ABSTRACT
The intracellular pathogen Salmonella enterica serovar Typhimurium has emerged as a major cause of foodborne illness, representing a severe clinical and economic concern worldwide. The capacity of this pathogen to efficiently infect and survive inside the host depends on its ability to synchronize a complex network of virulence mechanisms. Therefore, the identification of new virulence determinants has become of paramount importance in the search of new targets for drug development. BolA-like proteins are widely conserved in all kingdoms of life. In Escherichia coli, this transcription factor has a critical regulatory role in several mechanisms that are tightly related to bacterial virulence. Therefore, in the present work we used the well-established infection model Galleria mellonella to evaluate the role of BolA protein in S. Typhimurium virulence. We have shown that BolA is an important player in S. Typhimurium pathogenesis. Specifically, the absence of BolA leads to a defective virulence capacity that is most likely related to the remarkable effect of this protein on S. Typhimurium evasion of the cellular response. Furthermore, it was demonstrated that BolA has a critical role in bacterial survival under harsh conditions since BolA conferred protection against acidic and oxidative stress. Hence, we provide evidence that BolA is a determining factor in the ability of Salmonella to survive and overcome host defense mechanisms, and this is an important step in progress to an understanding of the pathways underlying bacterial virulence.
IMPORTANCE BolA has been described as an important protein for survival in the late stages of bacterial growth and under harsh environmental conditions. High levels of BolA in stationary phase and under stresses have been connected with a plethora of phenotypes, strongly suggesting its important role as a master regulator. Here, we show that BolA is a determining factor in the ability of Salmonella to survive and overcome host defense mechanisms, and this is an important step in progress to an understanding of the pathways underlying bacterial virulence. This work constitutes a relevant step toward an understanding of the role of BolA protein and may have an important impact on future studies in other organisms. Therefore, this study is of utmost importance for understanding the genetic and molecular bases involved in the regulation of Salmonella virulence and may contribute to future industrial and public health care applications.
KEYWORDS: BolA, Galleria mellonella, Salmonella, acidic stress, oxidative stress, pathogenesis, virulence determinants
INTRODUCTION
Foodborne diseases represent severe clinical and economic challenges worldwide. One of the most common etiological agents of food poisoning is the pathogenic bacteria Salmonella. Hence, members of the genus Salmonella have been extensively used as model organisms to establish new concepts in pathogenesis. Particularly, nontyphoidal serovars, such as Salmonella enterica serovar Typhimurium, have become a hot research topic due to their serious morbidity and mortality rates in both developed and developing countries. This intracellular pathogen causes various clinical symptoms ranging from self-limiting enteritis to life-threatening systemic infections (1).
The effectiveness of the infection depends on the ability of S. Typhimurium to synchronize several physiological adaptations in order to overcome host defense mechanisms. Bacterial virulence factors are molecules that enable microorganisms to replicate and disseminate within a host (2). Among the wide range of virulence factors, Salmonella pathogenicity islands (SPIs) play a main role in host-pathogen interactions. The accurate coordination of a huge plethora of virulence effectors is also crucial for bacterial invasion and proliferation during infection (reviewed in reference 3). Transcriptional regulatory networks play a central role in the control of the hierarchical expression and cross talk of virulence effectors, ensuring a progressive and efficient bacterial response (4). Several studies have already highlighted the critical contribution of transcriptional regulators in bacterial adaptation and survival (5–9). These studies underline the importance of the identification of new effector proteins to understand and overcome Salmonella pathogenesis strategies. Interestingly, some of our previous work has indicated that one of the potential effectors could be the transcription factor BolA, a protein that affects different pathways directly related to virulence (10, 11).
BolA, initially characterized in Escherichia coli (12), has received increasing attention in both prokaryotic and eukaryotic organisms. Particularly, in prokaryotic organisms this protein stands out because of its pleiotropic effects. In Gram-negative bacteria, bolA expression is induced at the onset of the stationary phase or in response to several stresses, including sudden carbon starvation and heat, acidic, or oxidative shock (13). Characterized as a transcription factor, BolA has an essential role in the regulation of cell morphology, outer membrane permeability, and the transition from a planktonic to sessile lifestyle (11, 14, 15). Moreover, very recently we have shown that BolA establishes an important cross talk with the second messenger c-di-GMP in the regulation of biofilm development (16). All of these adaptation processes confer specific advantages for bacterial survival and are often associated with virulence (3).
In the present work, our main goal was to unravel the role of BolA protein in the virulence of S. Typhimurium. For this purpose, we have chosen the greater wax moth Galleria mellonella as the infection model. G. mellonella has been extensively used as a model organism for a wide range of bacterial species (reviewed in reference 17), and it has already been shown to be an appropriate host to study S. Typhimurium pathogenicity (18, 19). The G. mellonella larvae offer great benefits as hosts, including cost-effectiveness and easy handling. Additionally, this system allows the study of many stages of infection at 37°C, the human body core temperature, and possesses remarkable similarities with the innate immune response of vertebrates (20).
In the manuscript, we have uncovered the importance of BolA for the success of S. Typhimurium infection, providing new clues to the pathogenesis strategies of this organism.
RESULTS
Identification of E. coli bolA homologs in S. Typhimurium.
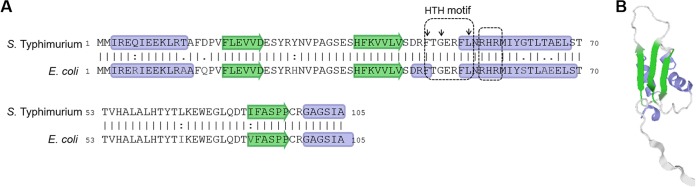
To search for BolA homologs among S. Typhimurium genomes, the E. coli protein sequence (GenBank accession no. APC50728.1) was used. A protein from Salmonella enterica serovar Typhimurium LT2 was identified (GenBank accession no. NP_459442). BolA homologs of both LT2 and SL1344 strains showed an overall identity of 99% (data not shown). The alignment of both homologs from S. Typhimurium and E. coli strains revealed high sequence conservation, with 91% identity and 95% similarity (Fig. 1). Moreover, a synteny analysis of the genomic regions in the neighborhood of bolA indicated the conservation of the neighboring genes across genera. Given such high levels of sequence conservation, it was not surprising to find that an S. Typhimurium BolA structure prediction matched the structure of the BolA protein from E. coli (PDB accession number 2DHM) as the most similar. Using this structure as the template, we obtained a three-dimensional (3D) model prediction (Fig. 1B) with a global model quality score of 0.9 in a maximum of 1. The predicted structure of S. Typhimurium BolA consists of an α1-β1-β2-α2-β3-α3 topology, with β1 and β2 as antiparallel sheets and β3 parallel to the β2 sheet, similar to the topology observed in other BolA-like proteins (21). The helix-turn-helix (HTH) signature FXGXXXL sequence, characteristic of BolA proteins (22) and typically implicated in protein-DNA interaction, is also present (Fig. 1B). Moreover, within the second helix we observed a highly conserved RHR signature sequence that is present in 41% of BolA sequences (23).
FIG 1.
In silico analysis of BolA protein from S. Typhimurium. (A) Pairwise sequence alignment of BolA from E. coli K-12 W3110 and a BolA homolog from S. Typhimurium LT2 obtained using EMBOSS Needle, version 6.6.0. The secondary structure prediction for both BolA proteins is presented; in green are the β-sheets, and in blue are the α-helixes. The small arrows above the sequence indicate the amino acids of the characteristic helix-turn-helix (HTH) signature FXGXXXL sequence. (B) Computer model of the 3D structure predicted for the S. Typhimurium LT2 BolA protein based on the crystal structure of BolA from E. coli K-12 W3110 (PDB accession number 2DHM).
BolA protects S. Typhimurium cells against acidic and oxidative stresses.
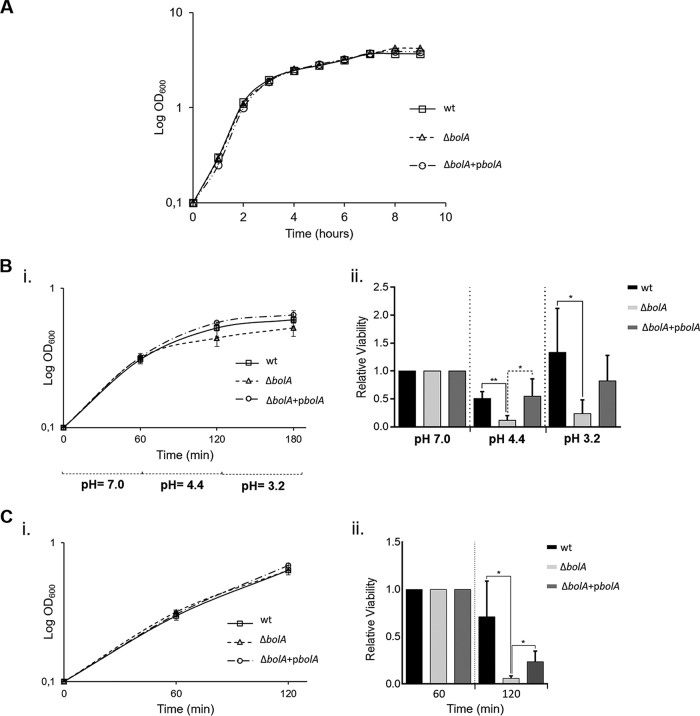
The S. Typhimurium LT2 strain presents an attenuated level of virulence due to a suboptimal translation of RpoS (σS) (24). Therefore, we have used a virulent S. Typhimurium strain (SL1344) to construct a BolA mutant strain. The deletion of BolA in E. coli was reported to slightly increase growth rates compared to the wild-type rate (10). However, in S. Typhimurium SL1344, BolA deletion did not significantly affect the growth rate relative to that of the wild-type strain, and the BolA mutant showed only a slight increase in the optical density at 600 nm (OD600) in stationary phase (Fig. 2A). Complementation of the SL1344 BolA mutant was achieved using a pRMA04 plasmid expressing BolA in trans (strain CMA821). The growth curve of the complemented strain was identical to that of the wild type (Fig. 2A). For growth under static conditions, a similar minor increase in the OD600 of the BolA mutant was registered (data not shown).
FIG 2.
The role of BolA in S. Typhimurium growth and the stress response. (A) Growth curve of S. Typhimurium SL1344 strains. Growth characteristics of the S. Typhimurium wild-type (wt), BolA mutant (ΔbolA), and complemented (ΔbolA+pbolA) strains, obtained by following the OD600 for 9 h. (B) Influence of BolA in the acid tolerance response of S. Typhimurium. Growth curves of S. Typhimurium SL1344 strains under acidic stress are shown (i). Cells were grown until they reached an optical density of 0.3 (corresponding to 60 min on the graph) in LB medium, pH 7, prior the adjustment to pH 4.4 for 1 h. The cells were then challenged by dropping the pH to 3.2 for 1 h. Viable cells were extrapolated by plating the appropriate dilutions on Luria agar plates (ii). (C) BolA impact on the S. Typhimurium oxidative response. Cultures were grown in LB medium and challenged for 1 h with 5 mM H2O2 at an optical density of 0.3 (corresponding to 60 min on the graph) (i). Viable cells were extrapolated by plating the appropriate dilutions on Luria agar plates (ii). The results were normalized and are presented relative to the number of viable cells obtained before stress induction (OD600 of 0.3). The experiments were performed in triplicate, and the results are represented as mean ± SD. *, P < 0.05; **, P < 0.01.
In E. coli, BolA is established as a general stress response transcription factor whose expression increases under imposition of diverse severe conditions, conferring protection to the cells (reviewed in reference 10). Therefore, we were interested in understanding whether S. Typhimurium BolA confers protection under acidic and oxidative stresses. For both stresses, growth curves were monitored by measuring the OD600 during the course of the experiment (Fig. 2B and C, graphs i). To evaluate the BolA role in the acid tolerance response, the bacterial cultures were first subjected to a preacidic adaptive shock stage (pH 4.4) for 1 h, followed by a pH fall (pH 3.2) during the same period (Fig. 2B). Our results revealed that although the bolA deletion strain is able to produce an acid tolerance response, it was about 5-fold more susceptible than the wild-type strain in both acidic challenges (Fig. 2B, graph ii). Similarly, under oxidative stress conditions, the absence of BolA produced a 7-fold decrease in viability compared to that of the wild-type strain (Fig. 2C, graph ii). Under both stresses, the effect obtained in the bolA deletion strain was at least partially reversed and wild-type levels were reached when a complemented strain was used, corroborating the previous results (Fig. 2B and C, graphs ii). These results indicate that S. Typhimurium BolA confers protection against acidic and oxidative stress.
The deletion of bolA decreases virulence of S. Typhimurium in a Galleria mellonella model of infection.
To evaluate the involvement of BolA in virulence, we used the preestablished insect model for Salmonella infection, G. mellonella, the greater wax moth (18). G. mellonella larvae were inoculated with 3 × 103 CFU/larva of the S. Typhimurium SL1344 wild-type and BolA mutant strains and incubated at 37°C, and their survival rate was daily registered over a period of 72 h. The effects of infection by the wild-type strain were rapidly seen after the first 24 h of infection, with a reduction of about 80% of the initial larval population (Fig. 3). In contrast, the deletion of the bolA gene was found to increase by 25% the larval survival rate in the course of infection (P < 0.05) compared with the rate of the wild-type strain (Fig. 3). Complementation of the mutant with the bolA gene in trans was also evaluated and was shown to restore the wild-type survival rates (Fig. 3). These results clearly show the involvement of BolA in the pathogenesis of S. Typhimurium in the model host G. mellonella.
FIG 3.
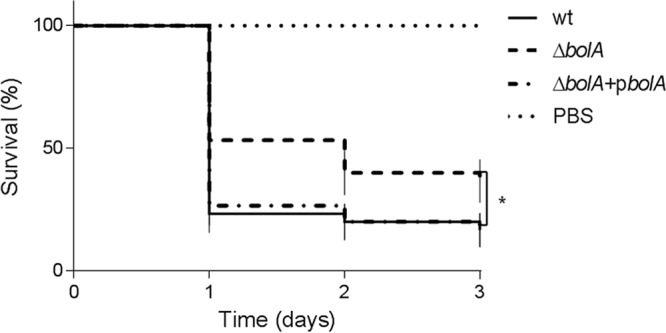
Survival of G. mellonella larvae following inoculation with S. Typhimurium strains. Kaplan-Meier survival curves represent larvae infected with 3 × 103 CFU/larva of the S. Typhimurium SL1344, BolA mutant (ΔbolA), and complemented (ΔbolA+pbolA) strains and PBS as a control. Larvae infected with the BolA mutant strain had enhanced survival compared to that of S. Typhimurium SL1344 or the complemented strain (*, P < 0.05). The data shown are means ± standard errors of the means, and differences in survival were calculated using a log rank test (Mantel-Cox). wt, wild type.
The bacterial load of the BolA mutant in hemolymph decreases during G. mellonella colonization.
In order to evaluate the proliferation of the S. Typhimurium BolA mutant within the insect hemocoel, we determined the viable bacterial load within the hemolymph of larvae in the course of infection (at 1, 4, 8, 12, and 24 h). To that end, larvae were challenged with 3 × 103 CFU/larva of S. Typhimurium SL1344 wild-type and BolA mutant strains, hemolymph from three living larvae was extracted and pooled, and the number of CFU was determined (Fig. 4). At the start of the infection, the three strains presented similar bacterial loads and were able to persist and proliferate in the larval hemolymph up to 12 h postinfection. However, the BolA mutant showed lower levels of proliferation (Fig. 4). Nevertheless, with the progression of the infection, a decrease of bacterial levels was noticed, especially for the ΔbolA strain, which reached values similar to those observed during the first hours of infection. In fact, at 24 h postinfection, the bacterial load in hemolymph was significantly lower for the bolA mutant than for the wild-type strain (P < 0.05). When the complemented strain was analyzed, behavior similar to that of the wild-type strain was observed. These results are in accordance with the results of the survival rates of infected larvae (Fig. 3) since at 24 h postinfection the wild-type infection led to a higher level of larval mortality.
FIG 4.
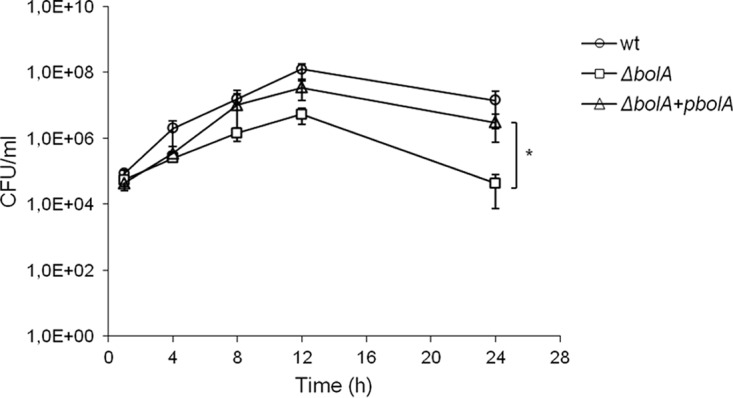
Hemolymph bacterial load in the course of S. Typhimurium infection. The viable bacterial load was determined in the hemolymph extracted from larvae infected with 3 × 103 CFU/larva of the S. Typhimurium SL1344, BolA mutant (ΔbolA), and complemented (ΔbolA+pbolA) strains during a period within the time course of infection. The bacterial load of larvae infected with the BolA mutant is significantly lower than the levels in larvae infected with wild-type (wt) strain and the complemented strain (*, P < 0.05; ANOVA and Tukey's test). Results (number of CFU per milliliter of hemolymph) represent means of three independent experiments.
The effect of BolA on the immune response of G. mellonella.
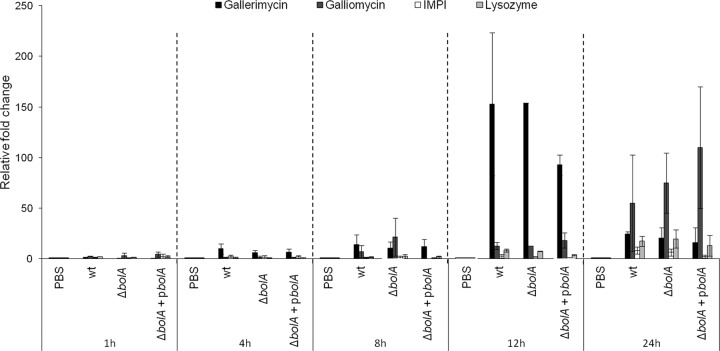
Host defense peptides are a crucial part of insect innate immunity, showing broad-spectrum microbicidal activity (17). In a previous study, we observed the induction of expression of four antimicrobial peptides upon infection of G. mellonella with S. Typhimurium SL1344 (18). In the present work, we examined whether the mutation in the bolA gene could either directly or indirectly interfere with the activation of the immune system of the insect host, leading to differences in the bacterial loads during infection. To evaluate this, larvae were infected with the wild-type, ΔbolA, and complemented strains. Gene expression levels of four selected antimicrobial peptides, namely, gallerimycin, galliomycin, lysozyme, and an inducible metallo-proteinase inhibitor (IMPI), were analyzed at 1, 4, 8, 12, and 24 h postinfection. Gene expression was determined by quantitative real-time PCR (RT-PCR) of the total RNA extracted for each time point. As shown in Fig. 5, no significant differences in the expression levels of immune-related peptides was observed in a comparison of the wild type with the BolA mutant strain.
FIG 5.
Immunogenic response of G. mellonella during infection with S. Typhimurium strains. Transcriptional activation of immune-responsive genes of G. mellonella was measured at 1, 4, 8, 12, and 24 h postinfection with 3 × 103 CFU/larva of the S. Typhimurium SL1344, BolA mutant (ΔbolA), and complemented (ΔbolA+pbolA) strains. The transcriptional levels of gallerimycin, galliomycin, IMPI, and lysozyme were determined by quantitative RT-PCR analysis and are shown relative to the expression levels in noninfected larvae injected with PBS. No significant differences were observed between strains. Results were normalized to the expression of the housekeeping gene actin and represent three independent experiments.
The deletion of bolA leads to lower levels of intracellular bacteria within hemocyte cultures.
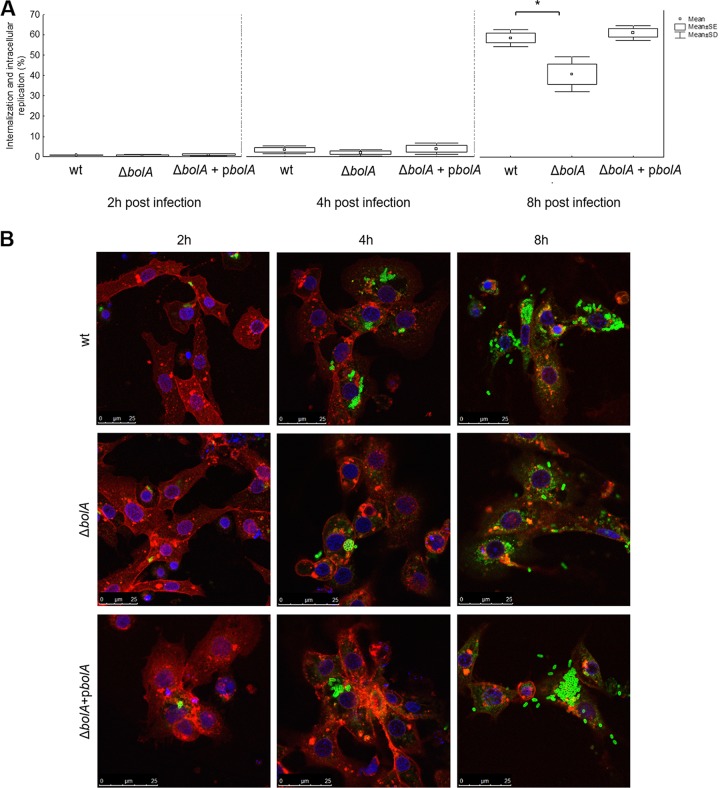
To further evaluate the effect of S. Typhimurium bolA deletion in the host-pathogen interaction in this insect model, hemocytes were extracted from larval hemolymph and used for in vitro cultures. The primary cell cultures of hemocytes were infected with the wild-type, BolA mutant, and complemented strains at a multiplicity of infection (MOI) of 50:1. Bacterial load was evaluated after 2, 4, and 8 h in a gentamicin protection assay and also visualized by fluorescence microscopy (Fig. 6). At 2 h, similar levels of phagocytic activity were observed for hemocytes infected with any of the three strains (Fig. 6A). These results were confirmed after 4 h of infection through the images of fluorescence microscopy in which only some hemocytes were infected with bacteria but the number of bacteria inside the hemocytes seemed similar for the three strains (Fig. 6B). Within the next 2 h of infection, the number of intracellular wild-type bacteria increased 4-fold, while the mutant strain increased 3-fold compared to the level at the previous time point. Nevertheless, this difference was not statistically significant (data not shown). However, in the course of the experiment, at 8 h postinfection, the difference between the wild type and the mutant was noteworthy, with the wild type reaching nearly 60% of the input of infection and the mutant having only 40% (P < 0.05). Complementation was achieved, and the phenotype was restored (Fig. 6A). Fluorescence microscopy images taken at 8 h postinfection indicate that the difference observed was related to higher intracellular bacterial division in hemocytes infected with the wild-type and complemented strains (Fig. 6B). In fact, some hemocytes showed large agglomerates of bacteria that were not observed in the mutant strain. Despite initial cellular uptake of BolA mutant bacteria at a level similar to that of the wild type, the wild type seemed to resist infection better than the mutant.
FIG 6.
In vitro infection of hemocytes with S. Typhimurium strains. (A) Primary hemocytes cell cultures were infected with the S. Typhimurium SL1344 wild-type, bolA mutant (ΔbolA), and complemented (ΔbolA+pbolA) strains (MOI of 50:1). Bacterial load of hemocytes was assessed using a gentamicin protection assay, and values of internalization and intracellular replication were obtained at 2, 4, and 8 h. Bacterial load of hemocytes infected with the mutant was significantly lower than that of larvae infected with the wild-type strain or the complemented strain at 8 h postinfection (*, P < 0.05, ANOVA and Tukey's test). Results are shown as percentages of the bacterial input and represent means of three independent experiments. (B) Fluorescence confocal microscopy images of the hemocytes infected with the S. Typhimurium SL1344 wild-type, bolA mutant (ΔbolA), and complemented (ΔbolA + pbolA) strains at 2, 4, and 8 h postinfection. Hemocyte membranes were labeled with WGA conjugated with Alexa 633 (red), while bacteria were stained with S. Typhimurium LPS antibody and Alexa 488 (green), and Hoechst dye was used for staining the cell's nucleus (blue). Experiments were repeated at least three times, and one representative experiment is shown.
DISCUSSION
Pathogenic bacteria have evolved a wide array of virulence mechanisms, essentially comprised by fine-tuned effectors that are critical for the success of infection. A proper coordination of these effectors is required for the efficiency of the virulence process. We have identified in E. coli a transcription factor, the BolA protein, that was shown to be involved in the regulation of several cellular processes related to virulence, such as membrane permeability, bacterial motility, and biofilm formation (11, 14–16). Moreover, we proposed that E. coli BolA is a general stress response transcription factor whose expression increases under diverse severe growth conditions, conferring protection to the cells (reviewed in reference 10). Homologs of this E. coli gene are found widely distributed across all cellular organisms. These facts prompted us to study the impact of BolA protein on the virulence capacity of Salmonella Typhimurium, one of the most common foodborne etiological agents.
When bacteria were cultured in a nutrient-rich medium (Luria-Bertani [LB]), the deletion of BolA in E. coli was reported to increase the growth rate compared to that of the wild-type strain (10). In contrast, in S. Typhimurium SL1344, BolA deletion did not significantly affect the growth rate relative to that of the wild-type strain, and only a slight increase in the OD600 of the BolA mutant at stationary phase was observed (Fig. 2A). However, when S. Typhimurium with a deletion of bolA was exposed to low pH or hydrogen peroxide, we observed significantly reduced bacterial survival, similar to what was observed for the E. coli bolA homolog (Fig. 2B and C). Interestingly, both stresses are important since they are used as antimicrobial weapons by mammalian cells in order to rapidly clear bacterial pathogens (25, 26). All of these results point to a possible role of BolA in S. Typhimurium virulence capacity.
In order to study the effects of BolA on S. Typhimurium proliferation and host survival in vivo, we chose the wax moth G. mellonella as the infection host, a model organism that has several advantages and has been increasingly used to study virulence mechanisms. The G. mellonella larva has previously been established as a successful model for S. Typhimurium infection (18, 19). Infection of G. mellonella larvae with the S. Typhimurium BolA mutant showed an attenuation of about 25% in virulence compared to the wild-type level (Fig. 3). Importantly, the complementation of the bolA deletion perfectly restored the wild-type phenotype, showing that BolA contributes to the virulence of this pathogen.
Following infection, the host immune response displays a set of coordinated and complex mechanisms in order to successfully immobilize or kill the pathogen. Hemolymph, which functions analogously to mammalian blood, comprises an innate immune response with remarkable similarities to the response in vertebrates. Analysis of S. Typhimurium replication in G. mellonella by direct bacterial enumeration in hemolymph demonstrated that bacteria persisted and increased by 1,000-fold during the first 12 h of infection. This result is in accordance with results of our previous study (18, 19). Moreover, the level of S. Typhimurium SL1344 replication appears to be consistent with that of the mouse model, in which this strain exhibited a 10,000-fold increase in the mouse spleen within 3 days (27). Nevertheless, in the progression of the insect infection we could see a decrease in bacterial levels. More noteworthy was the attenuated replication of the BolA mutant strain that, despite an initial increase in bacterial numbers, had values similar to those observed in the early stages of infection after 24 h of infection (Fig. 4). This result is consistent with the pronounced survival of G. mellonella after 24 h of infection with the mutant bacterial strain and supports the involvement of BolA in the successful colonization of S. Typhimurium (Fig. 3).
The innate immune responses of insects are of two major types, the cellular and the humoral. The cellular response is mediated by hemocytes that are involved in not only phagocytosis but also encapsulation and clotting (28, 29). The humoral response is composed of soluble effector molecules that immobilize or kill the pathogen and includes complement-like proteins, melanin, and antimicrobial peptides (30, 31). In order to deepen our understanding of the involvement of the humoral response in the differences between the survival rates of the BolA mutant and the wild-type strain, we investigated the differences in the expression levels of immune-related peptides. As seen in Fig. 5, wild-type and BolA mutant strains elicited analogous humoral responses, suggesting that this type of immune response is not relevant to the resolution of the infection. Nevertheless, it is worth noticing the inversion of the expression profiles in the course of infection. Up to 12 h of infection, gallerimycin had higher levels of expression; however, at 24 h mRNA levels of gallerimycin decreased, and those of galliomycin increased. This response profile is commonly observed in response to different pathogens (32–35).
In contrast to what happens regarding the G. mellonella humoral response, BolA seems to have a remarkable effect on S. Typhimurium evasion of cellular immunity. Despite the similar internalization capacities among the different strains, BolA substantially improves the capacity of bacteria to replicate inside hemocytes in later stages of infection (Fig. 6). Hemocyte-mediated immunity constitutes one of the most effective components of the G. mellonella defense mechanisms. Upon phagocytosis, bacteria are exposed to a wide range of antimicrobial effectors that rapidly identify and kill foreign pathogens (36, 37). Consistent with this, BolA promotes an increase of 5- and 7-fold in S. Typhimurium survival in response to oxidative stress and acidic burst. Therefore, we propose that BolA is a relevant player in S. Typhimurium virulence, contributing to the survival of the pathogen in a hostile environment during the course of infection. In this regard, our work constitutes an important advance in the understanding of pathogenic strategies and indicates that BolA contributes to Salmonella pathogenicity.
MATERIALS AND METHODS
Oligonucleotides, bacterial strains, and plasmids.
All bacterial strains and plasmids used in this study are listed in Table 1. The oligonucleotides used in this work were synthesized by STAB Vida and are listed in Table 2. All Salmonella strains used are isogenic derivatives of the wild-type S. Typhimurium strain SL1344. Restriction enzymes, T4 DNA ligase, and Phusion DNA polymerase were purchased from Thermo Fisher Scientific and used according to the supplier's instructions.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| SL1344 | hisG rpsL xyl Strr | 46 |
| CMA820 | SL1344 ΔbolA::Catr | This study |
| CMA821 | CMA820 carrying the pRMA04 plasmid | This study |
| Plasmids | ||
| pKD3 | Plasmid encoding FRT-flanked Cat cassette; pANTSγ, Ampr Catr | 38 |
| pKD46 | Temperature-sensitive λ-Red recombinase expression plasmid; oriR101 Ampr | 38 |
| pWKS29 | Low-copy-number plasmid; pSC101, Ampr | 47 |
| pRMA04 | pWSK29 carrying bolA, Ampr | This study |
TABLE 2.
Oligonucleotides used in this work
| Primer name (target) | Sequence (5′–3′) | Reference or source |
|---|---|---|
| RNMS003 | AGGGATGGGTGACATCACAGCGTTGTCGGAGGAGATTTTCATGATGATACGTGAGCAAATAGAAGAAAAAGTGTAGGCTGGAGCTGCTTC | This study |
| RNMS004 | AACATACTGGAAAAGCGGAGGTAGTTGCAAATCCGTTTTGCTACGCGATGCTTCCCGCGCCGCGACAAGGCATATGAATATCCTCCTTAG | This study |
| RNMS011 | GCTCTAGAGCGTCAACTCTACTCCGCAAGCAC | This study |
| RNMS012 | AAAGGTACCTTTGTGGTCAACGCAGGTAAATTCG | This study |
| P1RT (gallerimycin) | CGCAATATCATTGGCCTTCT | 48 |
| P2RT (gallerimycin) | CCTGCAGTTAGCAATGCAC | 48 |
| P1RT (IMPI) | AGATGGCTATGCAAGGGATG | 48 |
| P2RT (IMPI) | AGGACCTGTGCAGCATTTCT | 48 |
| P1RT (lysozyme) | TCCCAACTCTTGACCGACGA | 48 |
| P2RT (lysozyme) | AGTGGTTGCGCCATCCATAC | 48 |
| P1RT (actin) | ATCCTCACCCTGAAGTACCC | 48 |
| P2RT (actin) | CCACACGCAGCTCATTGTA | 48 |
| P1RT (galliomycin) | TCGTATCGTCACCGCAAAATG | 49 |
| P2RT (galliomycin) | GCCGCAATGACCACCTTTATA | 49 |
The λ-Red-mediated mutagenesis method (38) was used to obtain the bolA deletion strain. Briefly, a PCR fragment was obtained by amplification of a pKD3 plasmid using the primer pair RNMS003/RNMS004 (Table 2). The resulting fragment, carrying the chloramphenicol (Cat) cassette flanked by 70-nucleotide (nt) homologous extension regions adjacent to the bolA gene, was transformed in S. Typhimurium SL1344 electrocompetent cells containing pKD46 to allow recombination with the bacterial chromosome. P22 HT105/1 int-201 transduction was used to obtain the SL1344 ΔbolA strain (CMA820) in a fresh background (39).
The pRMA04 plasmid was constructed by inserting a PCR-amplified DNA fragment carrying the bolA coding sequence and promoter region (using primers RMNS011/RMNS012) (Table 2) into XbaI and KpnI sites of a pWSK29 vector. The resulting pRMA04 plasmid was transformed in CMA820 competent cells to obtain the complemented strain CMA821. All constructs were confirmed by DNA sequencing at STAB Vida.
Bacterial and insect growth conditions.
S. Typhimurium strains were grown in Luria-Bertani (LB) broth at 37°C and 220 rpm. For Galleria mellonella infection experiments, cultures were grown overnight in LB medium at 37°C under static conditions. When appropriate, antibiotics were used at the following concentrations: 100 μg/ml ampicillin, 90 μg/ml streptomycin, and 25 μg/ml chloramphenicol.
Galleria mellonella larvae were reared in our laboratory, from egg to last-instar larvae, at 25°C in darkness, as previously described (18, 40). We used the natural foods beeswax and pollen grains, and last-instar larvae weighing 225 ± 25 mg were selected for the experiments.
Sequence analysis.
The BolA homolog from S. Typhimurium was identified using the BLASTp tool at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov) with the BolA sequence from E. coli K-12 W3110 (GenBank accession no. APC50728.1) as the query. Homologs were aligned using EMBOSS Needle, version 6.6.0, a pairwise sequence alignment tool at the European Bioinformatics Institute (EMBL-EBI) website (http://www.ebi.ac.uk/). Synteny analysis was performed using standard gene visualization tools at NCBI. Protein structural prediction was obtained with Swiss model (https://swissmodel.expasy.org/) (41), based on the crystal structure of BolA from E. coli K-12 W3110 (PDB entry 2DHM).
Acidic and oxidative stress assays.
Bacterial cultures were diluted from overnight growth to an optical density of 0.1. The cultures were grown aerobically in LB medium (pH 7.0) at 37°C and 220 rpm for 1 h (corresponding to an optical density at 600 nm [OD600] of 0.3) before challenge under the selected stress conditions. Acidic stress was accomplished by adding 30% (vol/vol) HCl. Specifically, the medium pH was adjusted to 4.4 for 1 h, followed by a challenge at pH 3.1 for an additional hour. For oxidative stress, cells were challenged for 1 h with 5 mM H2O2. To evaluate viabilities, cells were diluted in phosphate-buffered saline (PBS), serially diluted, and inoculated on LB agar plates. The number of viable cells was extrapolated directly from the number of colonies obtained after 16 h of growth at 37°C and was expressed as the number of CFU/milliliter. The results were normalized by the number of cells obtained before stress induction.
Galleria mellonella killing assays.
G. mellonella killing assays were performed based on a previously described method, with some changes (18). Briefly, cultures of S. Typhimurium were grown overnight in LB broth under static conditions at 37°C. The OD of the cultures was measured at 600 nm, and the appropriate volume was collected to ensure that all the strains had the same OD. Cells were then harvested by centrifugation and suspended in PBS. Tenfold serial dilutions were performed to obtain 3 × 103 CFU per volume of injection. This concentration was confirmed by inoculating serial dilutions on LB agar plates. A micrometer was used to control the volume of a disposable hypodermic microsyringe, and a 3.5-μl aliquot of each bacterial dilution was injected into the larvae via the hindmost left proleg, which had been previously surface sanitized with 70% (vol/vol) ethanol. For each condition, we used 10 larvae, and as a control a set of larvae was injected with PBS. Following injection, larvae were placed in petri dishes and stored in the dark at 37°C.
Larval survival was followed over a period of 3 days, and caterpillars were considered dead when they displayed no movement in response to touch. Using results from three independent experiments, Kaplan-Meier survival curves were plotted, and differences in survival rates were calculated by using a log rank (Mantel-Cox) statistical test; all analyses were performed with GraphPad Prism, version 6, software.
Extraction of larval hemolymph.
To determine the number of viable bacteria in the hemocoel during the course of infection, hemolymph of infected larvae was collected as previously described (18). Briefly, three living larvae were anesthetized on ice and surface sterilized with ethanol at 1, 4, 8, 12, and 24 h after injection. The larvae were bled, and the outflowing hemolymph was immediately transferred into a sterile microtube containing a few crystals of phenylthiourea to prevent melanization. Bacterial quantification was done by preparing 10-fold serial dilutions of hemolymph in PBS and inoculating the mixture onto LB agar plates. Bacteria were enumerated by CFU counting after incubation at 37°C for 24 h, and results are presented as the number of CFU per milliliter of hemolymph.
In vitro cultivation of G. mellonella hemocytes.
To isolate G. mellonella hemocytes, hemolymph was extracted from last-instar larvae, previously anesthetized on ice and surface sterilized with ethanol, by puncturing the larval abdomen with a sterile needle (42). Hemolymph was immediately collected into anticoagulant buffer (98 mM NaOH, 145 mM NaCl, 17 mM EDTA, and 41 mM citric acid; pH 4.5) in a 1:1 proportion. Hemocytes were spun in a centrifuge at 250 × g for 10 min at 4°C, washed twice with PBS, and spun at 250 × g for 5 min at 4°C. Finally, hemocytes were gently suspended in 1 ml of Grace's insect medium (GIM) (Thermo Fisher Scientific) supplemented with 10% (vol/vol) fetal bovine serum, 1% (wt/vol) glutamine, and 1% (wt/vol) antibiotic/antimycotic solution (10,000 units of penicillin G, 10 mg of streptomycin, 25 mg/liter amphotericin B). Suspended hemocytes were counted with a hemocytometer and incubated at 26°C in 24-well plates at a concentration of 2 × 105 cell/ml. Monolayers of primary Galleria hemocytes were used for experiments the next day.
Gentamicin protection assay of hemocytes.
To determine the intracellular bacterial load of in vitro hemocytes during the course of infection, a protocol based on the gentamicin protection assay was used (43). Prior to infection with bacteria, Galleria hemocyte monolayers were washed with PBS, and medium was replaced with GIM without antibiotics. Cultures of S. Typhimurium SL1344 were grown as described above, and the appropriate volume was collected to obtain 4 × 103 bacteria/ml in each well. After 1 h of infection at 37°C, the hemocytes were carefully washed twice with PBS, followed by the addition of GIM containing 100 mg/liter of gentamicin to kill extracellular bacteria. After 1 h of incubation, supernatants were inoculated in LB agar plates to confirm the effectiveness of antibiotic treatment, and medium was replaced with GIM containing 10 mg/liter of gentamicin. The quantification of viable intracellular bacteria was performed at 2, 4, and 8 h after infection. For that, infected cell monolayers were lysed with 0.5% (vol/vol) Triton X-100 for 20 min, and the number of CFU was determined by inoculating 10-fold dilutions of cell lysates on LB agar plates, followed by incubation at 37°C for 24 h.
Confocal microscopy of hemocytes.
For microscopy analysis, in vitro hemocytes were isolated and infected as described above; however, glass coverslips were placed into the 24-well plates before hemocyte seeding. After 2, 4, and 8 h of infection, wells were washed with PBS, and samples were fixed with 3.7% (wt/vol) paraformaldehyde for 20 min, quenched with 50 mM NH4Cl for 10 min, immersed in 0.2% (vol/vol) Triton X-100 for 5 min, and saturated with 5% (wt/vol) bovine serum albumin for 30 min. The bacterial immunostaining was performed using a monoclonal antibody to S. Typhimurium anti-lipopolysaccharide (LPS) (1:500; Abcam) followed by the secondary polyclonal goat anti-mouse serum coupled to Alexa 488 (1:500; Santa Cruz Biotechnology). Cells were stained using wheat germ agglutinin (WGA) conjugated with Alexa 633 (1:200; Thermo Fisher Scientific). Finally, coverslips were mounted in Vectashield (Vector Laboratories) containing Hoechst 33342 fluorescent dye (Thermo Fisher Scientific) for nuclear staining. All samples were examined on a Leica TCS SP5 (Leica Microsystems CMS GmbH) inverted microscope (model no. DMI6000) with a 63× water apochromatic objective (44).
G. mellonella RNA extraction.
Sets of 20 larvae were infected with S. Typhimurium strains at a concentration of 3 × 103 CFU/larva, as previously described for the survival assays. At 1, 4, 8, 12, and 24 h after injection, three living larvae per set were cryopreserved, sliced, and homogenized in 1 ml of TRIzol reagent (Sigma-Aldrich). Whole-animal RNA was extracted according to the manufacturer's protocol. After extraction, RNA was treated with an RNase-free DNase set (Qiagen). The purified RNA was quantified spectrophotometrically (NanoDrop ND-1000).
Quantitative real-time PCR.
The transcriptional levels of genes encoding the G. mellonella antimicrobial peptides gallerimycin, galliomycin, inducible metalloproteinase inhibitor (IMPI), and lysozyme were determined with a Rotor-Gene 3000 (Corbett) system using a SensiFast SYBR kit (Bioline) according to the supplier's instructions. cDNA was synthesized from 1 μg of purified RNA with a SensiFast cDNA synthesis kit (Bioline). The primers used are listed in Table 2. All samples were analyzed in triplicate, and the amount of mRNA detected was normalized using the mRNA value of the housekeeping gene actin. Relative quantification of gene expression was calculated by using the ΔΔCT (CT is threshold cycle) method (45).
Statistical analysis.
Data are expressed as mean values of a minimum of three independent experiments ± standard deviations (SD). Statistical analysis was carried out using Statistica, version 7, software (StatSoft). One-way analysis of variance (ANOVA) and Tukey's multiple-comparison test with an unequal group sample size were performed to determine statistically significant differences. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by Fundação para a Ciência e Tecnologia (FCT), Portugal with fellowships to D. Mil-Homens (SFRH/BPD/91831/2012), S. Barahona (PD/BD/113983/2015), R. N. Moreira (SFRH/BPD/84080/2012), I. J. Silva (SFRH/BPD/84086/2012), and S. N. Pinto (SFRH/BPD/92409/2013). The work at Instituto de Tecnologia Química e Biológica was financially supported by Project PTDC/BIA-MIC/4046/2014, by Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020, Programa Operacional Competitividade e Internacionalização. The work at Institute for Bioengineering and Biosciences was supported by FCT, Portugal UID/BIO/04565/2013, and Programa Operacional Regional de Lisboa 2020 (project no. 007317).
REFERENCES
- 1.Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, Aarestrup FM. 2011. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis 8:887–900. doi: 10.1089/fpd.2010.0787. [DOI] [PubMed] [Google Scholar]
- 2.Cross AS. 2008. What is a virulence factor? Crit Care 12:196. doi: 10.1186/cc7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabrega A, Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen S, Fang FC. 2012. Integrated stress responses in Salmonella. Int J Food Microbiol 152:75–81. doi: 10.1016/j.ijfoodmicro.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 6.Darwin KH, Miller VL. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol 181:4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garmendia J, Beuzon CR, Ruiz-Albert J, Holden DW. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149:2385–2396. doi: 10.1099/mic.0.26397-0. [DOI] [PubMed] [Google Scholar]
- 8.Feng X, Oropeza R, Kenney LJ. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol Microbiol 48:1131–1143. doi: 10.1046/j.1365-2958.2003.03502.x. [DOI] [PubMed] [Google Scholar]
- 9.Olekhnovich IN, Kadner RJ. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol 189:6882–6890. doi: 10.1128/JB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guinote IB, Moreira RN, Barahona S, Freire P, Vicente M, Arraiano CM. 2014. Breaking through the stress barrier: the role of BolA in Gram-negative survival. World J Microbiol Biotechnol 30:2559–2566. doi: 10.1007/s11274-014-1702-4. [DOI] [PubMed] [Google Scholar]
- 11.Dressaire C, Moreira RN, Barahona S, Alves de Matos AP, Arraiano CM. 2015. BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio 6:e02352-. doi: 10.1128/mBio.02352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aldea M, Hernandez-Chico C, de la Campa AG, Kushner SR, Vicente M. 1988. Identification, cloning, and expression of bolA, an ftsZ-dependent morphogene of Escherichia coli. J Bacteriol 170:5169–5176. doi: 10.1128/jb.170.11.5169-5176.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos JM, Freire P, Vicente M, Arraiano CM. 1999. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol Microbiol 32:789–798. doi: 10.1046/j.1365-2958.1999.01397.x. [DOI] [PubMed] [Google Scholar]
- 14.Santos JM, Lobo M, Matos AP, De Pedro MA, Arraiano CM. 2002. The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol Microbiol 45:1729–1740. doi: 10.1046/j.1365-2958.2002.03131.x. [DOI] [PubMed] [Google Scholar]
- 15.Freire P, Vieira HL, Furtado AR, de Pedro MA, Arraiano CM. 2006. Effect of the morphogene bolA on the permeability of the Escherichia coli outer membrane. FEMS Microbiol Lett 260:106–111. doi: 10.1111/j.1574-6968.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- 16.Moreira RN, Dressaire C, Barahona S, Galego L, Kaever V, Jenal U, Arraiano CM. 2017. BolA is required for the accurate regulation of c-di-GMP, a central player in biofilm formation. mBio 8:e00443-. doi: 10.1128/mBio.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai CJ, Loh JM, Proft T. 2016. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7:214–229. doi: 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viegas SC, Mil-Homens D, Fialho AM, Arraiano CM. 2013. The virulence of Salmonella enterica serovar Typhimurium in the insect model Galleria mellonella is impaired by mutations in RNase E and RNase III. Appl Environ Microbiol 79:6124–6133. doi: 10.1128/AEM.02044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bender JK, Wille T, Blank K, Lange A, Gerlach RG. 2013. LPS structure and PhoQ activity are important for Salmonella Typhimurium virulence in the Galleria mellonella infection model [corrected]. PLoS One 8:e73287. doi: 10.1371/journal.pone.0073287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desbois AP, McMillan S. 2015. Paving the way to acceptance of Galleria mellonella as a new model insect. Virulence 6:410–411. doi: 10.1080/21505594.2015.1036218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasai T, Inoue M, Koshiba S, Yabuki T, Aoki M, Nunokawa E, Seki E, Matsuda T, Matsuda N, Tomo Y, Shirouzu M, Terada T, Obayashi N, Hamana H, Shinya N, Tatsuguchi A, Yasuda S, Yoshida M, Hirota H, Matsuo Y, Tani K, Suzuki H, Arakawa T, Carninci P, Kawai J, Hayashizaki Y, Kigawa T, Yokoyama S. 2004. Solution structure of a BolA-like protein from Mus musculus. Protein Sci 13:545–548. doi: 10.1110/ps.03401004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchko GW, Yee A, Semesi A, Myler PJ, Arrowsmith CH, Hui R. 2015. Solution-state NMR structure of the putative morphogene protein BolA (PFE0790c) from Plasmodium falciparum. Acta Crystallogr F Struct Biol Commun 71:514–521. doi: 10.1107/S2053230X1402799X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roret T, Tsan P, Couturier J, Zhang B, Johnson MK, Rouhier N, Didierjean C. 2014. Structural and spectroscopic insights into BolA-glutaredoxin complexes. J Biol Chem 289:24588–24598. doi: 10.1074/jbc.M114.572701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilmes-Riesenberg MR, Foster JW, Curtiss R III. 1997. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun 65:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen R, van der Straaten T, van Diepen A, van Dissel JT. 2003. Responses to reactive oxygen intermediates and virulence of Salmonella typhimurium. Microbes Infect 5:527–534. doi: 10.1016/S1286-4579(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 26.Ren J, Zhao M, Yao Y. 2014. Acid stress response of Salmonella and its relationship with virulence—a review. Wei Sheng Wu Xue Bao 54:367–375. (In Chinese.) [PubMed] [Google Scholar]
- 27.Brandt R, Petersen A, Brix S, Licht TR, Frokiaer H. 2013. Epithelial entry rather than the ensuing systemic immune response determines the pathogenicity of two Salmonella enterica serovar Typhimurium strains in a mouse model. Microbes Infect 15:911–919. doi: 10.1016/j.micinf.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Bergin D, Reeves EP, Renwick J, Wientjes FB, Kavanagh K. 2005. Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect Immun 73:4161–4170. doi: 10.1128/IAI.73.7.4161-4170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browne N, Heelan M, Kavanagh K. 2013. An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence 4:597–603. doi: 10.4161/viru.25906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browne EP, Littman DR. 2009. Myd88 is required for an antibody response to retroviral infection. PLoS Pathog 5:e1000298. doi: 10.1371/journal.ppat.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seitz V, Clermont A, Wedde M, Hummel M, Vilcinskas A, Schlatterer K, Podsiadlowski L. 2003. Identification of immunorelevant genes from greater wax moth (Galleria mellonella) by a subtractive hybridization approach. Dev Comp Immunol 27:207–215. doi: 10.1016/S0145-305X(02)00097-6. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee K, Altincicek B, Hain T, Domann E, Vilcinskas A, Chakraborty T. 2010. Galleria mellonella as a model system for studying Listeria pathogenesis. Appl Environ Microbiol 76:310–317. doi: 10.1128/AEM.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mil-Homens D, Bernardes N, Fialho AM. 2012. The antibacterial properties of docosahexaenoic omega-3 fatty acid against the cystic fibrosis multiresistant pathogen Burkholderia cenocepacia. FEMS Microbiol Lett 328:61–69. doi: 10.1111/j.1574-6968.2011.02476.x. [DOI] [PubMed] [Google Scholar]
- 34.Bergin D, Murphy L, Keenan J, Clynes M, Kavanagh K. 2006. Pre-exposure to yeast protects larvae of Galleria mellonella from a subsequent lethal infection by Candida albicans and is mediated by the increased expression of antimicrobial peptides. Microbes Infect 8:2105–2112. doi: 10.1016/j.micinf.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Wojda I, Taszlow P. 2013. Heat shock affects host-pathogen interaction in Galleria mellonella infected with Bacillus thuringiensis. J Insect Physiol 59:894–905. doi: 10.1016/j.jinsphys.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Craig M, Slauch JM. 2009. Phagocytic superoxide specifically damages an extracytoplasmic target to inhibit or kill Salmonella. PLoS One 4:e4975. doi: 10.1371/journal.pone.0004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathman M, Sjaastad MD, Falkow S. 1996. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect Immun 64:2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmieger H. 1971. The fate of the bacterial chromosome in P22-infected cells of Salmonella typhimurium. Mol Gen Genet 110:238–244. doi: 10.1007/BF00337836. [DOI] [PubMed] [Google Scholar]
- 40.Mil-Homens D, Ferreira-Dias S, Fialho AM. 2016. Fish oils against Burkholderia and Pseudomonas aeruginosa: in vitro efficacy and their therapeutic and prophylactic effects on infected Galleria mellonella larvae. J Appl Microbiol 120:1509–1519. doi: 10.1111/jam.13145. [DOI] [PubMed] [Google Scholar]
- 41.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 42.Brivio MF, Mastore M, Nappi AJ. 2010. A pathogenic parasite interferes with phagocytosis of insect immunocompetent cells. Dev Comp Immunol 34:991–998. doi: 10.1016/j.dci.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Wu G, Zhao Z, Liu C, Qiu L. 2014. Priming Galleria mellonella (Lepidoptera: Pyralidae) larvae with heat-killed bacterial cells induced an enhanced immune protection against Photorhabdus luminescens TT01 and the role of innate immunity in the process. J Econ Entomol 107:559–569. doi: 10.1603/EC13455. [DOI] [PubMed] [Google Scholar]
- 44.Pinto SN, Silva LC, de Almeida RF, Prieto M. 2008. Membrane domain formation, interdigitation, and morphological alterations induced by the very long chain asymmetric C24:1 ceramide. Biophys J 95:2867–2879. doi: 10.1529/biophysj.108.129858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 47.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 48.Altincicek B, Vilcinskas A. 2006. Metamorphosis and collagen-IV-fragments stimulate innate immune response in the greater wax moth, Galleria mellonella. Dev Comp Immunol 30:1108–1118. doi: 10.1016/j.dci.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Wojda I, Kowalski P, Jakubowicz T. 2009. Humoral immune response of Galleria mellonella larvae after infection by Beauveria bassiana under optimal and heat-shock conditions. J Insect Physiol 55:525–531. doi: 10.1016/j.jinsphys.2009.01.014. [DOI] [PubMed] [Google Scholar]