on other produce types (Fig. 5a); the within-group distance between bacterial communities on broccoli was significantly lower than the between-group distance between those on broccoli and each of the other produce types (Kruskal-Wallis test, P < 0.001) (Fig. 5b), indicating the uniqueness of bacterial communities on broccoli florets. Bacterial communities on broccoli were more similar to those of vegetables (lettuce, alfalfa, spinach, mung bean, and pepper) than those of fruits (strawberry, apple, peach, and grape); among vegetables, bacterial communities on broccoli were closer to those on vegetables typically grown at ground level (lettuce, alfalfa, and spinach) than to those on tree vegetables (mung bean and pepper) (Kruskal-Wallis test, P < 0.001) (Fig. 5b), implying that proximity to the ground may be an important factor affecting bacterial communities on the surface of fresh produce. With respect to vegetables, the high abundances of the families Pseudomonadaceae (Pseudomonas and unclassified genera) and Oxalobacteraceae (unclassified genus) and low abundance of Enterobacteriaceae were the defining features of broccoli-specific bacterial communities (Fig. 5c).
FIG 5.
Global comparisons of bacterial communities on broccoli with those on other types of fresh produce. The bacterial communities on broccoli were compared with those on 11 fruits and vegetables using principal-coordinate analysis (a) and average UniFrac distance comparison (b). Statistical significance was evaluated using the Kruskal-Wallis test with Dunn's post hoc test (the presence of different lowercase letters above different bars indicates statistically significant differences between groups). (c) Dominant genera (average relative abundance ≥3% in at least one type of fresh produce) were visualized on a heat map.
DISCUSSION
Understanding the ecology of the phyllosphere microbiota on leafy green vegetables is an important step toward improved food quality and safety. A few previous studies characterizing the bacterial communities residing on preharvest leafy vegetables have underscored their uniqueness and adaptation (11, 13, 28, 29). The present study provides a comprehensive description of bacterial community composition and diversity on the floret surfaces of field-grown broccoli using a combination of culture-independent methods, and it highlights their variability in relation to environmental and host-associated factors. Our findings illustrate differences in bacterial composition and abundance according to farming region and host physiology, and they identify the meteorological and spatial factors correlated with the bacterial communities on the floret surfaces of field-grown broccoli.
The results show the dominance of Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes phyla on broccoli florets, as seen on other fresh vegetables (10, 11, 30). Such bacterial diversity of the broccoli phyllosphere is greater than that predicted by culture-based analysis, which detects only a few bacterial species (31), and suggests that the majority of phyllosphere bacteria on broccoli florets are novel. Regardless of farming region and host growth, OTU-based comparison discriminated between bacterial communities on broccoli and other fresh vegetables and fruits, underscoring the interactions between the broccoli plant and bacterial populations. In contrast to fruits, members of the Enterobacteriaceae family are specific to the bacterial communities present on vegetables (9). For example, Enterobacteriaceae are more abundant on lettuce, spinach, sprouts, tomatoes, and peppers than on apples, peaches, and grapes. Enterobacteriaceae were abundant on broccoli, but much less so than Pseudomonadaceae. In addition, the families Oxalobacteraceae and Moraxellaceae were comparable (in terms of abundance) to the family Enterobacteriaceae. On alfalfa, the family Moraxellaceae was dominant. On field-grown lettuce, salad, and spinach, any of these three families may be codominant with the Enterobacteriaceae family (11, 12). This finding suggests that vegetable type-specific host-bacterium interactions may result in variations in the abundance of four families and that these bacterial families may be key taxa that discriminate the bacterial communities on different vegetables.
The four core bacterial families showed clear regional variations. Regional differences in family abundance may reflect weather conditions in each farming region. The phyllosphere is considered hostile to bacterial colonization and survival due to high variability in nutrient and water availability and solar radiation (7). Thus, bacterial populations on plant surfaces are highly influenced by weather conditions (32). Our data show that insolation (an index of UV exposure) and relative humidity (atmospheric moisture) made the greatest contribution to the observed variability in bacterial community composition. This finding is consistent with those of two previous studies of the lettuce phyllosphere, which described seasonal climate-based variation in bacterial community composition (10) and the impact of water availability on bacterial diversity (13). Since high relative humidity induces metabolic activity in phyllosphere bacteria (33); this factor may be one reason why the bacterial communities in region A were different from those in other regions. Solar radiation is also considered a critical environmental stress, especially for nonpigmented bacteria present on leafy greens (34). Truchado et al. (35) showed that long-term exposure to solar radiation results in a high abundance of Gammaproteobacteria and low abundance of Betaproteobacteria on lettuce. Bacterial communities in region D under high insolation and low cloud cover were also dominated by Pseudomonadaceae, belonging to Gammaproteobacteria, which is UV tolerant (36); low insolation and high cloud cover in region C correlated with the dominance of Oxalobacteraceae (belonging to Betaproteobacteria) accompanied by the genus Pseudomonas. Thus, UV exposure and availability of free moisture may determine the structure of phyllosphere bacterial communities on broccoli florets.
In a comparison of different types of fresh vegetables, our data support evidence that the distance from the ground plays a role in shaping bacterial communities (9). Along a vertical axis, soil and atmosphere are considered the most likely sources of foliar microbiota (7). For example, Acidobacteria (abundant in soil) have been detected on plant leaves (37), and airborne dust-derived Geodermatophilaceae have been detected on lettuce leaves (10). Experimental studies show that foliar microbiota on lettuce differs between field-grown plants and those grown in an air-conditioned laboratory (14). Spatial variations in bacterial diversity and community composition along a vertical axis were even found in the same plant (38). Thus, exposure to surroundings along a vertical axis may drive the niche differentiation of phyllosphere bacteria seeded from the environment onto broccoli and other fresh vegetables.
The present study identified host growth and health status as two host-associated factors shaping bacterial communities. However, their effects on bacterial populations were very different. While host growth influenced bacterial composition and diversity, the state of host health mostly influenced bacterial abundance. Associations between bacterial diversity and host growth were reported previously. Williams et al. (13) monitored daily variations in bacterial communities on lettuce and found that bacterial diversity increased with leaf age. Leff et al. (38) found higher bacterial diversity in leaves from older tissues than in leaves from young tissues within the same individual tree. Similarly, we found higher bacterial diversity on mature than on immature broccoli plants, although different weather conditions were also a factor. Our data support the hypothesis that host maturity exerts a driving force on microbial succession. We also found that Gammaproteobacteria predominated in communities on immature broccoli, implying that members of this class might colonize plants during early life.
It was also suggested that the health status of the host plant was significantly associated with a loss of total bacteria, although the data were not sufficiently supported with geography-matched healthy samples. Broccoli plants in region C were damaged by black rot and downy mildew during cultivation. In this regard, it is noteworthy that the lowest abundance of viable bacteria was observed on disease-damaged broccoli florets, compared with the florets of field-grown broccoli from other regions and retail broccoli. This indicates a positive correlation between host health and microbial load. Differences in community composition, but not in diversity, were observed between unhealthy and healthy broccoli florets; however, it is further evaluated with geographic controls because region-specific patterns were not ruled out. The results suggest that host health may have a larger effect on total abundance of phyllosphere microbiota; the data may imply an important role for mutual host-microbe interactions in microbial succession within the phyllosphere of fresh vegetables. More researches need to be performed with geographic controls to clarify if host health status affects the phyllosphere microbiota of broccoli florets.
This study provides a detailed description of bacterial community composition and diversity on broccoli floret surfaces, and we examined field-associated ecological factors that contribute to the establishment of bacterial populations on leafy green vegetables. This study evaluated the importance of key environmental (geographical location, weather conditions, and proximity to ground) and host-associated (host growth) factors that affect phyllosphere microbiota on fresh vegetables. The data demonstrate that complex interactions between the host, environment, and microbes play a role in defining niches important for resident bacteria. It remains unclear how the fresh vegetable-associated microbiota changes during the food processing stages of handling, packaging, transport, and storage after harvest.
MATERIALS AND METHODS
Sample collection.
Broccoli (Brassica oleracea var. italic) samples (n = 66) were collected from 22 commercial farms in four regions of Jeju Island (six farms in Daejeong [region A], five farms in Hallim [region B], five farms in Jocheon [region C], and six farms in Seongsan [region D]) during the growing and harvest seasons of 2014 to 2015. Broccoli plants were grown using standard farming methods. Immature plants grown for 8 to 9 weeks after seedlings planted in the soil (weight, 80 to 120 g) were collected at one farm in each region, while mature plants grown for 15 to 16 weeks (weight, 250 to 400 g) were collected from the remaining farms (Fig. S1b). The Agricultural Research and Extension Services of Jeju reported that broccoli plants at five farms in region C were physically damaged by black rot and downy mildew in late 2014 before sampling. A piece (∼10 g) was cut from a large flower head (floret) on a tree using a sterile knife, and a 30-g sample was gathered from three trees in a sterile plastic bag (Fig. S1a). Three samples were taken from widely separated locations in each field (Fig. S1a). Samples were delivered to the lab within 2 h and mixed with 120 ml of 0.1% buffered peptone water (Difco, Becton Dickinson, Sparks, MD, USA), and microbial cells were removed by sonication at maximum power for 10 min (Powersonic420; Hwashin Technology, Daegu, South Korea). A total of 66 sample fluids were used further for DNA extraction and viable cell counting. Daily meteorological data (temperature, relative humidity, cloud cover, precipitation, and hours of sunshine) for 1 week before and after the sampling date were provided by the Korea Meteorological Administration (http://web.kma.go.kr/eng/index.jsp), and a mean value of five parameters were used for analysis (Table S2). Retail samples (n = 40) were collected from 10 local grocery stores located on Jeju Island and were used for counting of viable cells.
DNA extraction, 16S rRNA gene PCR, and 454 pyrosequencing.
Sample fluid (40 ml) was centrifuged at 16,000 × g at 4°C for 1 min to pellet microbial cells. Total DNA was extracted using a PowerSoil DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). The V5–V6 hypervariable regions of the 16S rRNA gene sequence were amplified with primers 799f and 1115r, which do not amplify chloroplast DNA (39). For multiplexing the sample preparation, the forward primer contained 454 Life Sciences primer A, decamer multiplex identifier sequences, and the bacterial primer 799f (5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG NNNNNNNNNN AAC MGG ATT AGA TAC CCK G-3′). The reverse primer contained 454 Life Sciences primer A and the bacterial primer 1115r (5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG AGG GTT GCG CTC GTT G-3′). The PCR mixture contained template DNAs, each primer at a concentration of 0.2 μM, and 2× PCR master mix solution (i-Taq; iNtRON Biotechnology, South Korea). The PCR conditions were as follows: 94°C for 3 min, followed by 28 cycles of 94°C for 15 s, 55°C for 45 s, and 72°C for 1 min, and a final extension step at 72°C for 8 min. Negative controls were set to detect possible contamination of the commercial DNA extraction kits and reagents (40). Nine samples (A3, A6, A7, A18, C33, C43, C48, D56, and D58) that failed PCR amplification were excluded from sequencing. Four amplicon replicates were prepared per sample and then combined to yield a single sample. The amplicons were purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) and quantified using the Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Invitrogen, Carlsbad, CA, USA). The amplicon replicates were combined in an equimolar amount and sequenced using a 454 pyrosequencing GS FLX Titanium apparatus (Roche 454 Life Sciences, Branford, CT, USA).
16S rRNA amplicon analysis.
The 16S rRNA amplicon data were analyzed as described by Kim and Bae (41). Raw sequences were filtered based on quality using the QIIME software package 1.9.0 to reduce any effects of poor sequence quality and/or sequencing errors (42). The sequences were denoised using Denoiser, and reverse primer sequences were trimmed away. Chimeric sequences were further excluded by UCHIME (version 7.0). High-quality sequences were clustered at 97% sequence identity using SortMeRNA_SUMACLUST. The representative sequences of the operational taxonomic units (OTUs) were aligned against prealigned reference sequences using PyNAST, and unaligned sequences were removed. A phylogenetic tree of the representative sequences was constructed using FastTree. The OTUs identified as chloroplasts or mitochondria or unassigned to a specific phylum were excluded to reduce the noise of the OTU table (Table S3). Samples were then even-depth rarefied to 300 sequences. The sample A11 was also excluded due to low depth of sequencing. Taxonomy was assigned against the Greengenes database (2013-08 release) using the UCLUST classifier. Species richness (Observed_OTUs) and biodiversity indices (PD_Whole_Tree) were estimated. Beta diversity was determined based on weighted UniFrac distance. The 16S rRNA gene sequence data set of bacterial communities on fresh fruits and vegetables (9) were downloaded from Qiita (https://qiita.ucsd.edu/). This was combined with the data generated herein and analyzed as previously described with the OTU table rarefied to 200 sequences, according to Leff and Fierer (9).
Enumeration of viable bacteria.
The numbers of viable bacterial cells were determined by counting CFU on the following complex and selective media: tryptic soy agar (TSA; Difco, Becton Dickinson) for viable bacteria, Enterobacteriaceae Petrifilm EB (3M Microbiology Products Co., St. Paul, MN, USA) for Enterobacteriaceae, and Escherichia coli Petrifilm EC (3M Microbiology) and E. coli/coliforms Petrifilm CC (3M Microbiology) for E. coli and coliforms. The sample fluid was serially diluted 10-fold to 10−7 and incubated at 37°C for 48 h. CFU were monitored at 24-h intervals. Colonies specific for the selective media were interpreted according to the instructions in the 3M sample plate book (3M Microbiology Products Co., St. Paul, MN, USA).
Statistical analysis.
A P value of <0.05 was considered significant. An unpaired Student's t test, one-way ANOVA with a Tukey's post hoc test, and the Kruskal-Wallis with Dunn's multiple-comparison test were performed using GraphPad Prism version 5.0 for Windows (GraphPad Software, CA, USA). PCoA and db-RDA were performed based on weighted UniFrac distance using the R package vegan (43). The statistical significance for four observed variations was assessed using the function Adonis, with 999 permutations. Five meteorological parameters were correlated with the ordination of db-RDA using the function envfit, with 999 permutations. The random forest algorithm implemented in the R package “randomForest” was used to identify discriminant bacterial taxa according to farming region and host growth (44). For model accuracy, the numbers of ntree and mtry values were set at the level giving the minimum out-of-bag error rate.
Accession number(s).
The EMBL-EBI accession number for the 16S rRNA gene sequences is PRJEB20114.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (grant 2014R1A1A1003104) and the Ministry of Education (grant 2015R1D1A4A01019807).
We sincerely thank the farmers who allowed us to sample their fields.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02837-17.
REFERENCES
- 1.Esser DS, Leveau JH, Meyer KM, Wiegand K. 2015. Spatial scales of interactions among bacteria and between bacteria and the leaf surface. FEMS Microbiol Ecol 91:fiu034. doi: 10.1093/femsec/fiu034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, Hand P, Frankel G. 2010. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ Microbiol 12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- 3.Flores GE, Bates ST, Caporaso JG, Lauber CL, Leff JW, Knight R, Fierer N. 2013. Diversity, distribution and sources of bacteria in residential kitchens. Environ Microbiol 15:588–596. doi: 10.1111/1462-2920.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, Karisola P, Auvinen P, Paulin L, Makela MJ, Vartiainen E, Kosunen TU, Alenius H, Haahtela T. 2012. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci U S A 109:8334–8339. doi: 10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmody RN, Gerber GK, Luevano JM Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. 2015. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vorholt JA. 2012. Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 8.Rastogi G, Coaker GL, Leveau JHJ. 2013. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol Lett 348:1–10. doi: 10.1111/1574-6968.12225. [DOI] [PubMed] [Google Scholar]
- 9.Leff JW, Fierer N. 2013. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One 8:e59310. doi: 10.1371/journal.pone.0059310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JHJ. 2012. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J 6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dees MW, Lysoe E, Nordskog B, Brurberg MB. 2015. Bacterial communities associated with surfaces of leafy greens: shift in composition and decrease in richness over time. Appl Environ Microbiol 81:1530–1539. doi: 10.1128/AEM.03470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson CR, Randolph KC, Osborn SL, Tyler HL. 2013. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol 13:274. doi: 10.1186/1471-2180-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams TR, Moyne AL, Harris LJ, Marco ML. 2013. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS One 8:e68642. doi: 10.1371/journal.pone.0068642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams TR, Marco ML. 2014. Phyllosphere microbiota composition and microbial community transplantation on lettuce plants grown indoors. mBio 5:e01564-. doi: 10.1128/mBio.01564-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whipps JM, Hand P, Pink D, Bending GD. 2008. Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol 105:1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- 16.Balint-Kurti P, Simmons SJ, Blum JE, Ballare CL, Stapleton AE. 2010. Maize leaf epiphytic bacteria diversity patterns are genetically correlated with resistance to fungal pathogen infection. Mol Plant Microbe Interact 23:473–484. doi: 10.1094/MPMI-23-4-0473. [DOI] [PubMed] [Google Scholar]
- 17.Teplitski M, Warriner K, Bartz J, Schneider KR. 2011. Untangling metabolic and communication networks: interactions of enterics with phytobacteria and their implications in produce safety. Trends Microbiol 19:121–127. doi: 10.1016/j.tim.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Gram L, Ravn L, Rasch M, Bruhn JB, Christensen AB, Givskov M. 2002. Food spoilage–interactions between food spoilage bacteria. Int J Food Microbiol 78:79–97. doi: 10.1016/S0168-1605(02)00233-7. [DOI] [PubMed] [Google Scholar]
- 19.Jahangir M, Kim HK, Choi YH, Verpoorte R. 2009. Health-affecting compounds in Brassicaceae. Compr Rev Food Sci Food Saf 8:31–43. doi: 10.1111/j.1541-4337.2008.00065.x. [DOI] [Google Scholar]
- 20.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strange RN, Scott PR. 2005. Plant disease: a threat to global food security. Annu Rev Phytopathol 43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- 22.Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J Food Prot 67:2342–2353. doi: 10.4315/0362-028X-67.10.2342. [DOI] [PubMed] [Google Scholar]
- 23.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg Infect Dis 11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.USDA. 2015. Crop Values Annual Summary from National Agricultural Statistics Service. U.S. Department of Agriculture, Washington, DC: http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do?documentID=1050. [Google Scholar]
- 25.MAFRA. 2012. Statistics on production of greenhouse vegetable and greenhouse facilities for vegetable. Ministry of Agriculture, Food and Rural Affairs, Sejong, South Korea. [Google Scholar]
- 26.GARES. 2012. Broccoli cultivation. Agricultural Research and Extension Services, Jeju, South Korea: http://www.agri.jeju.kr/jeju/technologycenter/technology/vegetable.htm?page=8&act=view&seq=26075. [Google Scholar]
- 27.Rastogi G, Tech JJ, Coaker GL, Leveau JH. 2010. A PCR-based toolbox for the culture-independent quantification of total bacterial abundances in plant environments. J Microbiol Methods 83:127–132. doi: 10.1016/j.mimet.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Velasco G, Welbaum GE, Boyer RR, Mane SP, Ponder MA. 2011. Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J Appl Microbiol 110:1203–1214. doi: 10.1111/j.1365-2672.2011.04969.x. [DOI] [PubMed] [Google Scholar]
- 29.Yashiro E, Spear RN, McManus PS. 2011. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. J Appl Microbiol 110:1284–1296. doi: 10.1111/j.1365-2672.2011.04975.x. [DOI] [PubMed] [Google Scholar]
- 30.Margot H, Stephan R, Tasara T. 2016. Mungo bean sprout microbiome and changes associated with culture based enrichment protocols used in detection of Gram-negative foodborne pathogens. Microbiome 4:48. doi: 10.1186/s40168-016-0193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padaga M, Heard GM, Paton JE, Fleet GH. 2000. Microbial species associated with different sections of broccoli harvested from three regions in Australia. Int J Food Microbiol 60:15–24. doi: 10.1016/S0168-1605(00)00329-9. [DOI] [PubMed] [Google Scholar]
- 32.Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beattie GA. 2011. Water relations in the interaction of foliar bacterial pathogens with plants. Annu Rev Phytopathol 49:533–555. doi: 10.1146/annurev-phyto-073009-114436. [DOI] [PubMed] [Google Scholar]
- 34.Jacobs JL, Sundin GW. 2001. Effect of solar UV-B radiation on a phyllosphere bacterial community. Appl Environ Microbiol 67:5488–5496. doi: 10.1128/AEM.67.12.5488-5496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truchado P, Gil MI, Reboleiro P, Rodelas B, Allende A. 2017. Impact of solar radiation exposure on phyllosphere bacterial community of red-pigmented baby leaf lettuce. Food Microbiol 66:77–85. doi: 10.1016/j.fm.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 36.Sundin GW. 2002. Ultraviolet radiation on leaves: its influence on microbial communities and their adaptations. In Lindow SE, Hecht-Poiner EI, Elliot VJ (ed), Phyllosphere microbiology. APS Press, St; Paul, MN. [Google Scholar]
- 37.Bodenhausen N, Horton MW, Bergelson J. 2013. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8:e56329. doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leff JW, Del Tredici P, Friedman WE, Fierer N. 2015. Spatial structuring of bacterial communities within individual Ginkgo biloba trees. Environ Microbiol 17:2352–2361. doi: 10.1111/1462-2920.12695. [DOI] [PubMed] [Google Scholar]
- 39.Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. 2010. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim MS, Bae JW. 2016. Spatial disturbances in altered mucosal and luminal gut viromes of diet-induced obese mice. Environ Microbiol 18:1498–1510. doi: 10.1111/1462-2920.13182. [DOI] [PubMed] [Google Scholar]
- 42.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 44.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.