Abstract
Oligoadenylate synthetase (OAS) protein family is the major interferon (IFN)-stimulated genes responsible for the activation of RNase L pathway upon viral infection. OAS-like (OASL) is also required for inhibition of viral growth in human cells, but the loss of one of its mouse homolog, OASL1, causes a severe defect in termination of type I interferon production. To further investigate the antiviral activity of OASL1, we examined its subcellular localization and regulatory roles in IFN production in the early and late stages of viral infection. We found OASL1, but not OASL2, formed stress granules trapping viral RNAs and promoted efficient RLR signaling in early stages of infection. Stress granule formation was dependent on RNA binding activity of OASL1. But in the late stages of infection, OASL1 interacted with IRF7 transcripts to inhibit translation resulting in down regulation of IFN production. These results implicate that OASL1 plays context dependent functions in the antiviral response for the clearance and resolution of viral infections.
Keywords: anti-viral response, OASL1, stress granule, type I interferon
INTRODUCTION
Stress granules (SGs) are dynamic RNA–protein complexes composed of stalled translation initiation complexes and host RNA-binding proteins, such as T-cell-restricted intracellular antigen-1 (TIA-1), TIA-1-related protein (TIAR), and Ras GTPase-activating protein-binding protein-1 (G3BP1) (Ohn and Anderson, 2010). SG formation is initiated by phosphorylation of eukaryotic translation initiation factor 2α (eIF2α), which can be triggered by activation of protein kinases such as protein kinase R (PKR), heme-regulated inhibitor (HRI), general control nondepressible 2 (GCN2), and PKR-like endoplasmic reticulum kinase (PERK) (Kedersha et al., 2002). When phosphorylated eIF2α inhibits cellular mRNA translation, stalled translation initiation complexes combine with dispersed SG key components to form SGs (Kedersha et al., 1999). Various cellular stresses induce SGs as part of the stress response, leading to global translational repression. When the stress is resolved, sequestered polysomes are released from SGs, and translation of cellular proteins promptly resumes. Therefore, SGs are regarded as temporal repositories of silenced cellular mRNAs that promote cellular recovery (Arimoto et al., 2008; Tsai and Wei, 2010). Viral infections in eukaryotic cells can also induce assembly of SGs (Kedersha et al., 2002). However, inhibition of SG formation lead to significant decrease of type I IFN secretion following viral infection (Oh et al., 2016b; Onomoto et al., 2012; Yoo et al., 2014). These observations suggest that antiviral SGs (avSGs) play vital roles as immunological platforms for the interactions between antiviral proteins and their target RNAs, facilitating the expression of type I IFN and downstream interferon-stimulated genes (ISGs).
The 2′-5′ oligoadenylate synthetase (OAS) is a type I ISG that synthesizes short oligomers that activate RNase L to cleave cellular and viral RNAs promoting their rapid degradation. OASL (OAS-like) protein, a member of the OAS family, consists of one OAS domain and two ubiquitin-like (UBL) repeat domains. Humans have only one type of OASL protein, whereas mice have two homologs, OASL1 and OASL2, which have 74% and 49% amino acid sequence similarity to the human homolog, respectively (Eskildsen et al., 2002). Despite its lower sequence identity, OASL2 is functionally similar to OASL, which promotes RIG-I signaling via a UBL-caspase-recruitment activation domain (CARD) interaction (Zhu et al., 2014). On the contrary, OASL1 negatively regulates type I IFN synthesis by inhibiting translation of IRF7 (interferon-regulating transcription factor 7). This occurs by binding of OASL1 to the stem-loop structures in the 5′-untranslated region (UTR) of IRF7 mRNA (Kim et al., 2014b; Lee et al., 2013a). Loss of OASL1 results in elevated antiviral responses and prevents chronic viral infection by promoting T-cell effector functions (Lee et al., 2013b; Oh et al., 2016a). In addition, recent reports showed that OASL1 deficiency stimulates anti-cancer immune responses and autoimmune disease development by upregulating production of type I IFN (Choi et al., 2016; Sim et al., 2016). Taken together, these observations suggest that OASL1 has a distinct function such as downregulation of antiviral immunity. However, it remains unclear whether it is also required for stimulation of the antiviral immune response, as shown with human OASL homologs.
In this study, we investigated OASL1 functions in cells subjected to immunological challenge and explored in detail the mechanisms underlying the antiviral response. Synthetic poly (I:C), the viral dsRNA analog, was transfected to investigate subcellular localization of OASL1 and anti-viral proteins examined at different time-points to investigate OASL1 function at both early and late stages of virus infection. We found that OASL1 translocated into avSGs quickly upon viral infection or treatment with poly(I:C) and interacted with MDA5, a cytosolic viral sensor, within avSGs promoting type I IFN signaling. These results suggested that OASL1 promotes antiviral response via stress granules at the early stage, but eventually inhibit the translation of IRF7 as its transcripts accumulates in the late stage of infection.
MATERIALS AND METHODS
Cell culture, primary cell preparation, and in vitro treatment
NIH-3T3 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) with 10% fetal bovine serum (FBS, Gibco) and 100 U/ml penicillin-streptomycin (pen-strep, Gibco). Bone marrow-derived macrophages were obtained from primary bone marrow cells cultured for 7 days in DMEM supplemented with 20% L929 culture supernatant, 10% FBS, and 100 U/ml pen-strep. We tried to use minimum mice to obtain bone marrow cells and protocol was approved by the Institutional Animal Care and Use Committee of the Yonsei Laboratory Animal Research Center at Yonsei University.
For OASL1 overexpression, plasmids were mixed with Lipofectamine 2000 (Invitrogen) and incuated at 37°C in 5% CO2 for 24 h. For stimulation, cells were treated with 10 μg/ml poly(I:C) (GE Healthcare), 100 ng/ml LPS (InvivoGen), 100 ng/ml Pam3CSK4 (InvivoGen), 10 μg/ml poly(dA:dT) (InvivoGen), and 1 μg/ml 5′ppp-dsRNA (InvivoGen) for the indicated times. For cytoskeleton inhibition experiments, cells were pre-treated with 10 ng/ml cytochalasin D (Sigma) and 100 μM colchicine (Tocris) for 30 min before poly(I:C) stimulation. To prevent autophagy assembly, cells were treated with 100 nM Bafilomycin A (Sigma), 5 mM 3-MA (3-methyladenine) (Sigma), and 50 μM CQ (chloroquine di-phosphate salt) (Sigma) along with poly(I:C).
Virus infection
NIH-3T3 cells were infected with H1N1 influenza virus (New Caledonia/20/99) at a MOI of 10 for 2 h. After two washes in phosphate-buffered saline (PBS), fresh culture media was added, and the cells were cultured for an additional 16 h before preparation for immunocytochemistry (ICC).
Construction of plasmids for protein overexpression
EGFP-OASL1 (Oasl1, NP_660210.1) and EGFP-OASL2 (Oasl2, NP_035984.2) were generated by inserting the Oasl1 and Oasl2 DNA fragments from pCS4 3xFLAG OASL1 and pCS4 3xFLAG OASL2 into plasmid EGFP-C1 using appropriate restriction sites. Construction of OASL1 mutants was described previously (Lee et al., 2013a).
Immunofluorescence staining and microscopy
Cells grown on glass coverslips in 24-well plates were fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature (RT) and washed twice with PBS (pH 7.4). The fixed cells were permeabilized for 5 min at RT with 0.1% Triton X-100 solution or 1× BD Perm/Wash (BD Bioscience) diluted in triple distilled water (TDW), and then washed twice with PBS containing 0.5% Tween-20 (PBST). Blocking was performed with 3% bovine serum albumin (BSA) in PBST (blocking solution) for 1 h at RT, and the cells were then incubated with a primary antibody [rabbit α-KDEL (NOVUS Biologicals), rabbit α-RCAS (Cell Signaling), rabbit α-LAMP1 (Sigma), rabbit α-PMP70 (Abcam), goat α-TIAR (Santa Cruz Biotechnology), rabbit α-eIF4E (Cell Signaling), rabbit α-S6 (Cell Signaling), rabbit α-G3BP1 (Santa Cruz Biotechnology), rabbit α-MDA5 (Abcam), MitoTracker® Red (Invitrogen), rabbit α-LC3b (MBL International), or rabbit α-p62 (MBL International)] for 1 h at RT or overnight at 4°C. All primary antibodies were diluted in blocking solution. Next, cells were washed twice with PBST, and incubated for 1 h at RT with a secondary antibody [donkey α-goat Alexa 594 (Invitrogen), goat α-rabbit FITC (Sigma), or goat α-rabbit TRITC (Sigma)] diluted in blocking solution. Nuclei were stained with 4.6-diamidino-2-phenylindole (DAPI, Sigma) for 1 min at RT. After two washes, coverslips were mounted with mounting solution (Dako) on slide glass. To visualize intracellular poly(I:C), 1 μg/ml poly(I:C) (HMW) Rhodamine (InvivoGen) was used. Samples were examined by confocal microscopy on a LSM 510 or 700 and analyzed using the Carl Zeiss ZEN 2.3 lite software.
Fluorescence in situ hybridization (FISH) assay
RNA-protein double labeling was performed as described by Kim et al. (2014a) with a slight modification. The probe for visualization of endogenous Irf7 mRNA was designed to detect the 1218–2018 bp and 1668–2467 bp region of the Irf7 transcript. Cells were permeabilized with 80% acetone for 10 min at −20°C. A mixture of Irf7 mRNA probes (diluted 1:1000) was added, and the samples were incubated for 22 h at 37°C. ICC was performed to investigate intracellular localization of TIAR using goat α-TIAR and Alexa Fluor 647–conjugated donkey α-goat antibody (Invitrogen). Cells were examined by LSM 700 confocal microscopy.
Immunoprecipitation and western blotting
NIH-3T3 cells expressing 3xFLAG and SBP-tagged constructs were collected by centrifugation at 2000 g for 3 min at 4°C. After two washes with cold PBS followed by centrifugation, whole-cell lysate was obtained using lysis buffer [50 mM Tris HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100] containing freshly added protease inhibitor (Roche). Cell debris was removed by centrifugation at 14000 g for 10 min at 4°C, and the resultant supernatant was used for immunoprecipitation. Total cellular proteins were sequentially pulled down with streptavidin–Sepharose high performance beads (GE Healthcare) and anti-FLAG M2 affinity gel (Sigma). TBS (50 mM Tris HCl, with 300 mM NaCl, pH 7.4) was used for a harsh wash between each step, and the precipitated proteins were eluted with 3× FLAG peptide (Sigma). Equal amounts of immunoprecipitated proteins were subjected to SDS-PAGE, and then transferred to nitrocellulose membranes using a wet transfer system (Bio-Rad). The membranes were blocked with 5% w/v skim milk in TBS with 0.1% Tween-20 (TBST) for 1 h at RT, and then probed with primary antibodies diluted in blocking solution: rabbit α-OASL1 (Lee et al., 2013a), rabbit α-PKR (Santa Cruz), mouse α-HSP70 (Enzo Life Sciences), rabbit α-eIF4E (Cell Signaling), rabbit α-IRF7 (Invitrogen), or rabbit α-GAPDH (Santa Cruz). The blots were then rinsed with TBST, followed by incubation for 1–2 h at RT with horseradish peroxidase (HRP)-conjugated secondary antibodies. The membranes were developed with Amersham ECL reagents (GE Healthcare), and signals were detected on a LAS 4000 system. Signal intensity was measured using ImageJ software.
Design and synthesis of small double-stranded RNA
The small double-stranded RNA (dsRNA) used in this study, which contained 60% G/C base pairs, had the sequence GGAGUCCACGACUUCGCAGGCUCGUUACGU; this dsRNA was originally described by Szymanski et al. (2011). Oligoribonucleotides were synthesized by ST PHARM, Korea. For annealing, the strands were incubated at 95°C for 2 min, and then slowly cooled to 25°C over 45 min in a thermal cycler.
Reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNA from cells was isolated using the TRIzol reagent (Invitrogen). cDNA was synthesized from 2 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) with oligo-dT primers. cDNA was diluted 1:10 with TDW, and 3 μl aliquots were subjected to quantitative real-time PCR (qRT-PCR) on a Bio-Rad CFX instrument. Levels of individual genes were measured using gene-specific primers, and then normalized against the level of the housekeeping gene Gapdh by the ΔΔCT method.
Statistical analysis
Statistical analysis was performed using the GraphPad PRISM 6 software. Data shown in the figures represent means ± SD.
RESULTS
OASL1 associates with cytosolic speckles upon poly(I:C) treatment
To determine whether mouse OASL1 has additional functions during the antiviral response, we examined the intracellular localization of enhanced green fluorescent protein (EGFP)-tagged OASL1 (EGFP-OASL1) expressed in NIH-3T3 cells following various types of immune-related stimulation. When the cells were stimulated with Pam3CSK4, LPS, 5′pppdsRNA, or poly (dA:dT), which activate Toll-like receptors (TLRs) or RLRs, EGFP-OASL1 was evenly distributed in the cytosol, similar to the pattern observed in unstimulated cells. However, intracellular poly(I:C) treatment stimulated the formation of EGFP-OASL1 speckles in the cytosol, which persisted for over 12 h after stimulation (Fig. 1A and Supplementary Fig. S1A). To investigate the specificity of speckle formation, we tested whether OASL2, the other mouse homolog of OASL, could induce speckles in a poly(I:C)-dependent manner. In contrast to EGFP-OASL1, EGFP-OASL2 did not form a speckle pattern following poly(I:C) treatment (Fig. 1B). These results suggested that OASL1, but not OASL2, plays a specific role in speckle formation during antiviral responses.
Fig. 1. OASL1 associates with stress granules upon poly(I:C) treatment.
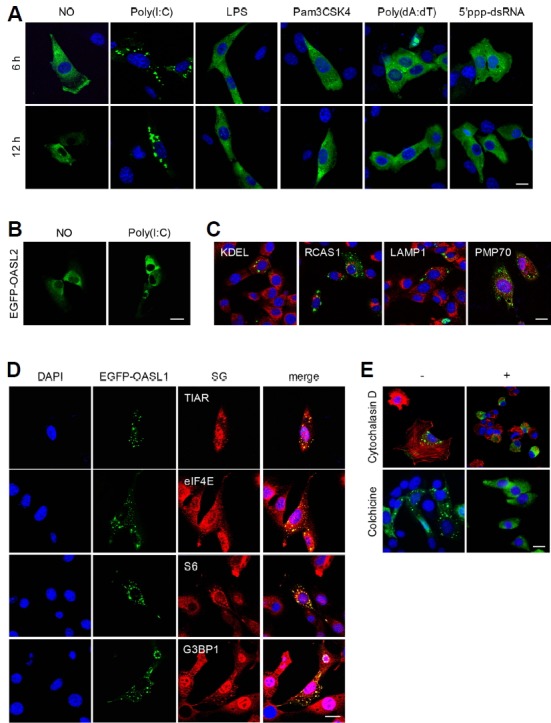
(A) OASL1 associated with cytoplasmic speckles specifically upon poly(I:C) treatment. NIH-3T3 cells expressing EGFP-OASL1 are shown following stimulation with various TLRs (indicated at the top) for 6 h (top panel) or 12 h (lower panel). (B) OASL2 did not associate with speckles upon poly(I:C) stimulation. EGFP-OASL2–overexpressing NIH-3T3 cells were stimulated with poly(I:C) for 6 h. (C) OASL1-containing speckles do not overlap with secretory vesicles. EGFP-OASL1–overexpressing NIH-3T3 cells were stained with specific antibodies against the indicated markers (KDEL, RCAS, LAMP1, and PMP70; shown in red). (D) OASL1-containing speckles colocalized with SG markers. EGFP-OASL1–expressing cells were stained with the indicated SG markers (TIAR, S6, eIF4E, and G3BP1) after stimulation with poly(I:C) for 6 h. (E) Actin and microtubules are required for formation of OASL1-containing speckles. EGFP-OASL1–xpressing cells were stained for β-actin after pre-treatment with Cytochalasin D or colchicine treatment followed by poly(I:C) stimulation. Nuclei were stained with DAPI (blue). Scale bars correspond to 10 μm. Images are representative of at least two independent experiments.
To determine the nature of the EGFP-OASL1 speckles, we investigated the colocalization of EGFP-OASL1 with various organelle-specific markers. We first tested components of the protein post-translational translocation system, such as the ER, Golgi, and lysosome, using the KDEL motif, RCAS1, and LAMP1 as markers, respectively. Immunostaining of EGFP-OASL1–expressing cells with antibodies against these marker proteins did not reveal any colocalization (Fig. 1C). In light of the role of the peroxisome in production of type III IFN, we also examined the PMP70 peroxisomal marker protein (Dixit et al., 2010; Wack et al., 2015), but again observed no colocalization with EGFP-OASL1 speckles. Recent work showed that SGs are immune-related complexes required for type I IFN regulation (Kedersha et al., 2013; Onomoto et al., 2012; Yoo et al., 2014). Therefore, we tested whether poly(I:C)-specific EGFP-OASL1 speckles localized at SGs during the antiviral response. Indeed, TIAR, a marker of SGs, colocalized with EGFP-OASL1 in most speckles, along with other SG components, including eukaryotic translation initiation factor 4E (eIF4E), ribosomal protein S6, and G3BP1, upon intracellular poly(I:C) stimulation (Fig. 1D). Next, we wondered whether OASL1 is required for SG formation following immune response. We investigated level of SG formation in Oasl1+/+ and Oasl1−/− bone marrow–derived macrophages (BMM), but there was no difference between the level of SG formation in both type of cells (Supplementary Fig. S1B). Because SG formation is dependent on the cytoskeleton (Chernov et al., 2009; Loschi et al., 2009), we examined the effects of the cytoskeleton inhibitors cytochalasin D and colchicine, which inhibit actin filaments and microtubule assemblies, respectively, on poly(I:C)-dependent EGFP-OASL1 speckle formation. Both inhibitors dramatically inhibited OASL1 speckle formation (Fig. 1E and Supplement Fig. S1C). These data indicated that the speckles were SGs, and that OASL1 translocates specifically into SGs during immune responses triggered by poly(I:C) treatment.
SGs are the major working platform for OASL1
Viral RNA is recruited to virally induced avSGs (Onomoto et al., 2012), promoting expression of type I IFN (Oh et al., 2016b; Yoneyama et al., 2016; Yoo et al., 2014). To determine whether the association with SGs is related to upregulation of RLR signaling, we analyzed the subcellular localizations of transfected dsRNAs and their cellular recognition components, such as MDA5, along with OASL1 upon poly(I:C) stimulation. To this end, we transfected rhodamine-conjugated poly(I:C), a mimic of viral RNAs, into OASL1-EGFP–expressing NIH-3T3 cells, and then monitored colocalization with EGFP-OASL1 and SGs. At approximately 1.5 h post-transfection, EGFP-OASL1 began to colocalize with intracellularly introduced rhodamine-poly(I:C) and 3 h post-transfection of poly (I:C), they started to interact with MDA5 and TIAR at SGs, indicating that OASL1 recognizes foreign RNAs and then interacts with the dsRNA recognition machinery at SGs (Fig. 2A). It is well known that MAVS-mediated RLR signaling is important for stimulating antiviral immune response genes (Lee et al., 2015; Seth et al., 2005; Yoneyama et al., 2015). And Several recent studies reported that intracellular RNA recognition by RIG-I and MDA5 at SGs promotes aggregation with MAVS in mitochondria, thereby activating type I IFN signaling (Yoneyama et al., 2016; 2015; Zhang et al., 2014). Consistent with this, MitoTracker® Red staining of poly(I:C)-treated cells revealed that EGFP-OASL1 speckles colocalized with mitochondria (Fig. 2A and Supplementary Fig. S2). This result suggested that OASL1 plays a role in the recognition of viral RNAs by RLRs at SGs.
Fig. 2. OASL1 interacts with RLR complex proteins in SGs along with its target RNAs.
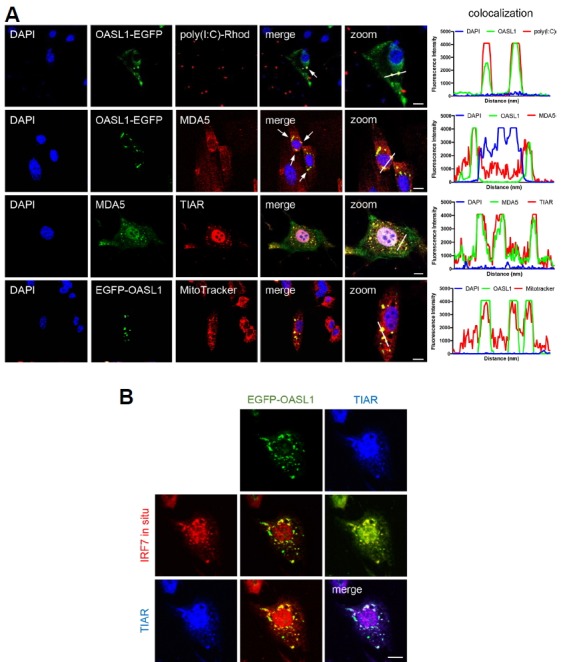
(A) OASL1 associated with MDA5 and poly(I:C) at SGs. Images show colocalization of the indicated markers in OASL1-EGFP–expressing cells after poly(I:C) stimulation. Co-localization intensity was quantified using ZEN 2.3 software. White line indicates the quantified region. Fluorescence intensity graph shows the level of co-localization and each line indicates corresponding fluorescence. (B) OASL1 colocalized with Irf7 RNA at several SGs. In cells stimulated with poly(I:C), endogenous Irf7 transcripts were visualized by FISH (red), followed by staining for TIAR (blue). Scale bars correspond to 10 μm. Images are representative of at least three independent experiments.
As OASL1 binding to Irf7 mRNA has been reported, we examined whether Irf7 mRNA is also located within EGFP-OASL1 speckles. We performed RNA fluorescence in situ hybridization (RNA FISH) followed by ICC to visualize Irf7 mRNA, EGFP-OASL1, and SGs. Six hours after transfection with poly(I:C), several Irf7 mRNA–containing speckles were colocalized with EGFP-OASL1 and TIAR in many SGs (Fig. 2B). These results indicate that SGs are the major sites for the proposed functions of OASL1 in antiviral responses.
The RNA-binding affinity of the OAS domain is critical for OASL1 recruitment to SGs
Because the OAS domain binds viral dsRNAs, we suspected that the poly(I:C) interacts with the OASL1 dsRNA-binding domain during translocation into SGs. To test this hypothesis, we used a series of OASL1 mutant constructs inserted into a 3xFLAG-containing vector: WT (OASL1), a UBL domain deletion mutant (OASL1-ΔUBL), an N-terminal 150–amino acid deletion mutant (OASL1-Δ150), an OAS domain deletion mutant (OASL1-Δ300), and an RNA binding–defective mutant containing the R192E, K196E, and K201E substitutions in the OAS domain (OASL1-RKK) (Fig. 3A) (Lee et al., 2013a). Following poly(I:C) treatment, the OAS domain deletion mutants (OASL1-Δ150 and OASL1-Δ300) were significantly reduced in colocalization at SGs (Fig. 3B and Supplementary Figs. S3A and S3B). However, the UBL domain deletion mutants (OASL1-ΔUBL) colocalized with TIAR at levels comparable to that of WT OASL1, suggesting that OASL1 association with SGs is dependent on RNA binding by OAS domains. In addition, the RNA binding–defective mutant (OASL1-RKK) completely failed to colocalize with OASL1 at SGs. Therefore, the RNA-binding affinity of OASL1 is a key determinant of proper translocation into SGs, and might also be important for interactions with other SG components.
Fig. 3. RNA-binding affinity of the OAS domain is required for OASL1 translocation into stress granules.
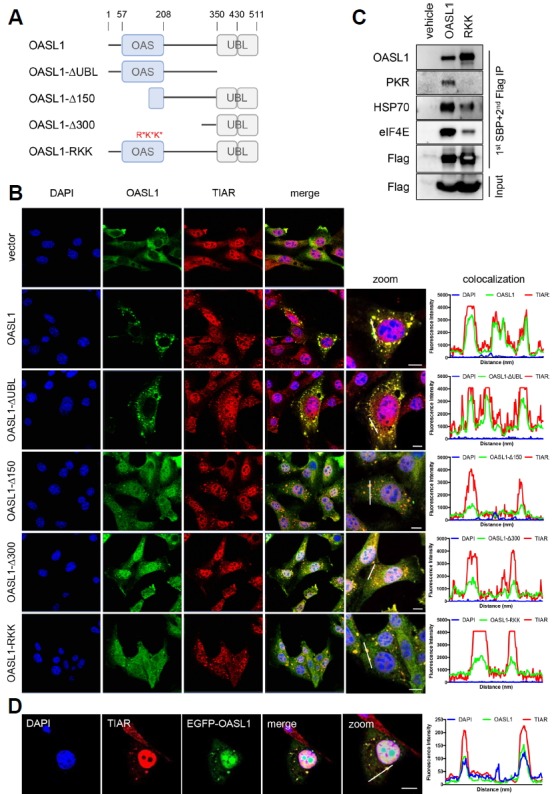
(A) Schematic diagram of full-length and mutant OASL1 constructs used in fluorescence imaging of cells and immunoprecipitation assays. The location of each domain is marked with amino acid position numbers (top). Point mutations that disrupt the RNA-binding activity of the OAS domain are indicated in red. (B) Localization of OASL1 mutant derivatives described in (A) along with the SG component TIAR (red) in cells stimulated with poly(I:C) for 6 h. (C) Immunoprecipitation of wild-type and RNA binding–defective OASL1. Cells expressing the indicated construct (top) were stimulated with poly(I:C) and analyzed by co-immunoprecipitation assay with the antibodies indicated at the left. This immunoblot is representative of two independent experiments. (D) OASL1 associated with SG upon viral infection. EGFP-OASL1–overexpressing cells were infected with H1N1 influenza virus (MOI=10) for 16 h. After infection, cells were stained with TIAR (red), and nuclei were stained with DAPI (blue). (B, D) Colocalization intensity was quantified by ZEN 2.3 software. White line indicates the quantified region. Fluorescence intensity graph shows the level of co-localization and each line indicates corresponding fluorescence. Scale bars correspond to 10 μm. Images are representative of at least two independent experiments.
We further confirmed the requirement of RNA-binding activity for the association of OASL1 with SGs using immunoprecipitation assays. FLAG and streptavidin-binding peptide (SBP) double-tagged WT OASL1 and RNA binding–defective OASL1 mutant (RKK) constructs were expressed in NIH-3T3 cells, and SG formation was induced by intracellular poly(I:C) treatment for 6 h. Sequential pulldown with SBP and anti-FLAG antibody coprecipitated components of SGs, including HSP70, eIF4E, and key antiviral responsive mediator, PKR (Onomoto et al., 2012; Park et al., 2011; Yoo et al., 2014). However, RNA binding–defective OASL1 (RKK) failed to interact with PKR and exhibited significantly weaker interactions with other SG components (Fig. 3C). These results indicated that the RNA-binding ability of OASL1 is critical for its association with SGs, and in particular with another antiviral dsRNA-binding protein, PKR.
Next, we investigated whether similar SG formation is induced during viral infection. To this end, we infected EGFP-OASL1–overexpressing cells with H1N1 influenza A virus. Sixteen hours after infection with influenza A virus, EGFP-OASL1 was associated specifically with SGs (Fig. 3D and Supplementary Fig. S3C), indicating that association of OASL1 with SGs may have a specific function in the promotion of antiviral immune responses.
OASL1-containing SGs promote type I IFN expression
The association of most OASL1 molecules with SGs prompted us to examine the effect of SG formation on the proposed functions of OASL1, i.e., activation of the MAVS pathway and inhibition of IRF7 translation. Treatment of poly(I:C) caused dose-dependent increases in the levels of inflammatory cytokines (OASL1, IFNα, IFNβ, and TNFα) and formation of OASL1-containing SGs (Supplementary Fig. S4). Small dsRNAs (sdsRNAs) induce proinflammatory cytokines and ISGs via RIG-I pathways (Kato et al., 2008), but are unable to induce SGs (Szymanski et al., 2011). We compared cytokine production between cells treated with sdsRNAs (30 nucleotides) or poly(I:C) (2–8 kb in average), and found that the mRNA levels of Tnfα and other ISGs (type I Ifn genes and Oasl1) were lower in cells treated with sdsRNA, even at high doses (Figs. 4A and 4B). This result indicates that OASL1- containing SG formation is required for activation of the MAVS pathway in NIH-3T3 cells. Similarly, treatment of BMMs with sdsRNA induced less productions of OASL1 and IRF7 than poly(I:C) treatment. However, the efficiency of IRF7 translational inhibition by OASL1 under sdsRNAs treatment condition was comparable to that of poly(I:C) treatment condition (Figs. 4C and 4D); indicating that inhibition of IRF7 translation by OASL1 was not affected in the absence of SGs (Figs. 4C and 4D). Therefore, stress granule formation was dispensable for inhibition of ribosome assembly by OASL1, but appears to play an important role in stimulating the MDA5–MAVS pathway.
Fig. 4. OASL1-containing SGs promote type I IFN production.
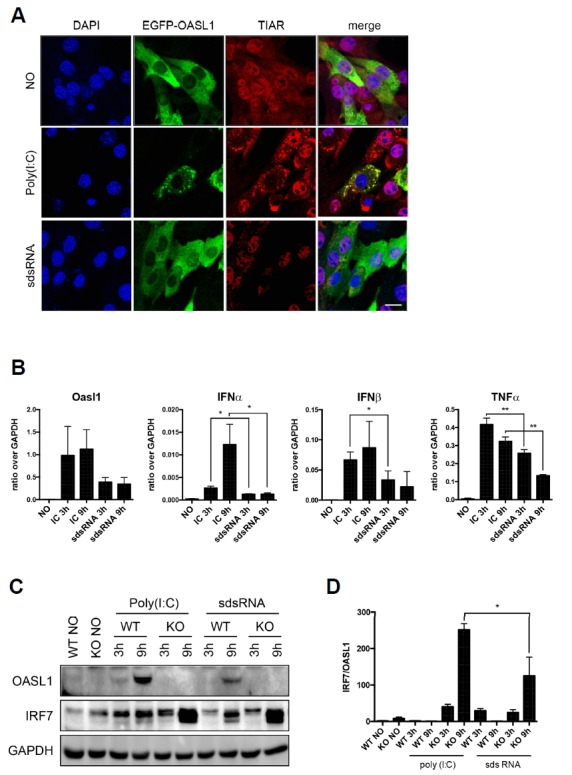
(A) Small dsRNA does not induce SG formation. EGFP-OASL1–expressing cells stimulated under the conditions indicated at the left [no stimulation, poly(I:C), or sdsRNA] were stained for SG with anti-TIAR antibody (red). Nuclei were stained with DAPI (blue). Scale bars correspond to 10 μm. (B) Reduced induction of cytokines by short double-strand RNA (sdsRNA). Wild-type BMMs were transfected with 1 μg/ml poly(I:C) or 100 nM sdsRNA for 3 or 9 h. Levels of Oasl1, Ifnα, Ifnβ, and Tnfα mRNAs were measured by qRT-PCR and normalized against the level of Gapdh mRNA in the same sample. Data represent means ± SD. Statistical significance was determined with the Student’s t-test. *P < 0.05, **P < 0.01. (C) Protein levels of OASL1 and IRF7 in Oasl1+/+ and Oasl1−/− BMMs stimulated with either poly(I:C) or sdsRNA. (D) The signal was measured by ImageJ software for each individual experiments and the graph represents the ratio of IRF7 signal intensity over OASL1 signal intensity. Each experiment was performed at least three times. Equal amounts of cytoplasmic extracts were analyzed by western analysis using the antibodies indicated at the left.
OASL1-containing SGs are cleared via a SG-autophagy clearance pathway
In eukaryotic cells, cytosolic SGs are often cleared by activation of autophagy (Buchan et al., 2013). Accordingly, we investigated whether autophagy is involved in the clearance of OASL1-containing SGs along with its target RNAs. Immunofluorescence revealed that microtubule-associated protein light chain 3b (LC3b) and p62, representative markers of autophagy, colocalized with EGFP-OASL1 speckles (Figs. 5A and 5B). Therefore, the dsRNA-binding affinity of OASL1 and its strong association with SGs during viral infection must be critically required in the clearance of viral and cellular dsRNAs from infected cells by autophagy. Together, these results indicate that autophagy-medicated clearance of dsRNA-OASL1 complex in SGs plays an important role in type I IFN response.
Fig. 5. OASL1-containing SGs colocalize with autophagy markers.
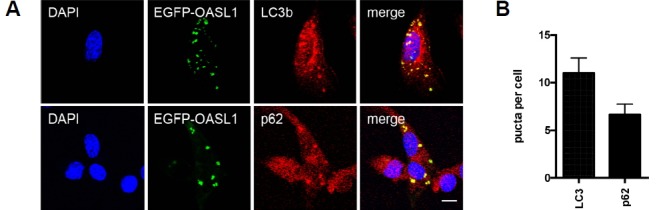
(A) Colocalization of OASL1-containing speckles with LC3 and p62. EGFP-OASL1–expressing cells were stained for LC3b and p62 (red) after 6 h of stimulation with poly(I:C). Nuclei were stained with DAPI (blue). Scale bars correspond to 10 μm. Images are representative of at least three independent experiments. (B) Number of puncta per cell from at least 5 different fields of images. (C) qRT-PCR analysis of type I IFN transcripts in Oasl1+/+ (WT) and Oasl1−/− (KO) BMMs stimulated with poly(I:C) in the presence of an autophagy inhibitor. Data represent means ± SD. Statistical significance was determined with the Student’s t-test. *P < 0.05, **P < 0.01.
DISCUSSION
SGs promote type I IFN expression by supporting the interactions of RIG-I and MDA5 with their target non-self RNAs (Narita et al., 2014; Oh et al., 2016b; Onomoto et al., 2012; Yoo et al., 2014). Our results revealed that OASL1 is a component of SGs during viral infection and contributes to type I IFN expression by trapping viral RNAs in SGs. When poly(I:C) was transfected into cells, OASL1 speckles colocalized with SGs at early time points. In SGs, OASL1 interacted with many components of SGs and the RNA-sensing receptor MDA5. The OAS domain, and specifically its RNA-binding ability, was required for the interaction with SGs, and the known target RNAs of OASL1 such as poly(I:C) and Irf7 mRNAs colocalized at SGs, which were targeted for removal by autophagy. Therefore, OASL1 in SGs appears to increase the sensitivity of innate immune receptors involved in dsRNA recognition and to promote clearance of the trapped dsRNAs (such as viral RNAs).
OASL and RIG-I localize at SGs upon Sendai virus (SeV) infection and stimulate RIG-I-dependent IFN induction (Zhu et al., 2014). OASL oligomerizes with RIG-I via its C-terminal UBL repeats, which mimic polyubiquitination. On the other hand, our results showed that the RNA-binding domain makes a more important contribution than the UBL repeats to the association with SGs. Furthermore, OASL1 interacted with MDA5 in SGs and did not assemble into SGs upon 5′ppp-dsRNA treatment, a RIG-I agonist. These differences could be due to the distinct ligand specificities of the two OASL homologs in different species. Antiviral responses against Respiratory syncytial virus (RSV), SeV, Herpes simplex virus-1 (HSV-1), West Nile virus (WNV), and Newcastle disease virus (NDV) are RIG-I–dependent and mediated by the human OASL and mouse OASL2 signaling pathways (Dhar et al., 2015; Zhu et al., 2014). However, picornaviruses, such as Mengovirus, EMCV, or poliovirus, are specifically sensed by MDA5, and this recognition is affected by Oasl1 mutation (Bruns and Horvath, 2015; Kato et al., 2006; Langereis et al., 2013; Triantafilou et al., 2012). Therefore, antiviral responses against these viruses may be mediated by OASL1. Although RIG-I and MDA5 share the MAVS-STING-TBK1 antiviral signaling pathway, the details of activation of the two major RLR sensors may differ. In addition, the role of MDA5 in SGs regarding regulation of type I IFN remains controversial, thus further study is needed.
Here, we propose that OASL1 plays a dual role, regulating type I IFN both positively and negatively depending on cellular circumstances. During the early phase of infection, OASL1 proteins are rarely present, but, as the initial type I IFN response progresses, more OASL1 proteins are produced, and they subsequently translocate into SGs with their target RNAs (for example, Irf7 transcripts and viral RNAs). This promotes interactions between target RNAs and the cellular RNA recognition machinery, resulting in translational arrest of Irf7 and elevated synthesis of type I IFN. In addition, the eventual activation of autophagy by SGs enables efficient clearance of both viral RNAs and abundant Irf7 RNAs from the cytosol. Thus, OASL1 plays both positive and negative roles in the antiviral response by sequestering target dsRNAs in SGs (Fig. 6).
Fig. 6.
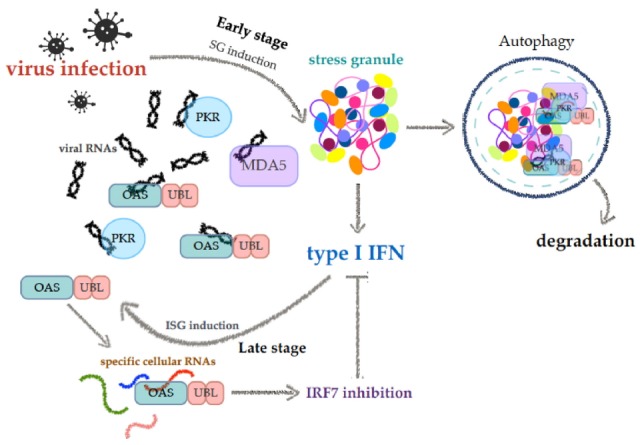
Proposed model of OASL1 functions
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program of the NRF, funded by the Ministry of Science, ICT & Future Planning (NRF-2017R1D1A1B06029547 to JSK-H), and the Bio and Medical Technology Development Program of the NRF, funded by the Ministry of Science, ICT & Future Planning (NRF-2012M3A9B4028272 to YJK). We thank SJL, JES and HRC for caring for the mice and for technical assistance.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- Bruns AM, Horvath CM. LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine. 2015;74:198–206. doi: 10.1016/j.cyto.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov KG, Barbet A, Hamon L, Ovchinnikov LP, Curmi PA, Pastré D. Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. J Biol Chem. 2009;284:36569–36580. doi: 10.1074/jbc.M109.042879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BY, Sim CK, Cho YS, Sohn M, Kim Y-J, Lee MS, Suh SW. 2′-5′ oligoadenylate synthetase-like 1 (OASL1) deficiency suppresses central nervous system damage in a murine MOG-induced multiple sclerosis model. Neurosci Lett. 2016;628:78–84. doi: 10.1016/j.neulet.2016.06.026. [DOI] [PubMed] [Google Scholar]
- Dhar J, Cuevas RA, Goswami R, Zhu J, Sarkar SN, Barik S. 2′-5′-oligoadenylate synthetase-like protein inhibits respiratory syncytial virus replication and is targeted by the viral nonstructural protein 1. J Virol. 2015;89:10115–10119. doi: 10.1128/JVI.01076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee ASY, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen S, Hartmann R, Kjeldgaard NO, Justesen J. Gene structure of the murine 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2002;59:1212–1222. doi: 10.1007/s00018-002-8499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee J, Lee S, Lee B, Kim-Ha J. Phylogenetic comparison of oskar mRNA localization signals. Biochem Biophys Res Commun. 2014a;444:98–103. doi: 10.1016/j.bbrc.2014.01.021. [DOI] [PubMed] [Google Scholar]
- Kim Y-M, Choi WY, Oh C-M, Han G-H, Kim Y-J. Secondary structure of the Irf7 5′-UTR, analyzed using SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) BMB Rep. 2014b;47:558–562. doi: 10.5483/BMBRep.2014.47.10.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis MA, Feng Q, van Kuppeveld FJ. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J Virol. 2013;87:6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kim B, Oh GT, Kim Y-J. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol. 2013a;14:346–355. doi: 10.1038/ni.2535. [DOI] [PubMed] [Google Scholar]
- Lee MS, Park CH, Jeong YH, Kim Y-J, Ha S-J. Negative regulation of type I IFN expression by OASL1 permits chronic viral infection and CD8+ T-cell exhaustion. PLoS Pathog. 2013b;9:e1003478. doi: 10.1371/journal.ppat.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N-R, Kim H-I, Choi M-S, Yi C-M, Inn K-S. Regulation of MDA5-MAVS antiviral signaling axis by TRIM25 through TRAF6-mediated NF-κB activation. Mol Cells. 2015;38:759–764. doi: 10.14348/molcells.2015.0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci. 2009;122:3973–3982. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita R, Takahasi K, Murakami E, Hirano E, Yamamoto SP, Yoneyama M, Kato H, Fujita T. A novel function of human pumilio proteins in cytoplasmic sensing of viral infection. PLoS Pathog. 2014;10:e1004417. doi: 10.1371/journal.ppat.1004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JE, Lee MS, Kim Y-J, Lee HK. OASL1 deficiency promotes antiviral protection against genital herpes simplex virus type 2 infection by enhancing type I interferon production. Sci Rep. 2016a;6:19089. doi: 10.1038/srep19089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S-W, Onomoto K, Wakimoto M, Onoguchi K, Ishidate F, Fujiwara T, Yoneyama M, Kato H, Fujita T. Leader-containing uncapped viral transcript activates RIG-I in antiviral stress granules. PLoS Pathog. 2016b;12:e1005444. doi: 10.1371/journal.ppat.1005444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Anderson P. The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip Rev RNA. 2010;1:486–493. doi: 10.1002/wrna.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K, Jogi M, Yoo J-S, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS ONE. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I-H, Baek K-W, Cho E-Y, Ahn B-Y. PKR-dependent mechanisms of interferon-α for inhibiting hepatitis B virus replication. Mol Cells. 2011;32:167–172. doi: 10.1007/s10059-011-1059-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim CK, Cho YS, Kim BS, Baek I-J, Kim Y-J, Lee MS. 2′-5′ Oligoadenylate synthetase-like 1 (OASL1) deficiency in mice promotes an effective anti-tumor immune response by enhancing the production of type I interferons. Cancer Immunol Immunother. 2016:1–13. doi: 10.1007/s00262-016-1830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski MR, Jezewska MJ, Bujalowski PJ, Bussetta C, Ye M, Choi KH, Bujalowski W. Full-length Dengue virus RNA-dependent RNA polymerase-RNA/DNA complexes: stoichiometries, intrinsic affinities, cooperativities, base, and conformational specificities. J Biol Chem. 2011;286:33095–33108. doi: 10.1074/jbc.M111.255034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K, Vakakis E, Kar S, Richer E, Evans GL, Triantafilou M. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- Tsai N-P, Wei L-N. RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell Signal. 2010;22:668–675. doi: 10.1016/j.cellsig.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wack A, Terczyńska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol. 2015;16:802–809. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Jogi M, Onomoto K. Regulation of antiviral innate immune signaling by stress-induced RNA granules. J Biochem. 2016;159:279–286. doi: 10.1093/jb/mvv122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Yoo J-S, Takahasi K, Ng CS, Ouda R, Onomoto K, Yoneyama M, Lai JC, Lattmann S, Nagamine Y, Matsui T, et al. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLoS Pathog. 2014;10:e1004012. doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Li Y, Xia J, He J, Pu J, Xie J, Wu S, Feng L, Huang X, Zhang P. IPS-1 plays an essential role in dsRNA-induced stress granule formation by interacting with PKR and promoting its activation. J Cell Sci. 2014;127:2471–2482. doi: 10.1242/jcs.139626. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, Ibsen MS, Schmid-Burgk JL, Schmidt T, Ganapathiraju MK, et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


