Abstract
Biliary atresia (BA) is a common cause of pediatric end-stage liver disease. While its etiology is not yet clear, evidence has suggested that BA results from interactions between genetic susceptibility and environmental factors. Disease relevant human cellular models of BA will facilitate identification of both genetic and environmental factors that are important for disease prevention and treatment. Here we report the generation of a human induced pluripotent stem cell line from a BA patient using episomal vectors. Patient-specific BA iPSC lines provide valuable tools for disease mechanism study and drug development.
Resource utility
We generated this patient-specific iPSC line to develop human cellular disease models of biliary atresia, a type of cholangiopathy possibly resulting from perturbed early development of the biliary system (Hartley et al., 2009). BA iPSC lines can be used to study molecular mechanisms underlying the disease and to discover contributing environmental factors (Tables 1 and 2).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal human pluripotent stem cell morphology | Not shown but available with author |
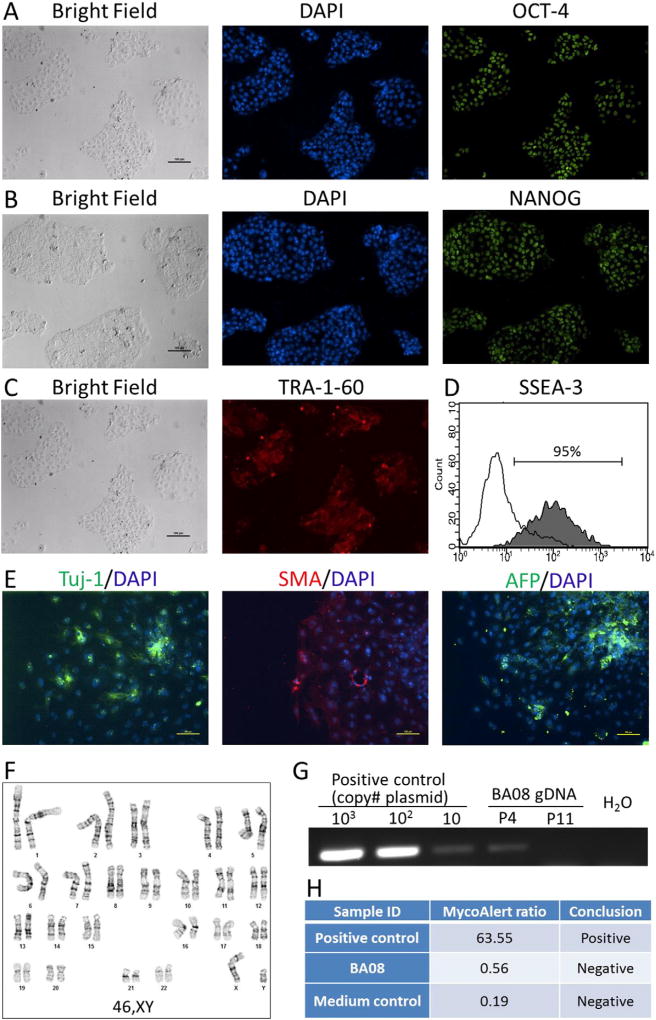
| Phenotype | Immunocytochemisty | Staining for expression of pluripotency markers: OCT4, NANOG and TRA-1-60 | Fig. 1 panel A– C |
| Flow cytometry | Assess cell surface marker expression: SSEA-3: 91% | Fig. 1 panel D | |
| Genotype | Karyotype (G-banding) and resolution | 46,XY | Fig. 1 panel F |
| Resolution 475– 525 | |||
| Identity | Microsatellite PCR (mPCR) | Not performed | – |
| STR analysis | PowerPlex 16HS (Promega) 16 sites. Match with donor | Submitted in archive with journal | |
| Mutation analysis (IF APPLICABLE) | Sequencing | – | – |
| Southern Blot OR WGS | – | – | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence. Negative. | Fig. 1 panel H |
| Differentiation potential | Embryoid body formation | Expression of genes in embryoid bodies: smooth muscle actin, Tuj-1 β-tubulin and α-feto protein. | Fig. 1 panel E |
| Donor screening (OPTIONAL) | HIV 1 + 2 hepatitis B, Hepatitis C | – | – |
| Genotype additional info (OPTIONAL) | Blood group genotyping | – | – |
| HLA tissue typing | – | – |
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
|
| |||
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency marker | Mouse anti-OCT4 | 1:200 | Millipore Cat# MAB4401, RRID:AB_2167852 |
| Pluripotency marker | Mouse anti-NANOG | 1:100 | BD Pharmingen Cat# 560109, RRID:AB_1645597 |
| Pluripotency marker | Mouse anti-TRA-1-60 | 1:200 | Millipore Cat# MAB4360, RRID:AB_11211864 |
| Pluripotency marker | Alexa Fluor 488 anti-human/mouse SSEA-3 | 1:20 | BioLegend Cat# 330306, RRID:AB_1279440 |
| Differentiation marker | Mouse anti-SMA IgG2a | 1:200 | Sigma-Aldrich Cat# A5228, RRID:AB_262054 |
| Differentiation marker | Mouse anti-Tuj-1 IgG, Alexa Fluor 488 labeled | 1:800 | Covance Research Products Inc. Cat# A488-435L, RRID:AB_10143904 |
| Differentiation marker | Rabbit anti-AFP IgG | 1:200 | Dako Cat# A0008, RRID:AB_2650473 |
| Secondary antibody | Alexa Fluor 488 goat anti-mouse IgG (H + L) | 1:500 | Invitrogen Cat #A11001, RRID:AB_2534069 |
| Secondary antibody | Alexa Fluor 555 goat anti-mouse IgM | 1:500 | Invitrogen Cat# A21426, RRID:AB_2535847 |
| Secondary antibody | AF555 Donkey anti-rabbit IgG | 1:500 | Thermo Fisher Scientific Cat# A-31572, RRID:AB_162543 |
| Secondary antibody | AF555 Goat anti-mouse IgG2a | 1:500 | Thermo Fisher Scientific Cat# A-21137, RRID:AB_2535776 |
| Primers | |||
| Target | Forward/reverse primer (5′– 3′) | ||
| Episomal Plasmids (qPCR) | EBNA-1 sequence in episomal plasmids | TTTAATACGATTGAGGGCGTCT/GGTTTTGAAGGATGCGATTAAG | |
Resource details
Peripheral blood cells isolated from a 2-year old biliary atresia patient were cultured under an erythroblast expansion condition before reprogramming. Non-integrating episomal plasmids expressing OCT4, SOX2, KLF4, c-MYC and BCL-XL were used to generate iPSCs. Five independent iPSC-like clones were picked and expanded. One iPSC line BA08.1 was expanded and characterized for its identity and stem cell property. When cultured in feeder-free conditions on plates coated with either matrigel or vitronectin, the iPSC line displays typical morphology of human pluripotent stem cells. The iPSC line expresses pluripotency-related transcription factors OCT4 (Fig. 1A) and NANOG (Fig. 1B) as well as cell surface markers TRA-1-60 (Fig. 1C) and SSEA3 (Fig. 1D). Pluripotency of BA08.1 was assessed by in-vitro embryoid body formation assay. Cells expressing markers for endoderm (α-feto-protein), mesoderm (smooth muscle actin) and ectoderm (Tuj-1 β III-tubulin) were detected in day-10 embryoid bodies (Fig. 1E). Karyotyping analysis of the cell line demonstrates a normal male karyotype (46,XY) (Fig. 1F). We also examined the presence of episomal plasmid DNA in the established iPSC lines using a pair of PCR primers specific to the EBNA sequence that is common in all three reprogramming plasmids. PCR amplifications of plasmid DNA controls and iPSC genomic DNA show that the level of vector DNA in BA08.1 cell line is below detection limit by passage 11, even though EBNA DNA can be detected at passage 4 (Fig. 1G). To confirm the identity of this iPSC line, DNA profiling was conducted using a short tandem repeat (STR) typing assay that includes 15 STR loci and amelogenin. Data from STR analysis demonstrate a complete match between BA08 iPSC line and fibroblastic cells isolated from the patient (data archived but not shown). The iPSC culture is also shown to be free from mycoplasma contamination by MycoAlert™ mycoplasma detection kit (Fig. 1H).
Fig. 1.
Characterization of biliary atresia patient-specific iPSC line BA08-1.
Materials and methods
Peripheral blood mononuclear cell expansion and reprogramming
Mononuclear cells were isolated from patient blood by Ficoll gradient centrifugation and cultured for 10 days in serum-free medium containing SCF, IL-3, EPO and transferrin (Chou et al., 2015). At the end of expansion, the cells were transfected with plasmids MOS (expressing OCT4 and SOX2, addgene plasmid #64120), MMK (expressing c-MYC and KLF4, addgene plasmid #64121) and GBX (expressing BCL-XL, addgene plasmid #64123) using 4D Nucleofector (Lonza) (Chou et al., 2015). The cells were then cultured in the same erythroblast expansion condition for two days before being plated onto vitronectin-coated (Life Technologies) plate in DMEM medium containing 10% FBS. Essential 8 medium was used to replace the serum-containing medium the next day and was used throughout the reprogramming process and for continued iPSC culture. Colonies with iPSC morphology were picked at day 14 and expanded as previously described (Chou et al., 2015).
Immunocytochemistry and flow cytometry
iPSC colonies grown on matrigel-coated (Corning) plates or EBs attached on gelatin-coated plates were fixed with 4% paraformaldehyde (Sigma) and washed with PBS. Fixed cells were incubated overnight with appropriate primary antibodies at 4 °C for immunocytochemistry. The next day, cells were washed twice with PBS and incubated with appropriate Alexa Flour 555 or 488 conjugated secondary antibodies (Invitrogen) in PBS at room temperature for 45 min followed by PBS wash. Cells were counterstained with DAPI before immunofluorescence analysis. Images were taken using the motorized Nikon Ti-E microscope and NIS-Elements software. Scale bars in Fig. 1 represent 100 µm. For SSEA3 flow cytometry analysis, cells were digested by Accutase and washed by PBS. 1 × 105 cells were incubated with Alexa 488-SSEA3 or isotype control antibody for 30 min at 4 °C. After PBS washing, the cells were analysed by a Guava EasyCyte Flow Cytometer (Millipore).
Embryoid body differentiation
EBs were formed using FBS-containing differentiation medium and cultured in suspension for 7 days. The resulting EBs were then plated on gelatin-coated 24-well plates for additional 3 days. The cells were fixed with 4% paraformaldehyde and stained for markers representing the three germ layers (Liu et al., 2010).
Karyotype analysis
Karyotyping of BA08.1 cell line at passage 7 was conducted by G-banding at WiCell Cytogenetics Lab (Madison, Wisconsin).
PCR detection of EBNA1 DNA sequence
Total DNA from cells was isolated using the Quick-DNA™ Miniprep Kit (Zymo Research). Serially diluted reprogramming plasmid MOS were used as positive controls for amplification. 100 ng of total iPSC DNA, positive and negative (H2O) controls were amplified for 40 cycles using Phusion DNA polymerase (NEB) and primers specific for EBNA1 (Chou et al., 2015).
Mycoplasma testing
Mycoplasma test was performed on antibiotics-free overnight culture medium using the MycoAlert Kit (Lonza) following manufacturer's instruction.
STR analysis
STR profiling was conducted at Johns Hopkins Genetic Resources Core Facility using PowerPlex 16 HS Kit (Promega). Markers included in the kit are amelogenin, CSF1PO, D13S317, D16S539, D21S11, D5S818, D7S820, THO1, TPOX, vWA, D18S51, D3S1358, FGA, D8S1179, Penta E and Penta D.
Resource table
| Unique stem cell line identifier | JHUi001-A |
|---|---|
| Alternative name(s) of stem cell line | BA08.1 |
| Institution | Johns Hopkins University, Baltimore, USA |
| Contact information of distributor | Yoon-Young Jang, yjang3@jhmi.edu |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info | Age: 2 year |
| Sex: M | |
| Cell source | Peripheral blood |
| Method of reprogramming | Integration-free, episomal plasmid transfection |
| Genetic modification | NO |
| Type of modification | N/A |
| Associated disease | Biliary Atresia |
| Gene/locus | N/A |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | N/A |
| Cell line repository/bank | N/A |
| Ethical approval | The Johns Hopkins Medicine Institutional Review Boards (approval number: IRB00083753) |
Acknowledgments
This work was supported by Maryland Stem Cell Research Fund, Colleen Mitchel BA 5K/Zachary Meehan Memorial Fund and the Johns Hopkins Pediatric Liver Center.
References
- Chou BK, Gu H, Gao Y, Dowey SN, Wang Y, Shi J, Li Y, Ye Z, Cheng T, Cheng L. A facile method to establish human induced pluripotent stem cells from adult blood cells under feeder-free and xeno-free culture conditions: a clinically compliant approach. Stem Cells Transl. Med. 2015;4:320–332. doi: 10.5966/sctm.2014-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Davenport M, Kelly DA. Biliary atresia. Lancet. 2009;374:1704–1713. doi: 10.1016/S0140-6736(09)60946-6. [DOI] [PubMed] [Google Scholar]
- Liu H, Ye Z, Kim Y, Sharkis S, Jang YY. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810–1819. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]