Abstract
Hypoxic culture is widely recognized as a method to efficiently expand human mesenchymal stem cells (MSCs) without loss of stem cell properties. However, the molecular basis of how hypoxia priming benefits MSC expansion remains unclear. In this report, our systemic quantitative proteomic and RT-PCR analyses revealed the involvement of hypoxic conditioning activated genes in the signaling process of the mitotic cell cycle. Introduction of screened two mitotic cyclins, CCNA2 and CCNB1, significantly extended the proliferation lifespan of MSCs in normoxic condition. Our results provide important molecular evidence that multipotency of human MSCs by hypoxic conditioning is determined by the mitotic cell cycle duration. Thus, the activation of mitotic cyclins could be a potential strategy to the application of stem cell therapy.
Keywords: cell proliferation lifespan, cyclin, multipotency, mitosis, human mesenchymal stem cell, hypoxia
INTRODUCTION
Various types of human mesenchymal stem cells (MSCs) reside in the hypoxic microenvironment, which seems to be conductive to stem cell longevity and the physiological niches (Davy and Allsopp, 2011; Tsai et al., 2011). Hypoxic conditions may be essential for the self-renewal and the maintenance of multipotency of human MSCs and hematopoietic stem cells (HSCs) (Rosova et al., 2008; Suda et al., 2011; Tsai et al., 2011). Indeed, the hypoxic culture of human MSCs inhibits cellular senescence, maintains MSCs properties, augments the differentiation capacity, and enhances their tissue regenerative potential, indicating that hypoxia increases the lifespan and the differentiation potential of MSCs (Mathieu et al., 2014; Rosova et al., 2008; Zhang et al., 2012).
In contrast to differentiated cells, stem cells mainly rely on glycolysis for their source of energy, which is very similar to cancer cells (Cairns et al., 2011; Mathieu et al., 2014). For example, HSCs generate energy mainly via anaerobic metabolism by maintaining a high rate of glycolysis for their function and long-term self-renewal (Suda et al., 2011). Moreover, MSCs also share the distinct metabolic properties of upregulated glycolytic genes, reduced mitochondria activity, and markedly increased lactate production (Mathieu et al., 2014; Varum et al., 2011; Yanes et al., 2010). Metabolic properties of stem cells appear to be important for their ability and long-term maintenance in the body (Greer et al., 2012; Rafalski et al., 2003), although the mechanics of these processes remain unclear.
Hypoxic culture is an efficient tool for the generation of MSCs with therapeutic properties (Das et al., 2012; Hu, 2014; Nagano et al., 2010; Suda et al., 2011; Tsai et al., 2011). Interestingly, similar to cancer cells, in hypoxic culture, MSCs have distinct metabolic requirements and their bioenergetics depend on a shift from oxidative to glycolytic metabolism (Cairns et al., 2011; Ito and Suda, 2014; Pattappa et al., 2011). The dependency of stem cells on glycolysis to produce ATP could be an adaptation to low-oxygen tension, given that hypoxia is a key feature of the stem cell niche (Mathieu et al., 2014; Mohyeldin et al., 2010; Suda et al., 2011). Although cellular adaptation to hypoxic conditions seems to be mediated mainly through the activation of hypoxic-inducible factors (HIFs), how hypoxic conditioning induces the metabolic switching to glycolysis and enhances differentiation potential remain unclear. Moreover, it is not yet clear whether the benefit of hypoxic conditioning is the expansion, cellular longevity, or multi-potent differentiation capacity of human MSCs. In this study, we found that hypoxic conditioning expands the mitotic cell cycle lifespan, which seems to confer the multipotency of differentiation lineage of MSCs.
MATERIALS AND METHODS
Cell culture
Human umbilical cord blood derived mesenchymal stem cells (hUCB-MSCs; PromoCell) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Hyclone) containing 10% fetal bovine serum (FBS; GIBCO) and 1% Penicillin/Streptomycin antibiotics at 37°C in a 5% CO2 incubator with 21% O2 (normoxia) or 1% O2 (hypoxia).
Cell proliferation assay
Cell proliferation was evaluated using a colorimetric method based on water-soluble tetrazolium salts (WST-1; CellVia, Abfrontier). HA-CCNA2 or HA-CCNB1 expressing recombinant adenovirus was infected in hUCB-MSCs with HP4 and infected cells were grown in normoxic conditions. 5 × 103 cells were seeded in 96-well culture plate. After 24 h incubation, 10 μl of CellVia was added and the cells were incubated for an additional 1 h at 37°C. Cells were measured using a microplate reader at a wavelength of 450 nm.
Differentiation assay
hUCB-MSCs were seeded in a 6-well culture plate with growth mediu. For adipogenesis, cells were cultured in adipogenic medium (low glucose DMEM, 10% FBS and 1% penicillin/streptomycin supplemented with 1 μM dexamethasone, 1 μM indomethacin, 10 μg/ml insulin and 500 μM IBMX) for 3 days, then transferred to an adipocyte maintenance medium (low glucose DMEM, 10% FBS and 1% penicillin/ streptomycin supplemented with 10 μg/ml insulin) for 1 day. This differentiation medium cycle was repeated for 2 weeks. For osteogenesis, cells were cultured for 4 weeks in an osteogenic medium (low glucose DMEM, 10% FBS and 1% penicillin/streptomycin supplemented with 0.1 μM dexamethasone, 50 μM L-ascorbate-2-phosphate and 10 mM β-glycerophsphate disodium). The osteogenenic medium was changed every 3 days.
Carboxyfluorescein succinimidyl ester (CFSE) assay
For assessment of the potential of cell proliferation, MSCs were trypsinized and washed once with phosphate buffered saline (PBS). CFSE (Invitrogen, 10 mM in PBS) was added to the cells and incubated at 37°C in the dark for 15 min. An equal volume of serum containing growth medium was added for quench the CFSE reaction. Cells were again incubated at 37°C in the dark for 5 min. CFSE-labeled MSCs were washed twice with growth medium and seeded in a culture plate.
RNA isolation and quantitative real-time PCR
Total RNA was extracted using an RNeasy Mini Kit (Qiagen) and quantified using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). 1 μg of total RNA was reverse-transcribed using an AccuPower RT premix (Bioneer). Real-time RT–PCR was performed using a LightCycler 480 system (Roche). The specific primers used were as follows: GAPDH (F 5′-CGA GAT CCC TCC AAA ATC AA-3′, R 5′-TGT GGT CAT GAG TCC TTC CA-3′); PPARgamma (F 5′-CTA AAG AGC CTG CGA AAG-3′, R 5′-TGT CTG TCT CCG TCT TCT TG-3′); LPL (F 5′-TCA ACT GGA TGG AGG AG-3′, R 5′-GGG GCT TCT GCA TAC TCA AA-3′); ALP (F 5′-CAA CAG GGT AGA TTT CTC TTG G-3′, R5′-GGT CAG ATC CAG AAT GTT CC-3′); Osteocalcin (F 5′-GGC AGC GAG GTA GTG AAG AG-3′, R 5′-CAG CAG AGC GAC ACC CTA GAC-3′)
RT2 profiler PCR array
RT2 profiler PCR array [Human Mesenchymal Stem Cells (PAHS-082ZA-2), Human Cell Cycle (PAHS-020ZA-2), Human Stem Cell Signaling (PAHS-047ZA-2)] (SA Biosciences) was performed using QuantStudio 6 flex Real-Time PCR (Life Technologies). Each array contained a panel of 84 primer sets related to the specific signaling genes plus five housekeeping genes and two negative controls. All analyses were performed using the web-based PCR Array Data Analysis Software available on the SA Biosciences website. Gene expression was normalized using a set of five housekeeping genes and fold regulation was used to analyze changes in gene expression. Gene expression detected with the RT2 SYBR Green qPCR master Mix (SA Biosciences).
Stable isotope labeling by amino acids in cell culture (SILAC)
Human UCB-MSCs were grown in 1% hypoxia with L-arginine-13C615N4 and L-lysine-13C615N2 supplemented growth medium (heavy weight medium) while control cells were grown in L-arginine-13C6 and L-lysine-D4 supplemented growth medium (light weight medium) in 20% normoxia for at least five doublings. Normoxia cells were labeled with supplemented light weight medium containing L-arginine and L-lysine without label. Pellets were lysed by NE buffer (20 mM HEPES pH 7.6, 250 mM NaCl, 1.5 mM MgCl2, 20% glycerol, 0.1% triton X-100, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail (P3100-010, GeneDepot).
Cell lysis and Western blotting
Cells were lysed by NE buffer and the lysates were clarified by centrifugation at 13,200 rpm for 30 min at 4°C. After being blocked in TBS-T (20 mM Tris, 137 mM NaCl, 0.1 % Tween-20, pH7.4) with 5% skim milk, membranes were incubated with the primary antibody at an appropriate dilution in TBS-T containing 5% skim milk or 3% bovine serum albumin for 3 h at room temperature or overnight in a cold room. Secondary antibody (anti-rabbit- or anti-mouse-conjugated horseradish peroxidase; Bethyl) was followed by ECL (AB Frontier) for visualization.
Bioinformatics analysis
The Uniprot accession IDs of the differentially regulated proteins were submitted to DAVID bioinformatics tool (http://david.abcc.ncifcrf.gov) or IPA (http://www.ingenuity.com) to perform functional annotation clustering and assign over-represented GO terms.
Fluorescence-activated cell sorting (FACS)
For cell cycle analysis using propidium iodide, cells were fixed with cold 70% ethanol overnight at −20°C. They were then washed once with PBS and incubated in PBS containing 50 μg/ml propidium iodide and 1 mg/ml RNase A for 30 min at room temperature. After staining, cells were washing with PBS and measured by flow cytometry.
Generation of recombinant adenoviruses
The human CCNA2 (KU009360) and human CCNB1 (BKU006297) clones were purchased from Korea Human Gene Bank (Medical Genomics Research Center, KRIBB, Korea). DNA fragments were excised and inserted into pShuttle vectors (Quantum). Adenoviral vectors were subsequently generated by recombination of the plasmids in BJ5183-AD-1 cells (Agilent Technologies). The resulting recombinant DNAs digested with PacI (NEB). Recombinant adenoviruses were recovered from 293A cells that were polyethylenimine (PEI)-transfected with the linearized recombinant adenoviral DNA. HP4, a type of cell penetrating protein transduction domain (PTD), was synthesized by PEPTRON, Inc. A mixture of recombinant adenovirus (rAdv) at a multiplicity of infection (MOI) of 100 plaque-forming units (PFU) per cell, and HP4 (100 nM) was incubated in a serum-free medium for 30 min at room temperature. The cells were washed and incubated with the rAdv and HP4 preparation. After 2 h, cells were washed and incubated with a serum-containing medium (Youn et al., 2008).
Statistical analysis
Data values were expressed as the mean ± SEM. The significant differences between groups were analyzed with Student t-test. A value of p < 0.05 was considered statistically significant.
RESULTS AND DISCUSSION
Hypoxia upregulates genes involved in cell cycle progression
To examine the protein expression profiles induced by hypoxic conditioning, human MSCs were isolated from human umbilical cord blood (UCB) by adhesion to tissue culture-coated plates in complete culture medium as previously described (Nagano et al., 2010). Human MSCs were cultured under normoxic (21% oxygen) or hypoxic (1% oxygen) conditions. When the culture became nearly confluent, the cells were trypsinized and subcultivated. As expected, hypoxia significantly accelerated the proliferation rate and extended the life span of human MSCs compared to normoxic cultures. Similar results were obtained with MSCs from two different donors (data not shown). Next, we employed the stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomic analysis was done (Fig. 1). Normoxic and hypoxic primed MSCs were cultured in both light and heavy SILAC media. The light-labeled cells were conditioned with normoxia and the heavy-labeled cells were conditioned with hypoxia (Fig. 1A). Cellular lysates were prepared and used to measure the hypoxia marker hypoxia inducible factor 1α (HIF1α). After in-solution protein digestion the proteins were identified and quantified by LC-MS/MS (Figs. 1B and 1C). For quantitative analysis of differences between paired experimental samples, a ratio of 1.5 or 2.0 was chosen as the threshold for screening significantly changed proteins (Figs. 1D and 1E). Using this criterion, we identified 1361 proteins that displayed significant changes in hypoxic-primed MSCs; 78 were up-regulated (log2 ratio ≥ 1) and 50 were down-regulated (log2 ratio ≤ 1) (Fig. 1E). Molecular function analysis established that the identified proteins were related with DNA replication, translation, cell cycle regulation, proliferation, glycolysis, oxidative metabolism, hypoxic responses, apoptotic signaling, mitochondrion, glutathione metabolism, anti-oxidation, and generation of precursor metabolite and energy (Fig. 1F). Importantly, the significantly up-regulated in hypoxic primed MSCs mostly mapped to the functional cellular network of cell cycle progression and proliferation (e.g., CCNA2, CCNB1, CTNNB1, HGF, EGFR, MCM6, and PCNA) and glycolytic metabolism (e.g., HK2, PFKP, PGK1, and PGAM1). The down-regulated proteins in these MSCs were mapped to apoptotic cell death (e.g., BID, CASP2, MAPK1, and PRDX1) and mitochondria metabolism (e.g., CDS2, ALDH1B1, GSTK1, MDH2, PRDX1, PPA2, and TGM2).
Fig. 1. Comparative analysis of protein expressions between normoxic and hypoxic primed MSCs by SILAC-based quantitative proteomics.
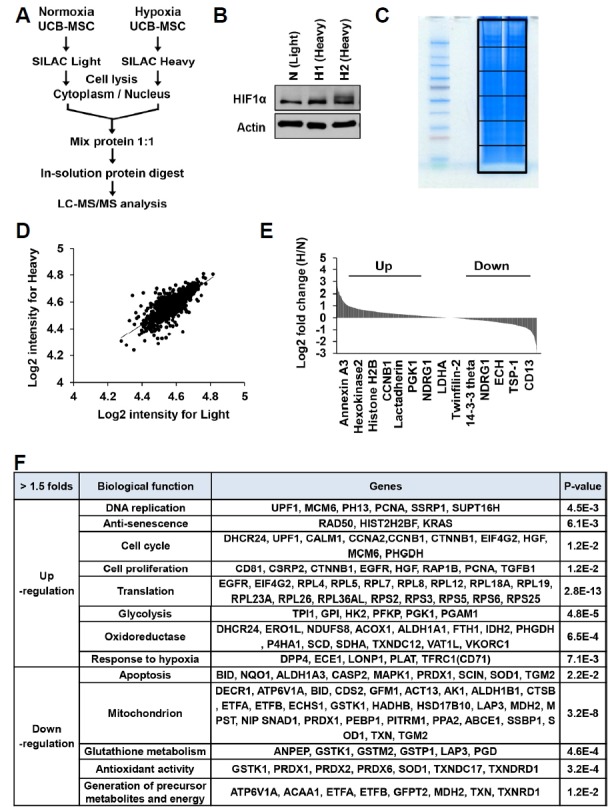
(A) Human umbilical cord blood-derived MSCs (MSCs) were cultured in normoxic (21% O2) or hypoxic (1% O2) conditions. Experimental scheme of SILAC labeling and LC-MS/MS analysis of normoxic and hypoxic primed MSCs at passage 3. (B) MSCs were grown in normoxic (light) and hypoxic (heavy) conditions, and analyzed by immunoblotting using anti-HIF1α antibody. (C) Both heavy and light samples were mixed, separated by SDS-PAGE, analyzed by in-gel-digestion, and then quantified by LC-MS/MS. (D, E) Ratio distribution of the 1361 differentially expressed proteins in hypoxic condition compared with the normoxic condition. Log2 SILAC ratios of 1.0 and −1.0 were the cut-offs for the regulated proteins (D). Representative upregulated (Up) and downregulated (Down) proteins by hypoxic conditioning were depicted (E). (F) List of proteins, which were upregulated or downregulated in hypoxic primed MSCs compared to the normoxic primed MSCs.
To further characterize the gene expression profiles of MSCs by hypoxic conditioning, mRNAs were prepared from normoxic and hypoxic primed MSCs at passage 3. Quantitative RT-PCR array analysis using gene expression profilers categorized the profiles into three different cellular signaling patterns: cell cycle, MSCs, and and stem cell. The expression levels of 12 of 84 cell cycle regulatory genes, 15 of 84 MSC marker genes, and 7 of 84 stem cell signaling genes were significantly up-regulated by hypoxic conditioning (≥ 1.5-folds) (Fig. 2A). Particularly, G1 cyclin (e.g., CCND2) and many cell cycle negative regulatory molecules (e.g., MDM2, GADD45, TP53, and CDKN3) were down-regulated in hypoxic primed MSCs (data not shown), whereas S and G2 cyclins (e.g., CCNA2) and key mitotic regulatory molecules (e.g., CDK1, CDC20, CCNB1, and AURKs) were clearly upregulated (Fig. 2B), indicating that hypoxia activates the cell cycle progression of MSCs. In addition, hypoxic conditioning promoted the expressions of MSC markers, such as OCT4 (Octamer-binding transcription factor) and BMP (bone morphogenic protein, which is part of the transforming growth factor-beta superfamily). Collectively, these data suggest that hypoxic conditioning significantly alters the expressions of genes involved in the signaling processes of cell cycle and proliferation, and in the maintenance of multipotency of MSCs.
Fig. 2. Comparision of gene ecpression pattern by hypoxic conditioning using RT2 profiler PCR array.
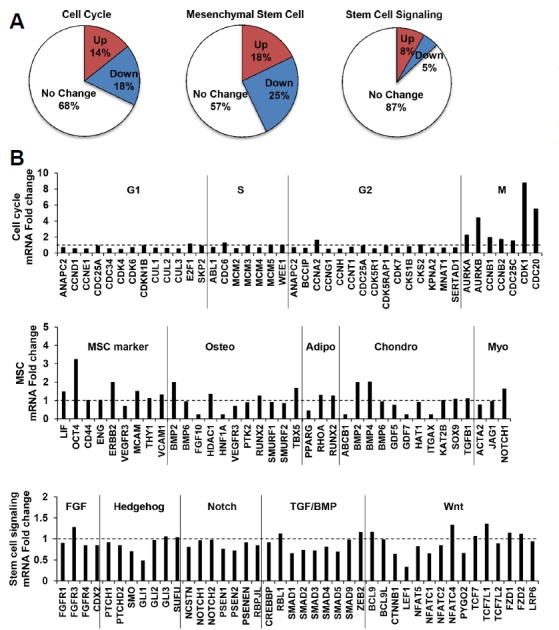
(A) Quantitative RT-PCR analysis of genes involved in the signaling processes of cell cycle, multipotency of MSC, and stem cell signaling cascade. The pie chart shows the percentages of upregulated and downregulated genes by hypoxic conditioning, and the categorization of genes as indicated (p < 0.05, 1.5-fold). (B) The graphs show the relative expression changes of genes involved in the cell cycle, multipotency of MSC, and stem cell signaling.
Introduction of mitotic cyclins extends the proliferation lifespan of human MSCs in normoxic condition
Since the functional characterization revealed that hypoxic conditioning significantly alters pathways involved in DNA replication, cell cycle regulation, and proliferation signaling, we particularly found that several proteins involved in the mitotic cell cycle progression were distinctly up-regulated in the hypoxic primed condition. Interestingly, hypoxic conditioning dramatically increased the protein levels of CCNA2 and CCNB1 (Fig. 3A). To examine whether the forced introduction of CCNA2 or CCNB1 could affect the proliferation potential of MSCs in the normoxic condition, we generated recombinant adenovirus expressing hemagglutinin (HA)-tagged CCNA2 (rAdv-HA-CCNA2) and CCNB1 (rAdv-HA-CCNB1), respectively (Fig. 3B). To determine the effect of CCNA2 or CCNB1 expression on proliferation rate, MSCs at passage 3 were infected with recombinant adenovirus and further cultured in normoxic and hypoxic condition for additional three passages. As shown in Figs. 3C and 3D, MSCs growth was quantified by randomly counting average cell numbers in each group. Interestingly, the overexpression of CCNA2 or CCNB1 significantly increased the proliferation rate of MSCs in normoxic condition. We further analyzed the proliferation rate by carboxyfluorescein succinimidyl ester (CFSE) labeling, which allow an assessment of the extent of multiple generations of cell division (Fig. 3E). This analysis also showed that the introduction of CCNA2 or CCNB1 increased the number of cell divisions in the normoxic condition; the majority number of cell division was 4 and 5 in the control MSCs, and 5 and 6 in CCNA2- or CCNB1-overexpressing MSCs, respectively (Figs. 3E and 3F). We used WST-1 assay to measure the proliferating potential of MSCs following additional four passages, and found that MCSs overexpressing CCNA2 or CCNB1 significantly increased the proliferating cell numbers in normoxic condition but not in hypoxic condition at day 8 (Fig. 3G).
Fig. 3. Overexpression of mitotic cyclins in normorxic condition activates the proliferation potential of MSCs.
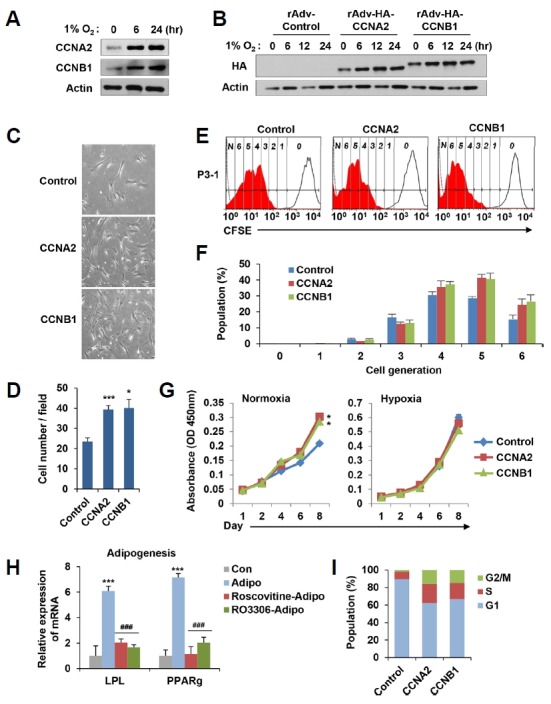
(A) MSCs were grown in hypoxic condition (1% O2) for the indicated time and immunoblotted with anti-CCNA2 or anti-CCNB1 antibody. (B) Generated recombinant adenovirus expressing hemagglutinin (HA)-tagged human CCNA2 (rAdv-HA-CCNA2) and CCNB1 (rAdv-HA-CCNB1) were infected into MSCs in conjugation with cell penetrating peptide PTD. Infected cells were grown in hypoxic condition (1% O2) and immunoblotted with anti-HA antibody. (C, D) rAdv-HA-CCNA2 and rAdv-HA-CCNB1 were infected to MSCs at passage 3, which were further cultured in normoxic condition for an additional three rounds of division. The figure shows the representative phase contrast images of MSCs (C). The graph showed the average number of cells per fields for each group. (D). (E, F) MSCs were infected with rAdv-HA-CCNA2 or rAdv-HA-CCNB1, labeled with CFSE, and further cultured in normoxic condition for an additional three rounds of division. An open histogram indicates a control MSCs (non CFSE labeled MSCs, 0), and the closed histogram indicates a proliferating MSCs (CFSE labeled MSCs, 1–6). Numbers (0 to 6) depicts the dividing number of MSCs according to serial 2-fold decreases of fluorescence intensity (E). The graphs show the percentage population of each dividing number of MSCs above (F). (G) rAdv-HA-CCNA2- or rAdv-HA-CCNB1-infected MSCs cells (at passages 4) were cultured in normoxic and hypoxic conditions, and at indicated days cell proliferation was determined by the WST-1 assay. (H) MSCs grown in hypoxic condition were forced to be differentiated into adipocytes (Adipo) in the presence of Roscovitine (10 μM, Adipo-Roscovitine) or RO3306 (10 μM, Adipo-RO3306) treatment for 2 weeks. The levels of LPL and PPARg mRNA were measured by qRT-PCR. ‘Cont’ indicates undifferentiated MSCs. (I) MSCs were infected with rAdv-HA-CCNA2 or rAdv-HA-CCNB1, cultured in normoxic condition, and harvested for propidium iodide staining to measure their DNA contents. The graphs show the percentage population of each phase of cell cycle. The all results represent mean values ± SEM. *P < 0.05 and ***P < 0.001 versus control and ###P < 0.001 versus adipogenesis.
To further assess how CCNA2 or CCNB1 affected MSCs multipotency, MSCs were forced to be differentiated into adipocytes and osteoblasts, respectively (Fig. 3H and Supplementary Fig. S1). At passage 8, normoxic MSCs, which frequently showed senescent phenotypes, were cultured in adipogenic or osteogenic differentiation media at the same time in normoxic or hypoxic conditions (Supplementary Fig. S1). As expected, adipocytes markers (LPL and PPARg) were expressed in both conditions, but the expression of osteoblasts markers (Osteocalcin and ALP) were further augmented in hypoxic condition. These results confirm that hypoxic condition can improve MSCs multipotency. Next, we used selective Cdk inhibitors, Roscovitine and RO3306, to mimic the inhibition of CCNA2 or CCNB1 activity. In condition that the levels of LPL and PPARg mRNAs were sharply increased about 6.0-folds and 7.1-folds, respectively, by adipogenic differentiation compared to the control undifferentiation, treatment of Roscovitine or RO3306 dramatically decreased the levels of LPL (2.0-folds or 1.7-folds) and PPARg (1.2-folds or 2.0-folds) mRNA, respectively (Fig. 3H), indicating that CCNA2 or CCNB1 activity contributes to the differentiation potential of MSCs.
Next, we compared the cell cycle profiles of MSCs by overexpressing CCNA2 or CCNB1 (Fig. 3I). As expected, the introduction of CCNA2 or CCNB1 significantly accumulated cells at S and G2/M phases, but reduced the population of cell at G1 phase. Collectively, these results suggest that the expression of CCNA2 or CCNB1 in normoxic MSCs activates the proliferating potential, which likely confers the multipotency of MSCs.
Although different sources of MSCs show different functional characteristics by hypoxic conditioning, recent reports for in vitro cultures and following clinical application have recommended MSCs cultivation under hypoxic condition (1% to 10% O2) (Davy and Allsopp, 2011; Rosova et al., 2008; Mathieu et al., 2014; Zhang et al., 2012). However, hypoxia can induce cell cycle arrest of majority of mammalian cells (Goda et al., 2003). In this study, the application of hypoxia to MSCs stimulated their proliferating potential by triggering, particularly, mitotic cell cycle progression. This highlights the potential that the multi-lineage differentiation of MSCs by hypoxic conditioning is largely based on their proliferation capacity. However, we do not exclude the possibility that another signaling pathway is required to provide the proliferating potential of hypoxic MSCs coupled with their multipotency.
Supplementary Information
ACKNOWLEDGMENTS
We would like to thank Dongrim Kang for technical assistance and Ae-Kyeong Kim for supplying human mesenchymal stem cells. This study was supported by a National Research Foundation grant funded by the Korean government (MEST) (2014R1A2A1A10050775 and 2017R1A2B3006776).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Das R, Jahr H, van Osch GJ, Farrell E. The role of hypoxia in bone marrow-derived mesenchymal stem cells: considerations for regenerative medicine approaches. Tissue Eng Part B Rev. 2012;16:159–168. doi: 10.1089/ten.TEB.2009.0296. [DOI] [PubMed] [Google Scholar]
- Davy P, Allsopp R. Hypoxia: are stem cells in it for the long run? Cell Cycle. 2011;10:206–211. doi: 10.4161/cc.10.2.14535. [DOI] [PubMed] [Google Scholar]
- Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia-inducible factor 1 is essential for cell cycle arrest during hypoxia. Mol Cell Biol. 2003;23:359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Wu R, Jiang Z, Wang L, Chen P, Zhang L, Yang L, Wu Y, Chen H, Chen H, et al. Leptin signaling is required for augmented therapeutic properties of mesenchymal stem cells conferred by hypoxia preconditioning. Stem Cells. 2014;32:2702–2713. doi: 10.1002/stem.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15:243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Zhou W, Xing Y, Sperber H, Ferreccio A, Agoston Z, Kuppusamy KT, Moon RT, Ruohola-Baker H. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell. 2014;14:592–605. doi: 10.1016/j.stem.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Nagano M, Kimura K, Yamashita T, Ohneda K, Nozawa D, Hamada H, Yoshikawa H, Ochiai N, Ohneda O. Hypoxia responsive mesenchymal stem cells derived from human umbilical cord blood are effective for bone repair. Stem Cells Dev. 2010;19:1195–1210. doi: 10.1089/scd.2009.0447. [DOI] [PubMed] [Google Scholar]
- Pattappa G, Heywood HK, de Bruijn JD, Lee DA. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J Cell Physiol. 2011;226:2562–2570. doi: 10.1002/jcp.22605. [DOI] [PubMed] [Google Scholar]
- Rafalski VA, Mancini E, Brunet A. Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J Cell Sci. 2003;125:5597–5608. doi: 10.1242/jcs.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosova I, Dao M, Capoccia B, Link D, Nolta JA. Hypoxic preconditioning results in increased motility and improved therapeutic potential of human mesenchymal stem cells. Stem Cells. 2008;26:2173–2182. doi: 10.1634/stemcells.2007-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Chen YJ, Yew TL, Chen LL, Wang JY, Chiu CH, Hung SC. Hypoxia inhibits senescence and maintains mesenchymal stem cell properties through down-regulation of E2A-p21 by HIF-TWIST. Blood. 2011;117:459–469. doi: 10.1182/blood-2010-05-287508. [DOI] [PubMed] [Google Scholar]
- Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley, CA, 4th, Ramalho-Santos J, Van Houten B, Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PloS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O, Clark J, Wong DM, Patti GJ, Sánchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JI, Park SH, Jin HT, Lee CG, Seo SH, Song MY, Lee CW, Sung YC. Enhanced delivery efficiency of recombinant adenovirus into tumor and mesenchymal stem cells by a novel PTD. Cancer Gene Ther. 2008;15:703–712. doi: 10.1038/cgt.2008.45. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


