Abstract
Chloroplasts are present in organisms belonging to the kingdom Plantae. These organelles are thought to have originated from photosynthetic cyanobacteria through endosymbiosis. During endosymbiosis, most cyanobacterial genes were transferred to the host nucleus. Therefore, most chloroplast proteins became encoded in the nuclear genome and must return to the chloroplast after translation. The N-terminal cleavable transit peptide (TP) is necessary and sufficient for the import of nucleus-encoded interior chloroplast proteins. Over the past decade, extensive research on the TP has revealed many important characteristic features of TPs. These studies have also shed light on the question of how the many diverse TPs could have evolved to target specific proteins to the chloroplast. In this review, we summarize the characteristic features of TPs. We also highlight recent advances in our understanding of TP evolution and provide future perspectives about this important research area.
Keywords: chloroplast evolution, endosymbiosis, protein import into chloroplasts, transit peptide
INTRODUCTION
The chloroplast (a type of plastid) is a unique organelle found in organisms belonging to the kingdom Plantae that is responsible for photosynthesis, which produces the primary carbon source for these organisms. Many reactions and cellular processes occur in chloroplasts based on the availability of this primary carbon source. These metabolic reactions lead to the production of secondary metabolites, lipids, amino acids, and many plant hormones (Dempsey et al., 2011; Facchinelli and Weber, 2011; Kobayashi and Wada, 2016; Schaller and Stintzi, 2009). In addition, chloroplasts are major organelles involved in ROS production (Shapiguzov et al., 2012), and they play crucial roles in maintaining ion homeostasis, particularly Fe ions, which are crucial for metabolic reactions and photosynthetic electron transport chains (Nouet et al., 2011). Chloroplasts also play a crucial role in late embryogenesis (Kim et al., 2009).
To execute these many reactions and cellular processes, chloroplasts must contain a large number of proteins, often in large quantities. In fact, chloroplasts are thought to contain more than 3000 proteins (Leister, 2003). The chloroplast has its own genome, which encodes chloroplast proteins. However, the chloroplast genome encodes only approximately 100 genes, including chloroplast rRNA (Yagi and Shiina, 2014). Thus, the chloroplast must import most of its proteins from the cytosol. More than 95% of chloroplast proteins are encoded in the nuclear genome (Shi and Theg, 2013). These proteins are post-translationally delivered to chloroplasts after their translation on cytosolic ribosomes. Thus, protein targeting to chloroplasts is an essential process for chloroplast activity and for the normal functioning of plants. In this review, we will summarize our current understanding of how the protein-targeting mechanism might have evolved during the endosymbiotic conversion of cyanobacteria to chloroplasts, how this mechanism functions, and what is still unknown about this process by focusing on a single targeting signal, the transit peptide (TP).
ENDOSYMBIOTIC CONVERSION OF CYANOBACTERIA TO CHLOROPLASTS
Extensive research on the origin of chloroplasts suggests that chloroplasts evolved from cyanobacteria via endosymbiosis. This hypothesis is based on the similarity between various aspects of chloroplasts and cyanobacteria, including the amino acid sequences of their proteins, their photosynthetic machinery and mechanisms, and their genomes (Gould et al., 2008; McFadden, 2014). However, exactly how this occurred and the underlying mechanism of endosymbiotic conversion remain a mystery. Endosymbiosis might have occurred via a process that included at least three crucial steps: 1) certain genes were laterally transferred from the endosymbiont to the host genome; 2) the transferred genes were transcribed and translated in the host cell; and 3) after translation, these proteins were sent back to the endosymbiont from the host cell. Of these, the first step, lateral gene transfer, might have been the initial but not the determining step for endosymbiosis, as lateral gene transfer between organisms occurs frequently. Instead, step 3, the delivery of proteins from the host cytosol to the endosymbiont, might have been the most pivotal step in endosymbiotic conversion. The delivery of specific protein to the endosymbiont might have been possible due to the development of a new protein targeting mechanism to the endosymbiont. Such successful protein delivery might have facilitated further gene transfer from the endosymbiont to the host genome, leading to a severe reduction in the endosymbiont’s genome, which eventually ended up losing its independence as an organism. However, these ideas are still speculative, and the exact processes remain elusive.
DELIVERING PROTEINS FROM THE CYTOSOL TO THE CHLOROPLAST
A specific protein-targeting mechanism is required for the delivery of proteins from the cytosol to an organelle, which consists of two parts: cargo proteins and the molecular machinery. Cargo proteins carry specific information for targeting to a particular organelle. The molecular machinery usually contains many components that recognize this targeting information and execute the targeting process. If chloroplasts indeed evolved from endosymbiotic bacteria, the targeting mechanism should have developed during organellogenesis (Garg and Gould, 2016; Zimorski et al., 2014). However, how protein-targeting mechanisms developed remains unclear. Cyanobacteria are Gram-negative bacteria with two envelope membranes. Thus, in order for a protein to be imported from the cytosol to an endosymbiont undergoing organellogenesis, a specific signal sequence might have initially been attached to the protein, specifically a nucleus-encoded, chloroplast-destined protein. Subsequently, molecular machinery might have developed that allows the import process to occur more efficiently and more specifically. Thus, the development of the targeting signal must have been the most crucial event leading to the development of this protein-targeting mechanism. Currently, most proteins imported into chloroplasts contain an N-terminal cleavable signal sequence, the TP (Bruce, 2000; Lee et al., 2006).
The entire process of protein import into chloroplasts can be divided into several different steps (Fig. 1) (Jarvis, 2008; Li and Chiu, 2010; Paila et al., 2015). The first step involves specific sorting of chloroplast proteins in the cytosol after translation. After the sorting step, the proteins navigate through the cytosol. In contrast to other organelles, chloroplast-destined proteins exist transiently in an unfolded form in the cytosol. Thus, an important question is how the precursor proteins pass through the aqueous cytosol to the chloroplast in an unfolded form. The cytosol contains a protein quality control mechanism to remove unfolded or mis-folded proteins, which are highly prone to forming cytotoxic non-specific protein aggregates in the cell (Lee et al., 2009b; 2013; 2016). Thus, the unfolded chloroplast precursors must somehow be excluded from the cytosolic quality control system while they are navigating through the cytosol to chloroplasts. These precursor proteins then bind to the surface of the chloroplast. After binding to chloroplasts, the proteins are imported into these organelles via translocation through the two envelope membranes, followed by delivery into the chloroplast stroma (Fig. 1) (Lee et al., 2013). Finally, the TP is cleaved from the mature region of the protein by SPP (stromal processing peptidase). Depending on their final destination, proteins imported into the stroma are further targeted to suborganellar locations within the chloroplast (Lee et al., 2017).
Fig. 1. The multiple steps of preprotein import into chloroplasts.
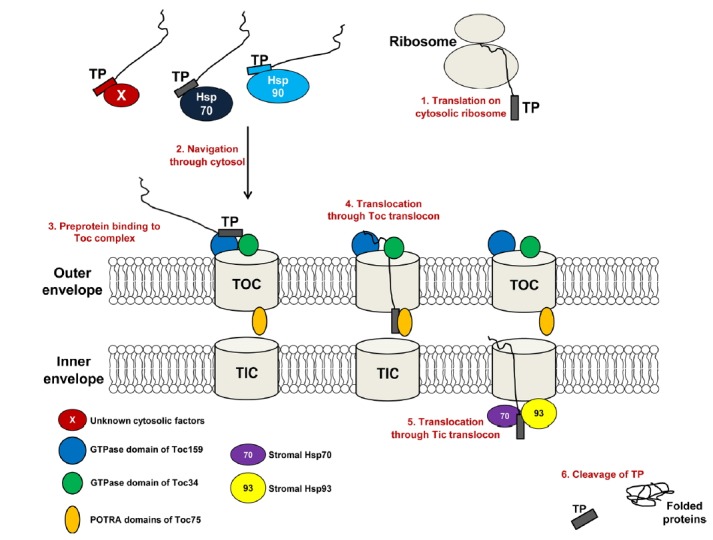
The transit peptide (TP) is a signal sequence in chloroplast interior proteins. In the cytosol, the TP is recognized by cytosolic chaperones such as Hsp70, Hsp90, or factors yet to be identified, which leads to the targeting of preproteins to the chloroplast. At the outer envelope membrane of the chloroplast, the TP interacts with the GTPase receptors Toc159 and Toc34 of the Toc complex, which initiates the translocation of the preprotein through the import channel at the outer envelope membrane. The TP of the preprotein is released from the Toc complex and recognized by the POTRA domains of the Toc75 channel, which coordinate the transfer of the preprotein to the Tic complex. The TP is released from the Tic complex and interacts with stromal chaperone Hsp70 or Hsp93, which has ATPase activity, allowing them to pull the preprotein into the chloroplast. The TP is cleaved off by a stromal processing peptidase during or after the import process.
INFORMATION ENCODED BY AND CHARACTERISTIC FEATURES OF TPs
The N-terminal TP is necessary and sufficient for the import of TP-containing proteins into chloroplasts, suggesting that TPs contain all the information needed for the entire process, from sorting in the cytosol to processing of the TP in the stroma (Chotewutmontri et al., 2017; Lee et al., 2006). However, certain TP-containing proteins are diverted to the outer or inner envelope membranes during translocation through the import channels, suggesting that some additional information may override TP-mediated translocation through the import channels into the chloroplast stroma (Lee et al., 2017; Okawa et al., 2014; Viana et al., 2010). Many studies have been conducted to elucidate the sequence information encoded by the TPs. Recent studies clearly demonstrated that TPs contain TP-specific, critical sequence motifs that are distributed throughout the entire long region of the TP (Chotewutmontri et al., 2017; Lee et al., 2006; 2008; 2009a; 2015). However, TPs show great diversity in terms of primary structure (Lee et al., 2008; 2015; Li and Teng, 2013). Despite their sequence diversity, certain features are common among the many TPs. First, in general, moderate hydrophobicity at the N-terminal regions of TPs is critical for their efficient import into chloroplasts (Chotewutmontri and Bruce, 2015; Lee et al., 2006; 2008). Moreover, the segment containing hydrophobic residues at the N-termini of TPs is thought to be part of a sequence motif recognized by stromal Hsp70, which is crucial for preprotein translocation into chloroplasts (Chotewutmontri and Bruce, 2015; Chotewutmontri et al., 2012). Second, TPs tend to have the basic residues Lys and Arg, but they lack acidic residues in the middle of their sequences (Bruce, 2000). Substituting the basic residues in TPs with alanines or acidic residues adversely affects preprotein import into chloroplasts, although the mechanism by which these basic residues contribute to their efficient import into chloroplasts is largely unknown (Razzak et al., 2017). Third, TPs contain numerous proline residues (Lee et al., 2018; Zybailov et al., 2008). Prolines in TPs are thought to be important for the unstructured nature of TPs by acting as helix breakers (Rensink et al., 2000). We recently obtained evidence that the prolines in TPs are involved in a later step of protein import, where they are crucial for efficient translocation through the chloroplast envelope membranes (Lee et al., 2018). Fourth, specific sequence motifs in TPs are recognized by cytosolic AtHsc70-4, which mediates proteasomal degradation of unimported preproteins in the cytosol to avoid cellular damage (Lee et al., 2009b; 2016). Finally, TPs contain specific motifs for import regulation, depending on the age of the plant (Teng et al., 2012). These findings suggest that the unusually long TPs contain numerous sequence motifs that support many steps involved in the long journey from the cytosol to the chloroplast stroma, as well as motifs for cytosolic quality control and import regulation.
DESIGN PRINCIPLES OF DIVERSE TPs
As described above, a great deal of research has revealed important information about the sequences of TPs. However, there is still long-lasting confusion about the information carried by TPs due to their high degree of sequence diversity. Consistent with this observation, the sequence motifs in one TP share little similarity with the sequence motifs in another TP (Lee et al., 2008; 2015; Li and Teng, 2013). Thus, it is important to explore what underlies such diversity in TPs and how they function in protein import into chloroplasts. Li and Teng (2013) proposed an interesting model for the design principle of TPs explaining how diverse TPs might have been generated and how they function. Consistent with this idea, we showed that functional hybrid TPs could be generated from the TPs of RbcS and Cab (chlorophyll a/b-binding protein). Thus, certain sequence motifs from one TP are fully interchangeable with those from another TP despite the difference in amino acid sequence (Lee et al., 2009a; 2015). In addition, we showed that whether sequence motifs are active depends on the overall context of the TPs; certain sequence motifs were active in both the original and newly generated hybrid TPs, whereas certain sequence motifs were active in the original context but became inactive in hybrid TPs, and vice versa. Moreover, we were able to generate a fully functional synthetic TP by incorporating three motifs from RbcS and Cab TPs into an N-terminal segment containing 80 amino acids from CPY (carboxypeptidase Y), a vacuolar protein (Lee et al., 2015). These results provide clues about how diverse TPs arose during chloroplast evolution and are consistent with the M&M (multi-selection and multi-order) model proposed by Li and Teng (2013). Based on these observations and the M&M model, we propose a design principle for a functional TP: in terms of molecular machinery, one or more molecular components are necessary at each step that are able to interact with multiple sequence motifs; in terms of signal sequences, a TP should have at least one binding site for each component for efficient import, which can be generated through selective assembly of multiple potential sequence motifs that can interact with each component of the molecular machinery during each step (Fig. 2) (Lee et al., 2015; Li and Teng, 2013). The presence of multiple components at a particular step and the ability of components to interact with multiple sequence motifs could underlie the diversity of TP sequences.
Fig. 2. Design principle of the diverse transit peptides.
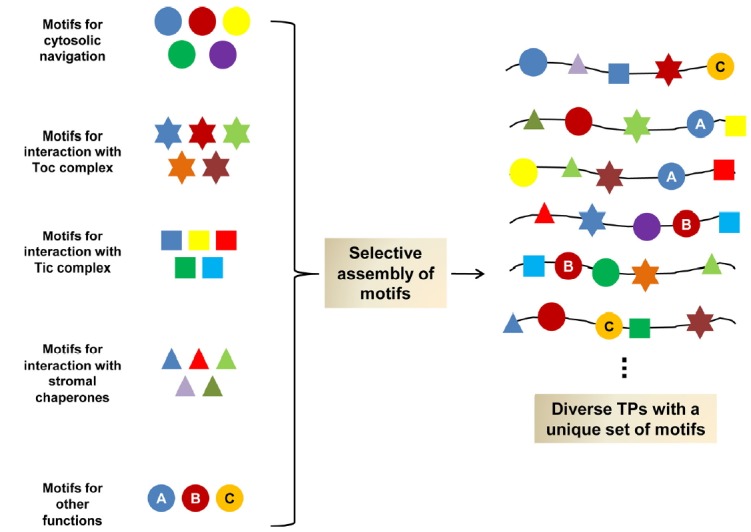
During chloroplast evolution, the diverse transit peptides (TPs) might have arisen via selective assembly of a large pool of potential motifs responsible for the interaction with import factors that function at various steps in the import process in the cytosol and at the chloroplast. We propose that one or multiple molecular factors are involved in each step of protein import and that each of these factors is able to interact with multiple motifs.
MOLECULAR MACHINERY THAT RECOGNIZES THE TP DURING PROTEIN IMPORT INTO CHLOROPLASTS
Many protein factors have been shown to play a role in protein import into chloroplasts. In the cytosol, Hsp90 and a guidance complex composed of Hsp70 and 14-3-3 recognize TPs and facilitate protein targeting to chloroplasts (May and Soll, 2000; Qbadou et al., 2006) (Fig. 1). Hsp70 and Hsp90 differentially recognize various TPs and coordinate the docking of preproteins to the Toc complex (May and Soll, 2000; Qbadou et al., 2006). In addition, Hsc70-4 and At-BAG 1 (Arabidopsis thaliana Bcl2-associated athanogene 1) play important roles in preventing the accumulation of unimported precursors in the cytosol via the ubiquitin-mediated 26S proteasome pathway (Lee et al., 2009b; 2016). AtHsc70-4 specifically binds to the sequence motifs in the TP (Lee et al., 2009b). At the chloroplast surface, GTPases and Toc64 function as import receptors of preproteins. GTPases can be divided into two families, Toc159 and Toc34. Arabidopsis contains multiple isoforms in both the Toc34 and Toc159 families; the Toc159 family comprises AtToc159, AtToc132, AtToc120, and AtToc90, and the Toc34 family comprises AtToc33 and AtToc34 (Constan et al., 2004; Ivanova et al., 2004; Kubis et al., 2004). These import receptors also recognize specific sequence motifs at the TP (Fig. 1) (Chotewutmontri et al., 2012; Holbrook et al., 2016; Lee et al., 2009a). The β-barrel protein, Toc75, functions as a translocation channel at the outer envelope membrane. TPs that translocate through the Toc75 channel are recognized by the polypeptide transport-associated (POTRA) domains of Toc75 at the intermembrane space (Fig. 1), which coordinates the transfer of TPs to the Tic complex located at the inner envelope membrane (Paila et al., 2016). The exact nature of the Tic translocon remains controversial. However, Tic20 is generally regarded as the major translocation channel at the inner envelope of the chloroplast (de Vries et al., 2015; Kikuchi et al., 2009; 2013; Nakai, 2015). In the stroma, multiple chaperones such as cpHsc70, Hsp93, and Hsp90C pull preproteins into the chloroplast (Fig. 1) (Inoue et al., 2013; Liu et al., 2014; Su and Li, 2010). The multiple proline residues in TPs may be recognized by these chaperones for efficient translocation through the outer and inner import channels (Lee et al., 2018). Another important factor is SPP, which cleaves off the TP during or after translocation (Richter and Lamppa, 1999; Trosch and Jarvis, 2011).
CONCLUSION
We now have a much deeper understanding of the nature of the information encoded by the TP and how this sequence information is decoded by the molecular machinery. This information sheds new light on the processes involved in endosymbiotic conversion. However, many questions remain regarding the exact mechanisms of action during the import process. These questions include how the TPs are actually translocated through the import channel and how they are specifically sorted in the cytosol, particularly for proteins with hydrophobic TMDs, which, if not properly sorted, might possibly be targeted to the ER (endoplasmic reticulum). It will be challenging to answer these questions, but the answers will certainly increase our understanding of the complicated mechanisms for protein import into chloroplasts.
Another interesting question about protein targeting to chloroplasts is how chloroplast proteins are specifically delivered to chloroplasts but not to mitochondria in plant cells. Eukaryotic cells can be divided into two groups based on the presence of two endosymbiotic organelles: chloroplasts and mitochondria. Animals and fungi contain only mitochondria, whereas plants and algae contain both organelles. Thus, it remains elusive whether plant cells require a mechanism to ensure specific targeting between these organelles and if such mechanism exists, how it functions. This question arises because TPs share a high degree of similarity with the targeting signals of mitochondrial proteins (Bhushan et al., 2006; Schleiff and Becker, 2011). In both organelles, more than 90% of organellar proteins are imported from the cytosol after translation. Like chloroplast proteins, most mitochondrial preproteins also contain a long, cleavable N-terminal signal sequence, termed the presequence (Lee et al., 2012). Moreover, presequences and TPs have similar amino acid compositions, including a high degree of hydroxylated amino acids, high proline residue contents, and a lack of acidic residues (Bhushan et al., 2006). Of course, presequences differ from TPs in some ways, such as their high propensity to form an amphiphilic α-helical structure (Abe et al., 2000). It is currently unclear how the targeting mechanisms of chloroplast and mitochondrial proteins have become similar to each other. One possibility is that evolutionary interactions occurred between the two organelles during the development of the targeting mechanisms; the targeting mechanisms of the organelle that evolved earlier might have contributed to those of the other organelle. These issues need to be addressed in the future to fully understand how proteins are specifically targeted to chloroplasts by TPs.
ACKNOWLEDGMENTS
This research was carried out with the support of the “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ010953012018)”, Rural Development Administration, Republic of Korea. Dong Wook Lee was supported by grant no. NRF-2017R1C1B1006784 from the National Research Foundation, the Ministry of Science, and Information and Computer Technology, Korea.
REFERENCES
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Bhushan S, Kuhn C, Berglund AK, Roth C, Glaser E. The role of the N-terminal domain of chloroplast targeting peptides in organellar protein import and miss-sorting. FEBS Lett. 2006;580:3966–3972. doi: 10.1016/j.febslet.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Bruce BD. Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 2000;10:440–447. doi: 10.1016/s0962-8924(00)01833-x. [DOI] [PubMed] [Google Scholar]
- Chotewutmontri P, Bruce BD. Non-native, N-terminal Hsp70 molecular motor recognition elements in transit peptides support plastid protein translocation. J Biol Chem. 2015;290:7602–7621. doi: 10.1074/jbc.M114.633586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Reddick LE, McWilliams DR, Campbell IM, Bruce BD. Differential transit peptide recognition during preprotein binding and translocation into flowering plant plastids. Plant Cell. 2012;24:3040–3059. doi: 10.1105/tpc.112.098327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Holbrook K, Bruce BD. Plastid Protein Targeting: Preprotein Recognition and Translocation. Int Rev Cell Mol Biol. 2017;330:227–294. doi: 10.1016/bs.ircmb.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Constan D, Patel R, Keegstra K, Jarvis P. An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 2004;38:93–106. doi: 10.1111/j.1365-313X.2004.02024.x. [DOI] [PubMed] [Google Scholar]
- de Vries J, Sousa FL, Bolter B, Soll J, Gould SB. YCF1: A Green TIC? Plant Cell. 2015;27:1827–1833. doi: 10.1105/tpc.114.135541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF. Salicylic Acid biosynthesis and metabolism. Arabidopsis Book. 2011;9:e0156. doi: 10.1199/tab.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinelli F, Weber AP. The metabolite transporters of the plastid envelope: an update. Front Plant Sci. 2011;2:50. doi: 10.3389/fpls.2011.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg SG, Gould SB. The role of charge in protein targeting evolution. Trends Cell Biol. 2016;26:894–905. doi: 10.1016/j.tcb.2016.07.001. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Holbrook K, Subramanian C, Chotewutmontri P, Reddick LE, Wright S, Zhang H, Moncrief L, Bruce BD. Functional analysis of semi-conserved transit peptide motifs and mechanistic implications in precursor targeting and recognition. Mol Plant. 2016;9:1286–1301. doi: 10.1016/j.molp.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Inoue H, Li M, Schnell DJ. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Pro Natl Acad Sci USA. 2013;110:3173–3178. doi: 10.1073/pnas.1219229110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova Y, Smith MD, Chen K, Schnell DJ. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol Biol Cell. 2004;15:3379–3392. doi: 10.1091/mbc.E03-12-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008;179:257–285. doi: 10.1111/j.1469-8137.2008.02452.x. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Oishi M, Hirabayashi Y, Lee DW, Hwang I, Nakai M. A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell. 2009;21:1781–1797. doi: 10.1105/tpc.108.063552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Bedard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science. 2013;339:571–574. doi: 10.1126/science.1229262. [DOI] [PubMed] [Google Scholar]
- Kim C, Lee KP, Baruah A, Nater M, Gobel C, Feussner I, Apel K. (1)O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid. Proc Natl Acad Sci USA. 2009;106:9920–9924. doi: 10.1073/pnas.0901315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Wada H. Role of lipids in chloroplast biogenesis. Subcell Biochem. 2016;86:103–125. doi: 10.1007/978-3-319-25979-6_5. [DOI] [PubMed] [Google Scholar]
- Kubis S, Patel R, Combe J, Bedard J, Kovacheva S, Lilley K, Biehl A, Leister D, Rios G, Koncz C, et al. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell. 2004;16:2059–2077. doi: 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Lee S, Lee GJ, Lee KH, Kim S, Cheong GW, Hwang I. Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol. 2006;140:466–483. doi: 10.1104/pp.105.074575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kim JK, Lee S, Choi S, Kim S, Hwang I. Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell. 2008;20:1603–1622. doi: 10.1105/tpc.108.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Lee S, Oh YJ, Hwang I. Multiple sequence motifs in the Rubisco small subunit transit peptide independently contribute to Toc159-dependent import of proteins into chloroplasts. Plant Physiol. 2009a;151:129–141. doi: 10.1104/pp.109.140673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, Jurgens G, Hwang I. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell. 2009b;21:3984–4001. doi: 10.1105/tpc.109.071548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DW, Yoo YJ, Duncan O, Oh YJ, Lee YJ, Lee G, Whelan J, Hwang I. Mitochondrial targeting of the Arabidopsis F1-ATPase gamma-subunit via multiple compensatory and synergistic presequence motifs. Plant Cell. 2012;24:5037–5057. doi: 10.1105/tpc.112.105361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Jung C, Hwang I. Cytosolic events involved in chloroplast protein targeting. Biochim Biophys Acta. 2013;1833:245–252. doi: 10.1016/j.bbamcr.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Lee DW, Woo S, Geem KR, Hwang I. Sequence motifs in transit peptides act as independent functional units and can be transferred to new sequence contexts. Plant Physiol. 2015;169:471–484. doi: 10.1104/pp.15.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Kim SJ, Oh YJ, Choi B, Lee J, Hwang I. Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins. Mol Plant. 2016;9:1428–1431. doi: 10.1016/j.molp.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Lee DW, Lee J, Hwang I. Sorting of nuclear-encoded chloroplast membrane proteins. Curr Opin Plant Biol. 2017;40:1–7. doi: 10.1016/j.pbi.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Lee DW, Yoo YJ, Razzak MA, Hwang I. Prolines in transit peptides Are crucial for efficient preprotein translocation into chloroplasts. Plant Physiol. 2018;176:663–677. doi: 10.1104/pp.17.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. Chloroplast research in the genomic age. Trends Genet. 2003;19:47–56. doi: 10.1016/s0168-9525(02)00003-3. [DOI] [PubMed] [Google Scholar]
- Li HM, Chiu CC. Protein Transport into Chloroplasts. Ann Rev Plant Biol. 2010;61:157–180. doi: 10.1146/annurev-arplant-042809-112222. [DOI] [PubMed] [Google Scholar]
- Li HM, Teng YS. Transit peptide design and plastid import regulation. Trends Plant Sci. 2013;18:360–366. doi: 10.1016/j.tplants.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Liu L, McNeilage RT, Shi LX, Theg SM. ATP Requirement for Chloroplast Protein Import Is Set by the K-m for ATP Hydrolysis of Stromal Hsp70 in Physcomitrella patens. Plant Cell. 2014;26:1246–1255. doi: 10.1105/tpc.113.121822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Soll J. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–64. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. Origin and evolution of plastids and photosynthesis in eukaryotes. Cold Spring Harb Perspect Biol. 2014;6:a016105. doi: 10.1101/cshperspect.a016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M. YCF1: A green TIC: response to the de Vries et al. Commentary Plant Cell. 2015;27:1834–1838. doi: 10.1105/tpc.15.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouet C, Motte P, Hanikenne M. Chloroplastic and mitochondrial metal homeostasis. Trends Plant Sci. 2011;16:395–404. doi: 10.1016/j.tplants.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Okawa K, Inoue H, Adachi F, Nakayama K, Ito-Inaba Y, Schnell DJ, Uehara S, Inaba T. Targeting of a polytopic membrane protein to the inner envelope membrane of chloroplasts in vivo involves multiple transmembrane segments. J Exp Bot. 2014;65:5257–5265. doi: 10.1093/jxb/eru290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila YD, Richardson LGL, Schnell DJ. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J Mol Biol. 2015;427:1038–1060. doi: 10.1016/j.jmb.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila YD, Richardson LG, Inoue H, Parks ES, McMahon J, Inoue K, Schnell DJ. Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. Elife. 2016;5 doi: 10.7554/eLife.12631. pii: e12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qbadou S, Becker T, Mirus O, Tews I, Soll J, Schleiff E. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J. 2006;25:1836–1847. doi: 10.1038/sj.emboj.7601091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzak MA, Lee DW, Yoo YJ, Hwang I. Evolution of rubisco complex small subunit transit peptides from algae to plants. Sci Rep. 2017;7 doi: 10.1038/s41598-017-09473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink WA, Schnell DJ, Weisbeek PJ. The transit sequence of ferredoxin contains different domains for translocation across the outer and inner membrane of the chloroplast envelope. J Biol Chem. 2000;275:10265–10271. doi: 10.1074/jbc.275.14.10265. [DOI] [PubMed] [Google Scholar]
- Richter S, Lamppa GK. Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J Cell Biol. 1999;147:33–43. doi: 10.1083/jcb.147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Stintzi A. Enzymes in jasmonate biosynthesis - structure, function, regulation. Phytochem. 2009;70:1532–1538. doi: 10.1016/j.phytochem.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Becker T. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol. 2011;12:48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- Shapiguzov A, Vainonen JP, Wrzaczek M, Kangasjarvi J. ROS-talk - how the apoplast, the chloroplast, and the nucleus get the message through. Front Plant Sci. 2012;3 doi: 10.3389/fpls.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM. The chloroplast protein import system: from algae to trees. Biochim Biophys Acta. 2013;1833:314–331. doi: 10.1016/j.bbamcr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Su PH, Li HM. Stromal Hsp70 Is Important for Protein Translocation into Pea and Arabidopsis Chloroplasts. Plant Cell. 2010;22:1516–1531. doi: 10.1105/tpc.109.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YS, Chan PT, Li HM. Differential age-dependent import regulation by signal peptides. Plos Biol. 2012;10:e1001416. doi: 10.1371/journal.pbio.1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trosch R, Jarvis P. The stromal processing peptidase of chloroplasts is essential in Arabidopsis, with knockout mutations causing embryo arrest after the 16-cell stage. PLoS One. 2011;6:e23039. doi: 10.1371/journal.pone.0023039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana AA, Li M, Schnell DJ. Determinants for stop-transfer and post-import pathways for protein targeting to the chloroplast inner envelope membrane. J Biol Chem. 2010;285:12948–12960. doi: 10.1074/jbc.M110.109744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Shiina T. Recent advances in the study of chloroplast gene expression and its evolution. Front Plant Sci. 2014;5:61. doi: 10.3389/fpls.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF, Gould SB. Endosymbiotic theory for organelle origins. Curr Opin Microbiol. 2014;22:38–48. doi: 10.1016/j.mib.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]


