Abstract
Background
Liquid biopsy involves the analysis of cell-free nucleic acids, mainly circulating free DNA (cfDNA), in bodily fluids such as blood. The obtaining of specimens is easier for patients and less invasive than tissue biopsy, but the method has certain limitations.
Method
This review is based on pertinent publications retrieved by a selective literature search.
Results
Because the concentration of cfDNA in plasma is less than 0.001%, special amplifying techniques must be used to enable a search for specific mutations. Liquid biopsy can be used in patients with non–small cell lung cancer (NSCLC) if no tissue is available for biopsy; when performed for this indication, it has 67% sensitivity and 94% specificity. If liquid biopsy does not reveal a mutation, this may be due either to the absence of the mutation in the tumor or to the inadequate sensitivity of the measuring technique. This uncertainty associated with negative findings can be reduced by the simultaneous demonstration of reference mutations derived from a primary tumor tissue analysis. In comparison to tissue studies, the search for tumor-specific mutations by liquid biopsy is 70% sensitive and 69% specific; this corresponds to a positive predictive value of 86% and a negative predictive value of 46%.
Conclusion
Liquid biopsy and tumor tissue analysis are complementary, rather than alternative, techniques for therapeutically relevant genetic investigation of tumors. Comparative studies are needed so that further indications can be determined for liquid biopsy in the diagnostic evaluation of cancer.
Liquid biopsy is a term used to describe a diagnostic approach relying on cell-free nucleic acids from bodily fluids—usually circulating free deoxyribonucleic acids (circulating free DNA, cfDNA)—which are isolated from blood, enhanced and subsequently analyzed. The first description of cfDNA dates back to 1948 (1). However, only after 50 years cfDNA rose to high clinical relevance with the detection of diagnostic fetal DNA in maternal blood samples (2). As a result, liquid biopsy and cfDNA testing have become a standard method in prenatal diagnosis (3). In one pregnant women who underwent prenatal testing, cancer was accidentally diagnosed by means of mutation identification in cfDNA (2). This observation paved the way for the use of liquid biopsy in tumor genetic diagnosis (mutation analysis). This method was systematically advanced as an aid to cancer diagnosis (3). Today, liquid biopsy plays an important role to detect T790M resistance mutations in the epidermal growth factor receptor (EGFR) gene for the targeted treatment of non–small cell lung cancer (NSCLC).
Definitions
Circulating cell-free nucleic acids and circulating tumor cells
Liquid biopsy is primarily used to analyze cell-free nucleic acids in peripheral blood. However, this method can also be used to analyze cell-free nucleic acids from other bodily fluids, such as cerebrospinal fluid, urine and ascites fluid. In tumor diagnosis, cfDNA isolated from peripheral blood is typically used for genetic analysis (4). This cfDNA originates from tissues with high cell turn-over, i.e. a high rate of cell proliferation and cell death. Thus, sources of cfDNA other than tumors comprise normal regenerating tissue in various states, including hematopoiesis, immune reactions, inflammatory processes and necrosis caused by inadequate blood supply. CfDNA originating from tumor cells is also called circulating free tumor DNA (ctDNA). The release of ctDNA into the bloodstream (shedding) is influenced by tumor type, location, vascularization, and size (5, 6). The ctDNA of a colorectal cancer lesion weighing 100 g (approximately 3 × 1010 tumor cells) accounts for approximately 3.3% of a patient’s total cfDNA (7). The average plasma concentration of cfDNA is 180 ng/mL (0 to >1000 ng/mL) (8). The DNA fragments are very short, on average only 160 base pairs (4, 8), indicating that they originate from apoptotic cells (9).
Using cfDNA/ctDNA, somatic tumor genetic alterations, such as point mutations, deletions or amplifications can be detected. Detection of translocations (gene fusions) is best achieved on the level of messenger ribonucleic acid (mRNA) (10) and requires circulating free RNA (cfRNA) or circulating free tumor mRNA (ctRNA). cfRNA/ctRNA is so small (on average 40 base pairs) that it is normally unsuitable for detecting mutations (11). Consequently, other sources of material for RNA analyses, such as exosomes (12) or tumor-educated platelets (TEP), are being explored (13, 14). Since these materials have not yet become available for routine diagnosis, today it is not possible to reliably perform tumor genetic analyses for translocations based on liquid biopsies.
Circulating tumor cells (CTCs) are being considered as another potential target for tumor genetic diagnosis from peripheral blood. However, they only occur in small amounts (1–10 CTC/mL of blood) (15) and are difficult to enrich. As a result, CTCs currently play no role in tumor genetic analyses from blood (16).
Applications
Liquid biopsies can be used for a number of diagnostic applications, such as companion diagnostics, molecular monitoring and molecular staging (4, 5).
Tumor genetic analysis for targeted therapy (companion diagnostics)
The use of targeted treatments usually requires to determine the mutation status of a biomarker (companion diagnostics). An interesting indication for liquid biopsy is the detection of new or resistance-conferring mutations associated with tumor progression after completion of a treatment course to plan further therapy and to identify additional targeted treatment options.
Liquid biopsy can be used to detect mutations in cases where the number of tumor cells is insufficient for tumor genetic analysis. It is also recommended for difficult-to-reach tumors and in patients in whom invasive sampling is contraindicated, as in such cases no tissue or cell material can be obtained from the tumor (table 1). This applies to about 30% of patients with NSCLC (17, 18).
Table 1. Comparison of the advantages and disadvantages of tissue biopsy and liquid biopsy in the detection of mutations in molecular pathological assays.
| Advantage | Disadvantage | |
| Tissue biopsy | Morphological correlate (high diagnostic certainty) |
Lack of representativeness in the presence of tumor heterogeneity |
| Liquid biopsy | Representative analysis in the presence of multiple tumor foci |
No diagnostic value if no mutation is detected |
Both methods have their strengths and limitations. Thus, the combined use of the two complimentary methods represents the highest diagnostic standard for the patient.
Moreover, cancer progression is often associated with the presence of several tumor foci or metastatic lesions which may show heterogeneous tumor genetics (19). In this constellation, the advantage of liquid biopsy would be that it detects ctDNA shedded from all tumor lesions and thus can deliver representative results even in case of tumor genetic heterogeneity (table 1). Patients with NSCLC harboring an activating mutation in the EGFR gene who are undergoing targeted therapy with tyrosine kinase inhibitors (TKI) frequently develop resistance by a secondary T790M mutation in the EGFR gene. The administration of osimertinib (3rd generation TKI) in the follow-on treatment results in reduced morbidity and improved quality of life compared to platinum-based standard chemotherapy, with comparable overall survival (20– 23). In the pivotal trial, qualitative comparisons of liquid biopsy versus tissue biopsy in NSCLC showed that the results obtained based on liquid biopsy (mean sensitivity of 0.67 and specificity of 0.94) were significantly poorer (table 2) (24).
Table 2. Qualitative comparison of tissue biopsy and liquid biopsy.
| Tumor | Sample size | Sensitivity | Specificity | Positive predictive value | Negative predictive value | Reference |
| NSCLC | 216 | 0.70 | 0.69 | 0.86 | 0.46 | Oxnard 2016 (23) |
| NSCLC | 2012 | 0.67 | 0.94 | not stated | not stated | Luo 2014 (24) |
| mKRK | 98 | 0.90 | 0.93 | 0.94 | 0.90 | Schmiegel 2017 (25) |
Different tumor entities, such as non–small cell lung cancer (NSCLC) or metastatic colorectal cancer (mCRC) shed different amounts of circulating free deoxyribo?nucleic acids (ctDNA) into the bloodstream. Based on the statistical quality parameters sensitivity (correct positive rate), specificity (correct negative rate), positive (accuracy) and negative predictive value (discriminatory power), the benefits of liquid biopsies compared to tissue biopsies can be evaluated on an individual basis. This study shows that in NSCLC a complete transition from tissue biopsy to liquid biopsy would result in a substantial reduction in diagnostic care quality. Given its advantages and its high positive predictive value, liquid biopsy is complementary to tissue biopsy in this setting.
Based on this body of evidence, the European Medicines Agency (EMA) approved in 2015 for patients receiving the TKI gefitinib and in 2016 for patients receiving the TKI osimertinib the use of blood for detection of the required activating mutations (geftinib) or inhibiting T790M mutation in the EGFR gene (osimertinib), if no adequate quantities of tumor tissue are available (www.ema.europa.eu/ema). To date, this is the only approved indication for the use of liquid biopsies in cancer diagnosis. Since tumors of various entities release different quantities of ctDNA into the bloodstream (5), liquid biopsy is not suitable for all types of tumors in routine testing for tumor genetic gene mutations (table 2). In metastatic colorectal cancer (sensitivity: 0.90, specificity: 0.93) (table 2) (25), liquid biopsy appears to be a suitable source of material for tumor genetic diagnosis searching for mechanisms of resistance in patients with disease progression, if corresponding treatments are available and/or approved (4, 5, 26, 27). However, data especially from clinical pivotal studies—as in NSCLC (21, 23)—are required before liquid biopsies can be used universally in routine diagnosis.
Diagnosis of tumor recurrence (molecular monitoring)
If information about tumor-specific reference mutations is available from previous tumor genetic testing—for example, in the APC gene or KRAS gene in colorectal cancer—, this can be used for the liquid biopsy and, in case of inconclusive imaging studies, for the diagnosis of recurrence. In this way, it is also possible to obtain information which helps to differentiate between a recurrence of a known tumor and a secondary tumor in patients with newly diagnosed tumor foci (4, 5, 26, 27). Here, intensive clinical studies are needed to prove the clinical relevance of liquid biopsy in this respect. Thus, at present it is of experimental value only.
Determining a prognostic parameter (molecular staging)
Tumors already shedding cfDNA into the bloodstream at lower stages (unio internationalis contra cancrum [UICC]: I, II) have a poorer prognosis (5). Liquid biopsies may be used in the future to differentiate between cfDNA-positive and cfDNA-negative stages of cancer and thus provide information relevant to prognosis.
This enables molecular staging which informs the decision to administer adjuvant therapy. However, the feasibility of this approach still has to be evaluated in clinical studies.
Tumor genetic mutation analysis from liquid biopsies
Blood collection
Special tubes should be used for blood collection. Most standard blood collection tubes are made of positively charged plastic which adsorbs the negatively charged cfDNA so that it can no longer be detected during analysis. In addition, blood cells start to break down after blood collection, releasing genomic DNA which distorts any analysis of actual cfDNA. These two processes have an impact on the sensitivity and results of analyses from liquid biopsies. Consequently, only blood collection tubes used for prenatal genetic screening or tubes specifically designed for tumor genetic testing should be used. They are made of negatively charged materials and contain stabilizing reagents, preventing blood cell breakdown for a period of about 5 days (28) so that specimens can be shipped by mail without the need for special cold chains.
Furthermore, a sufficiently large gauge needle (at least 21G) should be used to ensure nucleus-containing blood cells remain intact. Blood-sampling systems used for blood cultures are ideally suited for liquid biopsies as they are designed to ensure blood cells remain intact and vital. Finally, it is important to fill the blood collection tubes to the maximum level to prevent the formation of foam. When foam bubbles burst, high shear forces occur at the interface with blood fluid, resulting in the destruction of blood cells. The reddish tint of plasma after centrifugation due to hemolysis provides a good indication of the degree of cellular damage.
Detection of tumor genetic mutations, digital polymerase chain reaction
The key problem associated with the use of ctDNA to test for tumor genetic mutations is that they are present only in very small quantities in cfDNA which may be <0.001% (23). Consequently, the analysis of liquid biopsies requires the use of highly sensitive detection systems. Standard polymerase chain reaction (PCR) approaches are less suitable, as they do not permit control over the number of ctDNA molecules used and PCR duplicates may distort measurements. Digital PCR systems (dPCR) enable the user to set the number of cfDNA molecules to be used. These systems include:
Beads amplification magnetics (BEAMing) (29)
Digital droplet PCR (ddPCR) (30)
Digital next-generation sequencing (dNGS) (31).
The sensitivity of these detection methods is <0.1% (5, 29– 32). With BEAMing and ddPCR, however, only few genes (gene loci) can be examined. Thus, these methods are suitable for detecting known tumor genetic alterations, for example, a T790M mutation in the EGFR gene. This mutation shows whether resistance has developed after TKI treatment and thus treatment of the tumor with the 3rd generation TKI osimertinib is indicated (21). Only dNGS allows for high-sensitivity detection of numerous unknown mutations, for example if a tumor develops resistance to therapy (26, 33).
Reference mutation
The fundamental problem with the use of ctDNA for tumor genetic analyses is the lack of information about its proportion in cfDNA. At present, no marker is available which can be used to reliably distinguish ctDNA from tumor cells from cfDNA from normal cells. Only the detection of a known mutation from the primary tumor provides certainty that the analysis has been successful and yielded meaningful test results (21, 23). If no known mutation (wild type, WT) was identified, there are two possible explanations: First, no ctDNA is present, or, second, the sensitivity of the method is inadequate to detect ctDNA. For these unresolvable methodological reasons, liquid biopsies will never reach the quality offered by tumor tissue–based or tumor cell–based testing (21, 23).
Liquid biopsies have a high positive, but low negative predictive value; these values may vary significantly, depending on the tested tumor entity (table 2) (23– 25). This diagnostic uncertainty inherent to liquid biopsy can be minimized by additional testing for a known mutation of the tumor. The known mutation acts as a reference value or control for the significance and reliability of the tumor genetic analysis from the liquid biopsy. Known mutations occurring early in carcinogenesis (truncal mutation), e.g. mutations in the APC (adenomatous polyposis coli) gene or the KRAS (Kirsten-rat sarcoma) gene in colorectal cancer are suitable for use as reference values (34).
These reference mutations are obtained by analyzing tissue from the primary tumor. In case of the resistance-conferring T790M mutation of NSCLC under TKI treatment, the primary activating EGFR gene mutation, in particular, represents a reliable reference value for liquid biopsy as it occurs in all cells harboring the T790M mutation. If the reference value mutation cannot be detected, it indicates that the amount of ctDNA in cfDNA is inadequate. This deficit occurs in one third of all tests from liquid biopsies (23). In such cases, tumor genetic analysis from a tissue biopsy should be pursued (figure 1).
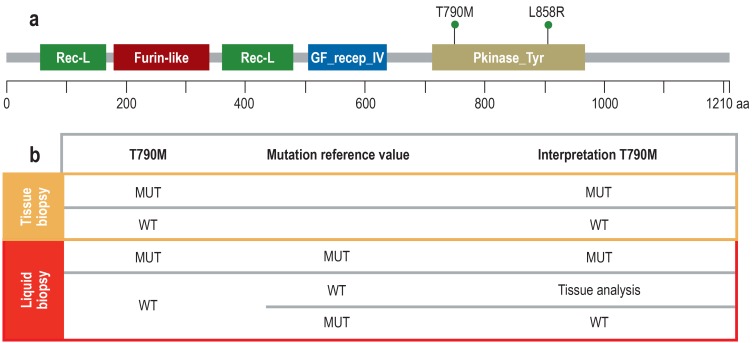
Figure 1.
Mutation reference value to assure the quality of liquid biopsy–based results
a) The diagram shows the epidermal growth factor receptor (EGFR) protein with domains (protein family [Pfam] nomenclature) and the two mutations L858R (activating mutation, exon 21) and T790M (resistance-mediated mutation, exon 20). The two mutations are located on the same deoxyribonucleic acids (DNA) strand (cis position) because the resistance-conferring T790M mutation only has an effect in the context of the activating mutation.
b) Since in tissue it is possible to exert histomorphological control over the number and proportion of tumor cells, the results of tumor genetic tests are reliable. In liquid biopsy, neither the absolute nor the relative proportion of ctDNA in the cfDNA is known; thus, any result is only reliable if a mutation (MUT) has been detected. If no mutation is detected, this can be due either to a wild type (WT) or to a sensitivity problem. However, in these cases it is helpful to analyze a mutation reference value. If this mutation can be detected, the amount of ctDNA present was adequate. Consequently, no T790M mutation (WT) is present. In the other case, a technical problem prevented mutation detection. Then, the tissue biopsy has to be analyzed to obtain a meaningful and clinically relevant test result. Since the T790M mutation represents a transition of cytosine (C) to thymine (T) (c.2369C>T), it can also occur by spontaneous deamination of C to uracil (U) which the mutation analysis would interpret as a mutation. Thus, it is important in this context too that the primary EGFR gene mutation is also tested for to be able to better distinguish between mutation and artificial transition.
Rec-L, receptor L-domain; Furin-like, furin-like cysteine rich region; GF recep_IV, growth factor receptor domain IV; Pkinase_Tyr, protein tyrosine kinase; aa, amino acid
Comparison of tissue biopsy and liquid biopsy
Despite numerous publications on liquid biopsy, so far only the pivotal study that led to the approval of osimertinib has provided reliable, tumor- and treatment-relevant data based on a large patient population and performed a head-to-head comparison of mutation detection in tissue biopsy and liquid biopsy (21, 23). In comparison with tissue-based testing, liquid biopsy detected tumor genetic mutations with a sensitivity of 0.7, a specificity of 0.69, a positive predictive value of 0.86 and a negative predictive value of 0.46.
Furthermore, liquid biopsy changed in 31% of cases (18 of 58) a negative tissue-based assay result to a positive mutation finding. By contrast, tissue analysis changed in 54% of cases (47 of 87) a negative liquid biopsy-based assay result to a positive tumor mutation finding (23).
These results clearly show that tumor genetic mutation results from the tissue-based assay and from the liquid biopsy complement each other in the detection of the T790M mutation in NSCLC and in combination provide the most reliable result for a patient. In addition, these data show that the tissue-based assay provides the correct result with a higher degree of certainty compared to liquid biopsy–based testing and should thus be given preference, whenever possible. If both a tissue biopsy and a liquid biopsy are available at the same time, initially the tumor genetic analysis of the tissue should be performed, and only in case of a negative result, the liquid biopsy should be analyzed.
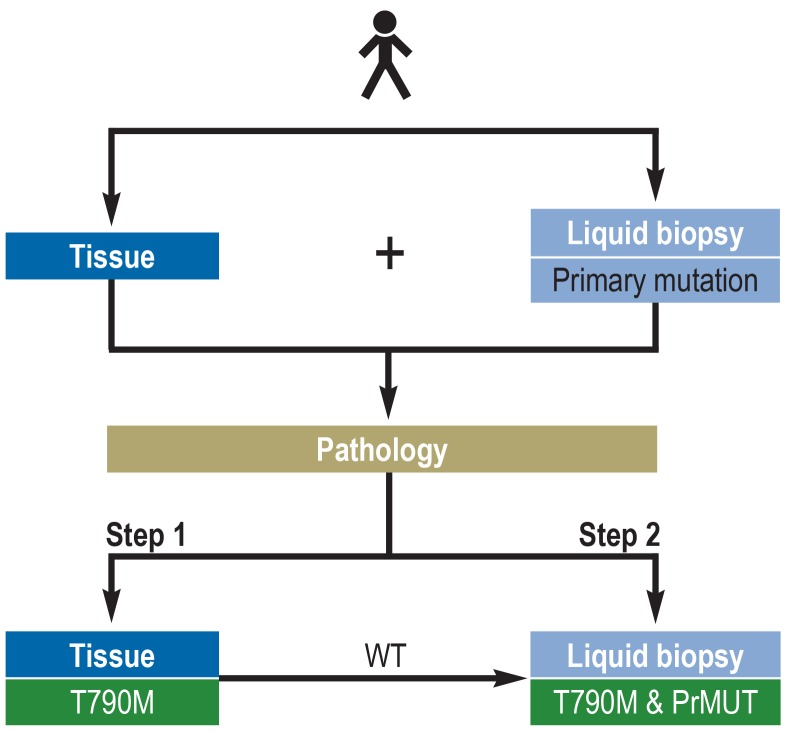
Given its low sensitivity and, above all, its low negative predictive value, the assay for T790M mutation should always also test for the known activating EGFR mutation as a reference value to obtain a meaningful test result (figure 2).
Figure 2.
Flowchart of the molecular pathological analysis of T790M resistance mutation
Ideally, both tissue biopsies and liquid biopsies are obtained from a tyrosine kinase inhibitor (TKI)–resistant lung cancer. Together with exact information about the primary mutation as a reference mutation, both specimens are analyzed in the pathology department. In line with quality control and the associated increased diagnostic certainty, mutation analysis of the tissue is performed first (Step 1). In case the T790M mutation is detected, the molecular pathological analysis can be stopped. However, if no T790M mutation is identified, the next step is to analyze in parallel the cfDNA from the liquid biopsy for T790M mutation and for the primary mutation (Step 2). With this approach, the detection rate can be increased by approximately 30% compared to performing only one analysis (only tissue or only liquid biopsy) (23). The complementary use of traditional tissue biopsy and liquid biopsy leads to the highest certainty of detection and thus provides the greatest benefit to the patient.
PrMUT, primary mutation; WT, wild type
Since the technical and time requirements for diagnosis from liquid biopsies and tissue biopsy are comparable, no significant increase in costs is to be expected.
KEY MESSAGES.
Liquid biopsy and tissue biopsy represent complementary methods for the detection of tumor genetic changes.
Successful mutation detection is of comparable value in the tissue biopsy and in the liquid biopsy (positive predictive value).
If no mutation is detected in the liquid biopsy, interpretation of this result is not possible due to the low negative predictive value of this method.
The search for mutations in liquid biopsies should include a known mutation of the tested tumor as a reference value or a reference mutation.
The European Medicines Agency has approved the use of liquid biopsies in non–small cell lung cancer (NSCLC) when material from tissue biopsies is not available in sufficient quantity.
Acknowledgments
Translated from the original German by Ralf Thoene, MD.
Footnotes
Conflict of interest statement
Prof. Jung received consultancy fees from Amgen, AstraZeneca, Biocartis, Bristol-Myers Squibb, Novartis, and Roche Pharma. He received reimbursement of travel expenses from AstraZeneca and Biocartis as well as lecture fees from AstraZeneca.
Prof. Kirchner received consultancy fees from Amgen, AstraZeneca, Bristol-Myers Squibb, Merck KGaA, MSD Sharp-Dome, Novartis, Pfizer, and Roche Pharma. He received lecture fees and reimbursement of travel expenses from AstraZeneca.
References
- 1.Mandel P, Metais P. The nucleic acids in blood plasma in humans. C R Seances Soc Biol Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 2.Bianchi DW, Chudova D, Sehnert AJ, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314:162–169. doi: 10.1001/jama.2015.7120. [DOI] [PubMed] [Google Scholar]
- 3.Bruckl WM, Wirtz RM, Bertsch T, Ficker JH, Jung A. [Liquid biopsy: detection of molecular markers for treatment decisions in lung cancer] Pneumologie. 2017;71:151–163. doi: 10.1055/s-0042-123803. [DOI] [PubMed] [Google Scholar]
- 4.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 5.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3007094. 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siravegna G, Bardelli A. Genotyping cell-free tumor DNA in the blood to detect residual disease and drug resistance. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res. 2016;14:898–908. doi: 10.1158/1541-7786.MCR-16-0044. [DOI] [PubMed] [Google Scholar]
- 10.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 11.Spornraft M, Kirchner B, Haase B, Benes V, Pfaffl MW, Riedmaier I. Optimization of extraction of circulating RNAs from plasma—enabling small RNA sequencing. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107259. e107259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taverna S, Giallombardo M, Gil-Bazo I, et al. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget. 2016;7:28748–28760. doi: 10.18632/oncotarget.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calverley DC, Phang TL, Choudhury QG, et al. Significant downregulation of platelet gene expression in metastatic lung cancer. Clin Transl Sci. 2010;3:227–232. doi: 10.1111/j.1752-8062.2010.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16:717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alix-Panabieres C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson DR, Dracopoli N, Petty BG, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med. 2012;10 138. doi: 10.1186/1479-5876-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilie M, Hofman P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res. 2016;5:420–423. doi: 10.21037/tlcr.2016.08.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mino-Kenudson M. Cons: Can liquid biopsy replace tissue biopsy?—the US experience. Transl Lung Cancer Res. 2016;5:424–427. doi: 10.21037/tlcr.2016.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenizia F, de Luca A, Pasquale R, et al. EGFR mutations in lung cancer: from tissue testing to liquid biopsy. Future Oncol. 2015;11:1611–1623. doi: 10.2217/fon.15.23. [DOI] [PubMed] [Google Scholar]
- 21.Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 22.Mao C, Yuan JQ, Yang ZY, Fu XH, Wu XY, Tang JL. Blood as a substitute for tumor tissue in detecting EGFR mutations for guiding EGFR TKIs treatment of nonsmall cell lung cancer: a systematic review and meta-analysis. Medicine (Baltimore) 2015;94 doi: 10.1097/MD.0000000000000775. e775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:3375–3382. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014;4 doi: 10.1038/srep06269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmiegel W, Scott RJ, Dooley S, et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol. 2017;11:208–219. doi: 10.1002/1878-0261.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–267. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norton SE, Lechner JM, Williams T, Fernando MR. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem. 2013;46:1561–1565. doi: 10.1016/j.clinbiochem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. BEAMing up for detection and quantification of rare sequence variants. Nat Methods. 2006;3:95–97. doi: 10.1038/nmeth850. [DOI] [PubMed] [Google Scholar]
- 30.Didelot A, Kotsopoulos SK, Lupo A, et al. Multiplex picoliter-droplet digital PCR for quantitative assessment of DNA integrity in clinical samples. Clin Chem. 2013;59:815–823. doi: 10.1373/clinchem.2012.193409. [DOI] [PubMed] [Google Scholar]
- 31.Sarwat M, Yamdagni MM. DNA barcoding, microarrays and next generation sequencing: recent tools for genetic diversity estimation and authentication of medicinal plants. Crit Rev Biotechnol. 2016;36:191–203. doi: 10.3109/07388551.2014.947563. [DOI] [PubMed] [Google Scholar]
- 32.Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA. 2015;112 E1317-25. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–481. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 34.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]