Abstract
Epigenetics, present a new discipline that attempts to explain significant differences in phenotypes among patients with the same disease. In contrast to the other epigenetic mechanisms that modulate gene transcription, non-coding RNAs act at the post-transcriptional level. They directly modulate the gene expression of mRNA genes leading to mRNA target cleavage and degradation and translation repression. Bioinformatic predictions indicate that non coding RNAs may be involved in the regulation of 60% of the coding genes and each non-coding RNA can have multiple target genes, and each gene may be regulated by more than one non-coding RNAs. In the last decade several studies have shown a significant role of non-coding RNAs in the regulation of bone metabolism and function of bone cells opening a new era in the understanding of bone biology in health and disease.
Keywords: Bone Metabolism, Epigenetics, Long Non Coding RNA, microRNAs
1. Introduction
Osteoporosis is a complex multifactorial disease characterized by low bone mass and impaired bone microarchitecture leading to increased fragility. Genetic susceptibility is an important factor, determining predisposition to fragility fractures. However, increased variability between phenotypes and large, sometimes unexplained, differences in response to treatment among individuals with similar severity of bone disease, based on low bone mass and the presence of fragility factures, created the need for identifying the link between individual genetic aspects and environmental influences. In the last decade a large amount of data have focused on the epigenetic factors that are strongly suspected to be involved in bone biology and bone diseases[1,2].
Epigenetic mechanisms refer to pathways that influence gene expression in postnatal life without altering the DNA sequence. These mechanisms include DNA methylation posttranslational modifications of histones, and post-transcriptional regulation by non-coding RNAs.
Alterations in epigenetic mechanisms have been associated with aging and estrogen- deficiency-induced bone loss[3,4]. The progressive understanding of the roles epigenetic mechanisms play in normal bone metabolism and in multifactorial bone disorders is critical for the comprehensive knowledge of bone biology in health and disease.
In this review we outline current literature on the role of non-coding RNAs-the so called ‘dark matter of DNA’ in bone remodeling and function of bone cells.
2.1 MicroRNAs
MicroRNAs (miRs) are small – approximately 21 nucleotides long – non-coding RNA molecules derived from intergenic or intronic genomic regions. Mature miRs are formed from premature larger forms of precursor microRNAs after cleavage with a double-stranded RNA endoribonuclease (Dicer). Mature forms are then incorporated into the miR induced silencing complex (miRISC)[5] in order to facilitate binding to the target messenger RNA (mRNA), usually within its 3′ untranslated regions (UTR)[6].
2.2 Molecular Mechanisms
Binding of miRISC to the specific UTR of mRNA molecules with complementary sites results in post-transcriptional gene repression through inhibition of translation or mRNA destabilization[7] (Figure 1). Absolute match between miR and the target-mRNA results in absolute degradation of the targeted transcript. In most cases, however, the mRNA-miR complementarity is not perfect, and results in translational repression without cleavage of the target mRNA. Generally, miRs are negative regulators of gene expression, although in some cases they may activate translation of target mRNAs[8].
Figure 1.
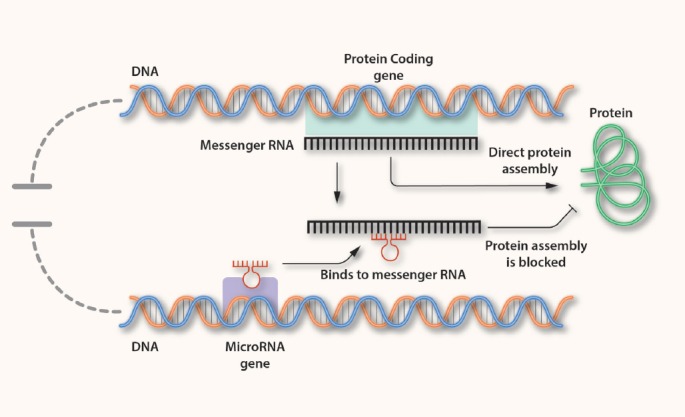
The circle of microRNA and mRNA interaction.
Interestingly a single miR can bind up to a hundred or more distinct mRNAs and 3’ UTR regions of most mRNAs contain binding sites for multiple miRs[9]. The complexity of this interaction allows the regulation of complex gene expression networks, following the paradigm of transcriptional factors[10]. Despite the appeared redundancy in miR-mRNA interactions, expression-alterations in single miR can alter significantly the cell phenotype.
A large amount of data of the last decade concerns the role of specific miR molecular signatures in various cancers leading the way for their potential use in the diagnostic and therapeutic field[1,11,12]. Increasing evidence has shown a similar role for miR in the regulation of osteogenesis and in metabolic bone diseases.
2.3 MicroRNAs and bone remodeling (Figure 2)
Figure 2.
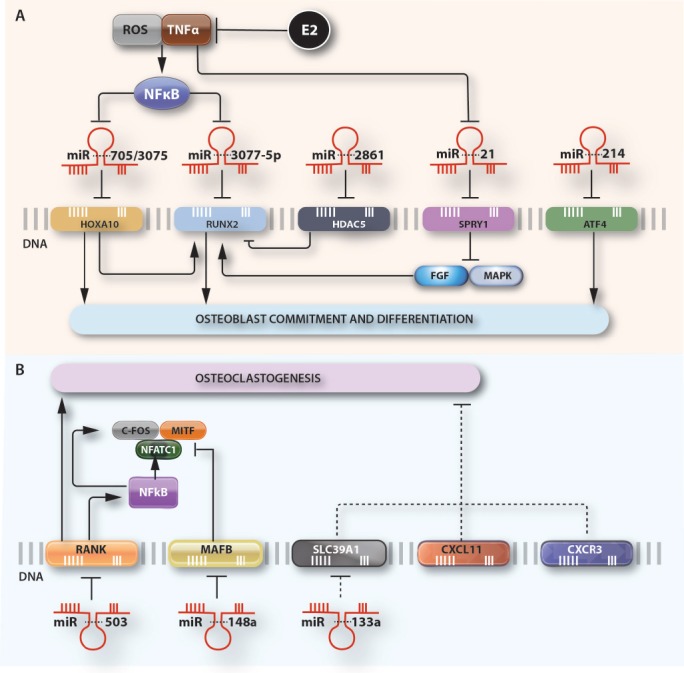
microRNAs affect the expression of specific genes during A) osteoblastogenesis and B) Osteoclastogenesis. MiR-3077-5p and miR-214 suppress osteogenic differentiation by directly inhibiting their respective target-genes. Conversely miR-705, miR 3075, miR-2861 and miR-21 directly target negative regulators of osteoblastogenesis resulting in enhanced osteoblast differentiation. The increased expression of miR-705 and miR-3077-5p and decreased levels of miR-21 are linked to high levels of TNFα and ROS, associated with estrogen deficiency. miR-21 suppresses Sprouty (SPRY) family of genes that antagonize the FGF signal transduction pathway in osteoblasts. Similarly, miR-503 inhibits osteoclastogenesis by directly targeting RANK gene while miR-148a and miR-133a promote osteoclastogenesis by directly targeting negative regulators of osteoclast differentiation. (Abbreviations: ROS, reactive oxygen species; TNFa, tumor necrosis factor; E2, estradiol; NFkb, nuclear factor-kappa-B; RUNX-2, Runt-related transcription factor 2; HOXA10, Homeobox A10; HDAC5, histone deacetylase 5; SPRY1, Protein sprouty homolog 1; ATF4, activating transcription factor 4; FGF, fibroblast growth factor; MAPK, mitogen activated protein kinase, MITF, Microphthalmia-associated transcription factor; NFATC1, Nuclear factor of activated T-cells, cytoplasmic 1; RANK, Receptor Activator of Nuclear Factor κ B; MAFB, V-maf musculoaponeurotic fibrosarcoma oncogene homolog B; SLC39A1, Solute Carrier Family 39 Member 1;CXCL11, C-X-C motif chemokine 11; CXCR3, C-X-C Motif Chemokine Receptor 3; miR, microRNA).
a) Micro-RNAs and osteoblast functions
Several studies have demonstrated significant effects of miRs in the regulation of genes that play a key role in osteoblast differentiation and function such as Runx-2, Osterix (Osx), bone morphogenetic proteins (BMPs) and components of the Wnt intracellular signaling pathway.
The Runx-2 transcription factor, which is considered the key master of osteogenesis, is both a direct and indirect target of several miRs that control its expression[13-22]. MiRs that bind directly to the Runx-2 complimentary sequence (namely miR-23a, miR-30a–d, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-211, miR-217, miR-335, miR-338, miR-433 and miR-3077-5p) negatively regulate the gene expression, while miR-2861 and miR-3960 that target the expression of Runx-2 suppressors, such as Histone deacetylase (HDAC) and Homeobox A2 (HOXA) genes respectively, stimulate the expression Runx-2 and thus enhance osteoblast differentiation[22,23]. In vivo the absence of miR-2861 in mice results in reduced BMD, bone formation, osteoblast number and Runx-2 protein levels while the protein expression of HDAC5 is increased and bone resorption and osteoclast activity, remain unaffected[23]. In line with in vitro and in vivo data in humans a homozygous mutation in pre-miR-2861 that blocks expression of miR-2861 was associated with primary osteoporosis in 2 related adolescents[23].
Downstream of Runx-2, another key regulator of osteoblastogenesis the zinc finger transcription factor Osterix (Osx), is also regulated by microRNAs. Osx promoter carries a Runx-2 responsive DNA element and Runx2 specifically transactivates Osx expression. It has been shown that miR-93 inhibits osteoblast mineralization by directly targeting and suppressing Osx expression[24].
Along with Runx-2 and Osterix, another osteoblast specific transcription factor, activating transcription factor 4 (ATF4), which is responsible for promoting osteocalcin expression, amino acid uptake and type I collagen synthesis, has been identified as a direct target of miR-214. High levels of miR-214 in bone tissue are associated with reduced bone formation with suppressed osteoblast activity and matrix mineralization in both human subjects and mouse models. Moreover, the osteoblast-specific inhibition of miR-214 significantly improves bone formation and bone mass in OVX- and hind limb-unloading induced osteoporosis in mice, leaving again bone resorption unaffected[25].
In earlier stages of osteoblast development miR-199a and miR-346, enhance commitment of mesenchymal stem cells (MSCs) in the osteoblast lineage by decreasing the expression of leukemia inhibitory factor (LIF) by MSCs[15]. These results demonstrate that differentiation of early stages of the osteoblast lineage is also regulated by miRs.
On the other hand, several studies have shown a complex regulatory feedback loop between genes related to osteogenesis and expression regulation of specific miRs. The miR-cluster 23a-27a-24-2 is negatively regulated by Runx-2 through a functional Runx-2 binding element. Interestingly, each of these miRs directly target the 3′ UTR of the gene that encodes a special AT-rich sequence binding protein -2 (SATB2), which act as a synergic co-factor with Runx2 in the nucleus to increase bone formation[19]. In addition Runx-2 binds to the promoter of the miR-3960/miR-2861 cluster and induces its expression controlling osteoblastogenesis through a positive regulatory feedback loop[22]. A regulatory feedback loop was also demonstrated for BMP-2. BMP-2 induces the expression of miRs that target muscle genes and down-regulates multiple microRNAs with osteogenic potential, pointing to the role of BMPs as selective inducers of tissue-specific phenotypes[14].
Wnt canonical intracellular signaling pathway is highly conserved in humans. Upon activation, the cytoplasmic protein β-catenin is translocated in the nucleus and forms dimers with the T-cell factor/lymphoid enhancer factor (TCF/LEF) group of transcription factors activating the transcription of target genes[26]. In bone tissue Wnt signaling has a critical role in both, skeletal development though actions in chondrocytes, and in bone remodeling throughout adult life. Studies of knockout and transgenic mice for Wnt pathway components have demonstrated that canonical signaling regulates most aspects of osteoblast physiology including commitment, differentiation, bone matrix formation/mineralization and apoptosis as well as coupling to osteoclastogenesis and bone resorption[26]. Several extracellular proteins act as agonists or antagonists to the Wnt membrane receptor complexes consisting of frizzled (Fz) G-protein-coupled receptors and the co–receptors low-density lipoprotein receptor-related protein 5 and 6 (LRP5/6) activating or suppressing Wnt signaling activity, respectively. Specific microRNAs, namely miR-29a, miR-218 and miR-335-5p have been demonstrated to directly target known Wnt antagonists such as Dickkopf-related protein 1 and 2 (DKK1 and DKK2), secreted Fz related protein-2 (SFRP2) and sclerostin (SOST) and enhance the Wnth/β-catenin signaling pathway and as a consequence osteoblast differentiation[27-29]. Apart from targeting the Wnt antagonists, activation of the Wnt/β-catenin signaling pathway by microRNAs is also mediated by suppressing the expression of adenomatosis polyposis coli (APC), an integral part of the β-catenin destruction complex in the cytoplasm[30], or catenin beta interacting protein 1 (CTNNBIP1), an inhibitor of β-catenin-mediated transcription[31]. Specifically miR-27 and miR-142-3p were shown to target directly APC gene in the mesenchymal precursor cell line, hFOB1.19[30], whereas, miR-29b directly down-regulates known inhibitors of osteoblast differentiation, such as CTNNBIP1, HDAC4, and tumor growth factor beta 3 (TGFβ3), proteins[31].
b) Micro-RNAs and osteoclast functions
Regulation of osteoclast differentiation is also targeted by microRNAs, although less extensively searched, compared to osteoblastogenesis. Among the key regulators of osteoclastogenesis, the receptor activator of nuclear factor-κB ligand/osteoprotegerin (RANKL/OPG) system, the transcription factors microphthalmia-associated transcription factor (MITF), C-Fos and PU.1 and the pro-apoptotic factors programmed cell death protein 4 (PDCD4) and FAS ligand (FasL)[32] have been identified as direct targets of specific miRs. The receptor of RANKL, RANK is identified as a direct target of miR-503[33]. Expression of miR-503 was decreased in circulating osteoclast progenitors (CD14 (+) peripheral blood mononuclear cells, PBMCs) from postmenopausal osteoporotic women compared with women without osteoporosis. Experiments with overexpression and silencing of miR-503 in CD14(+) PBMCs inhibited or enhanced, respectively RANKL-induced osteoclastogenesis. The results were confirmed in vivo with ovariectomy (OVX) mice models[33]. Silencing of miR-503 increased RANK protein expression, and decreased bone mass, whereas overexpression of miR-503 prevented bone loss in OVX mice. In line with preclinical data we demonstrated a significantly decreased expression of miR-503 and increased relative expression of RANK in the serum of osteoporotic women who sustained multiple vertebral fractures after discontinuation of denosumab, compared with osteoporotic women who did not experienced fractures after denosumab[34].
MiR-21 is also identified as a critical post-transcriptional regulator of osteoclastogenesis by both in vitro and in vivo studies. In vitro miR-21 regulated the expression of RANKL and its decoy receptor OPG in bone marrow mesenchymal stem cells from multiple myeloma[35], while PDCD4[36,37] and FasL[38] are also reported as a functional targets of miR-21 promoting osteoclastogenesis by suppressing osteoclasts-apoptosis. In vitro data were confirmed in miR-21 knockout mice[39]. In these mice which develop a normal skeletal phenotype, a postnatal miR-21 deficiency promoted trabecular bone mass accrual and prevented bone loss induced by OVX and aging due to decreased bone resorption and osteoclast function[39].
MiR-223 was shown to regulate osteoclastogenesis, both positively and negatively dependent on its expression levels[40,41]. The underlying molecular mechanism includes the suppression of the target-gene nuclear factor 1-A (NFI-A), an indirect inhibitor of the receptor of macrophage colony-stimulating factor (MCSF-R), which is one of the earliest pro-osteoclastogenic cytokines, thus stimulating osteoclast differentiation and the expression of other osteoclastogenic transcription factors such as MITF, c-Fos, and PU.1[40,41].
On the contrary, MITF is also a direct target for miR-155, that blocks activation of the osteoclast transcriptional program and thus osteoclastogenesis[42].
Studies with circulating CD14+ PBMCs have also reported a role for miR-148a[43] and for miR-133a[44] in osteoclastogenesis. ΜiR-148a promotes osteoclastogenesis in CD14+ PBMCs through suppression of V-maf musculoaponeurotic fibrosarcoma oncogene homolog B (MAFB), a transcription factor that negatively regulates RANKL-induced osteoclastogenesis. Silencing of miR-148a in mice resulted in increased bone mass through inhibition of bone resorption both in mice with OVX and in sham-operated controls. MiR-133a on the other hand was upregulated in circulating monocytes of postmenopausal women with low bone mass compared with age-matched women with high bone mass[44]. The bioinformatic target gene analysis in this study identified three potential osteoclast-related target genes, chemokine (C-X-C motif) ligand 11 and receptor 3 (CXCL11, CXCR3) and solute carrier family (zinc transporter), member 1 (SLC39A1)[44].
The majority of in vitro studies that investigated the role of miRs in bone metabolism were performed in mesenchymal and osteoblastic or osteoclastic cell lines of various stages of commitment predominantly of mouse origin, and less of human origin from bone marrow or CD14 PBMCs circulating osteoclasts. There are still no data available in osteocytes, although in one study miR-23a was reported to suppress Runx-2 in the terminally differentiated osteocyte, creating a feedback mechanism to attenuate osteoblast maturation[19].
The variability of the technical procedures, such as differentiation media used, RNA isolation methods and miR screening procedures, probably accounts for the increased number of different miRs that have been identified so far in bone cell cultures.
2.4 Circulating MicroRNAs in osteoporosis and fragility fractures
Based on in vitro and in vivo data demonstrating a role of specific miRs in bone remodeling several studies have investigated the association of circulating and tissue –expressed miRs with osteoporosis and fragility fractures in humans (Table 1).
Table 1.
Circulating microRNAs that have been identified in the serum, plasma or circulating monocytes of patients with bone disease.
| Study | Study Population | MicroRNAs Identified |
|---|---|---|
| Wang et al.[44] | Postmenopausal women with low BMD compared with postmenopausal women with high BMD | Upregulated: miR 133 |
| Seeliger et al.[45] | Patients with osteoporotic fractures compared with patients with non-osteoporotic fractures | Upregulated:miRNA-21, miRNA-23a, miRNA-24, miRNA-93, miRNA-100, miRNA-122a, miRNA-124a, miRNA-125b, miRNA- 148a |
| Weilner et al.[46] | Postmenopausal osteoporotic women with recent osteoporotic fractures compared with age-matched women without osteoporotic fractures |
Upregulated:miRNA-10a-5p, miRNA- 10b-5p, and miRNA-22-3p Downregulated:miRNA-133b, miRNA-328-3p, and let-7g-5p |
| Panach et al.[47] | Women with osteoporotic hip fracture compared with women with osteoarthritis | Upregulated:miRNA-21-5p, miRNA-122-5p, and miRNA-125-5b |
| Li et al.[48] | Postmenopausal women with osteoporosis and osteopenia compared with postmenopausal women with normal bone mass |
Upregulated:miRNA-133a Downregulated:miRNA-21 |
| You et al.[49] | Postmenopausal osteoporotic women compared with healthy premenopausal women | Downregulated:miRNA-27a |
| Heilmeier et al.[50] | Type 2 diabetic women with and without fractures and non-diabetic women with and without fractures |
Upregulated:miRNA-96- 5p, miRNA-181-5p, and miRNA-550a-5p, (diabetic patients) Downregulated: • miRNA-382-3p (diabetic and non-diabetic fractured patients) • miRNA-188-3p and miRNA-942 (osteoporotic patients) |
| Kocijan et al.[51] | Male and female patients with idiopathic or postmenopausal osteoporotic fractures compare with age- and gender- matched controls |
Upregulated:miRNA-152-3p, miRNA-335- 5p, and miRNA-320a Downregulated:let-7b-5p, miRNA-7-5p, miRNA-16-5p, miRNA-19a-3p, miRNA-19b-3p, miRNA- 29b-3p, miRNA-30e-5p, miRNA-93-5p, miRNA-140-5p, miRNA-215-5p, miRNA-186-5p, miRNA-324-3p, miRNA- 365a-3p, miRNA-378a-5p, miRNA-532-5p, and miRNA- 550a-3p |
| Yavropoulou et al.[52] | Postmenopausal osteoporotic women with and without fragility fractures | Downregulated:miR-21, miR-23a, and miR-29a |
| Anastasilakis et al.[53] | Postmenopausal women with osteoporosis treated with either denosumab or teriparatide |
Downregulated:
• miR-33 at 3 months of treatment with teriparatide compared with baseline values, • miR-133 at 12 months of treatment with teriparatide compared with baseline values |
In an interesting study by Seeliger et al., the serum miR profile was associated with the miR-expression in bone-tissue samples from patients with osteoporotic fractures[45] compared with patients with non-osteoporotic fractures. In particular, nine miRs (miR-21, miR-23a, miR-24, miR-93, miR-100, miR-122a, miR-124a, miR-125b, and miR- 148a) were found significantly upregulated in the serum of patients with osteoporotic fractures and five of them were also upregulated in the bone tissue of these patients (miR-21, miR-23a, miR-24, miR-100, and miR-125b).
MiR expression profile in the serum or plasma has been studied in different cohort studies with osteoporotic patients demonstrating inconsistent results. In one study which was performed in seven postmenopausal osteoporotic women suffering from recent osteoporotic fractures and seven controls (age-matched postmenopausal women without osteoporotic fractures), screening of 175 miRs led to the identification of 6 differentially expressed miRs, (namely miR-10a-5p, miR- 10b-5p, and miR-22-3p that were upregulated, and miR-133b, miR-328-3p, and let-7g-5p that were downregulated)[46]. Differential expression of miR-22-3p, miR-328-3p, and let-7g-5p were further validated in a larger sample (n=23). The second study compared women with osteoporotic hip fracture with women with osteoarthritis that had undergone surgery for hip implantation prosthesis[47]. In this study the researchers identified a different panel of miRs (miR-21-5p, miR-122-5p, and miR-125-5b) as biomarkers for osteoporotic fractures, that, however, were in line with the results previously reported by Seeliger et al[45].
In the third study researchers investigated the expression of three candidate miRs (miR-21, miR-133a, and miR-146a) in plasma samples from 120 Chinese postmenopausal women that were classified according to their BMD values in total hip, as osteoporotic, osteopenic or with normal bone mass[48]. ΜiR-133a was upregulated in circulating monocytes of osteoporotic and osteopenic women compared with the normal group, consistent with the results reported by Wang et al[44]. ΜiR-21 on the other hand was down-regulated in the group with osteoporosis and osteopenia compared with normal BMD values in contrast to what has been described in the Spanish cohort[47].
Serum miR-27a was also identified as being significantly downregulated in a cohort of 81 postmenopausal osteoporotic women compared with 74 healthy premenopausal women[49].
In 2 more recent studies serum miR signature was investigated in diabetic postmenopausal women with osteoporotic fractures[50] and in premenopausal women or men with idiopathic osteoporosis and fragility fractures[51].
The 375 miRs that were tested in the first study[50] demonstrated differential expression of miRs in type 2 diabetic women with and without fractures (48 microRNAs) as well as in nondiabetic women with and without fractures (23 microRNAs). Specifically, miR-382-3p, which was downregulated, was common between diabetic and nondiabetic fractured patients, miR-96- 5p, miR-181-5p, and miR-550a-5p, all upregulated, were specific among the diabetic patients and miR-188-3p and miR-942, both downregulated were specific among the osteoporotic patients. In the second study circulating miR signatures were investigated in male and female subjects with idiopathic or postmenopausal osteoporotic fractures[51]. Several miRs were differentially expressed (miR-152-3p, miR-335- 5p, and miR-320a were upregulated, and let-7b-5p, miR-7-5p, miR-16-5p, miR-19a-3p, miR-19b-3p, miR- 29b-3p, miR-30e-5p, miR-93-5p, miR-140-5p, miR-215-5p, miR-186-5p, miR-324-3p, miR- 365a-3p, miR-378a-5p, miR-532-5p, and miR- 550a-3p, were down-regulated) in fracture groups of men, premenopausal and postmenopausal women compared with their age- and gender- matched controls. Among these, eight (miR- 152-3p, miR-335-5p, miR-19a-3p, miR-19b-3p, miR-30e-5p, miR-140-5p, miR-324-3p, and miR-550a-3p) were reported to be of value in predicting fractures regardless of age and gender, with a higher predictive power than BMD or bone turnover markers. Although not all of the reported miRs had been yet related with bone remodeling, the results from this study provided evidence for the potential use of microRNA signatures as diagnostic tools of osteoporosis.
We have investigated the differential expression of specific miRs that were reported to correlate with bone metabolism both in serum and tissue samples and were linked with biological targets of miRs in humans in 48 postmenopausal osteoporotic women with and without fragility fractures compared with age-matched controls. In postmenopausal women with osteoporosis miR-124 and miR -2861 were significantly upregulated, and miR -21, miR -23a, miR -29a, miR -29b and miR -29c were downregulated compared with their age-matched controls. In the sub-group analysis of the osteoporotic women, miR -21, miR -23a, and miR -29a were significantly lower in those with at least one prevalent vertebral fracture compared with osteoporotic women without fractures[52]. In a more recent study, we searched for changes in the serum profile of miRs in postmenopausal osteoporotic women treated with either denosumab or teriparatide[53]. We have found that administration of teriparatide affects the relative expression of miRs related to the expression of the genes encoding the transcription factor Runx-2 (miR -33) and the antagonist of Wnt signaling Dkk-1 (miR -133). In particular miR -33-3p was significantly decreased at 3 months and miR -133a at 12 months of teriparatide treatment. Interestingly, we also found that changes of bone turnover markers (BTM) during treatment with denosumab and teriparatide were significantly correlated with changes in the serum expression of different miRs probably reflecting the different mechanism of action of the two anti-osteoporotic agents.
Specifically the relative expression of miR -24-3p and miR -27a was correlated with changes in BTM during teriparatide treatment and the relative expression of miR -21-5p, miR -23a-3p, miR -26a-5p, miR -27a, miR -222-5p and miR -335-5p with changes in BTM during treatment with denosumab[53].
Analysis of genome-wide association data, pointing on miR target sites and pre-miR coding sequences could also be of value in the quest for the pathogenetic mechanism of osteoporosis and other bone-related diseases. In a DNA-based study three polymorphisms in miR target sites in the 3′-UTR of the fibroblast growth factor 2 (FGF2) gene were found to be significantly associated with BMD of the femoral neck. The identified single nucleotide polymorphisms reside within predicted binding sites for nine miRs (miR -146a, miR -146b, miR -545, miR -25, miR -32, miR -92, miR -363, miR -367 and miR -92b) and possibly alter the binding affinity between FGF2 transcripts and miRs, resulting in higher levels of target-protein expression. Increased levels of FGF2, would stimulate osteoclast-induced bone resorption and thus increase bone loss[54].
3.1 Long non coding RNAs (lncRNAs)
Long ncRNAs (lncRNAs) have been defined as non-protein-coding nucleotide transcripts that are >200 nucleotides in length, allowing biochemical fractionation to exclude all known classes of small RNAs[55]. Depending on the anatomical properties of their gene loci, lncRNAs can be classified as: (1) sense or antisense: lncRNAs that are located on the same or the opposite strand of the nearest protein-coding genes; (2) divergent or convergent: lncRNAs that are transcribed in the divergent or convergent orientation compared to that of the nearest protein-coding genes; (3) intronic or intergenic: lncRNAs that locate inside the introns of a protein-coding gene, or in the interval regions between two protein-coding genes. There is no evidence, however, of any intrinsic difference directing their mechanism of action. The large number of these RNAs and, in many of them, the low evolutionary conservation and the low levels of expression, created a controversy about their true functionality[56,57]. Nonetheless, the continuing research provided considerable evidence that lack of primary sequence conservation in lncRNAs does not indicate lack of function[58,59], and many lncRNAs show evidence of structural conservation[60,61].
The loci that express lncRNAs show conservation of promoters[62], chromatin structure[63], and regulation by conventional transcription factors[64]. Furthermore, the lncRNAs were found to have a range of cellular half-lives as mRNAs[65] and they are differentially expressed in a tissue-specific manner[66,67]. Many lncRNAs are alternatively spliced, have isoforms that encode small proteins[68] and have intrinsic functions as trans-acting regulatory RNAs[69-71]. Furthermore, 3’UTRs can be separately expressed and convey genetic functions in trans[71], and may be further processed to produce subsidiary species[72]. lncRNAs are expressed in stem cells[73], muscles[74], T cells[75], mammary gland[76], and neurons[77], as well as in malignant neoplasms and other diseases[76,78-80]. This expression it seems to be partly controlled by conventional transcription factors[64,81].
The use of a bioinformatic method termed “Guilt by Association” allowed a general understanding of lncRNAs and protein coding genes that are tightly co-expressed and thus presumably co-regulated[82]. This method localizes protein coding genes and pathways significantly correlated with a given lncRNA using gene-expression analyses. This study revealed clusters of lncRNAs based on the pathways with which they associate. This approach has predicted diverse roles for lncRNAs, such as stem cell pluripotency, adipogenesis, osteogenesis and cancer.
3.2 Molecular Mechanisms of lncRNAs
lncRNAs participate in epigenetic regulations through recruitment of chromatin remodeling complexes to specific genomic loci[83,84]. Furthermore, they can regulate gene expression by interacting with proteins in various processes such as protein synthesis, imprinting, cell cycle control, alternative splicing, and chromatin structure regulation[82,85-91]. lncRNAs are also involved in enhancer-regulating gene activation (eRNAs), interacting directly with distal genomic loci[92]. Finally, certain lncRNAs serve as interacting partners or precursors for miRs or other short regulatory ncRNAs[93]. The ability of lncRNAs to bind to protein molecules provides them with several regulatory capacities. In the last few years several mechanistic modes of lncRNAs function have emerged. Four main themes that outline the main function are as follows: (Figure 3).
Figure 3.
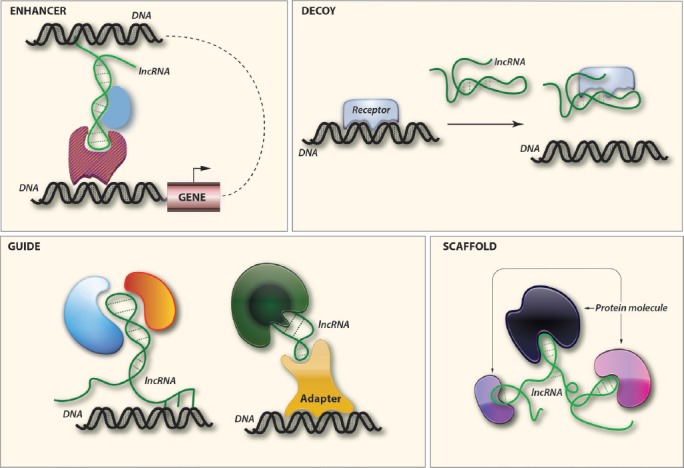
Emerging mechanistic modes of action for lncRNAs (See text).
a. Signal - enhancer
Since certain lncRNAs have been found to respond to various stimuli, they may act as molecular signals[94]. Studies using ChIP-Seq techniques, showed that the gene-activating enhancers produce lncRNA transcripts (eRNAs)[95], and their expression positively correlates with that of adjacent genes, indicating a possible role in the regulation of mRNA synthesis.
b. Decoy
lncRNA can function as molecular decoy to negatively regulate an effector. As an example Gas5 contains a sequence motif that resembles the DNA-binding site of the glucocorticoid receptor[96]. It can function as a decoy to release the receptor from DNA to prevent transcription of metabolic genes[97].
c. Guide
Binding with a target molecule, lncRNA may have the ability to guide it into the right position either in cis or in trans (on distantly located genes). The newly found eRNAs appear to exert their effects in cis by binding to specific enhancers and actively engaged in regulating mRNA synthesis[98].
d. Scaffold
Recent studies have found that several lncRNAs have the capacity to bind more than two protein molecules, where the lncRNAs serve as adaptors to form the functional protein complexes[98].
Despite the extensive research, it still remains difficult to understand the functions of lncRNAs. Unlike protein-coding genes whose mutations may result in altered phenotypes, mutations in lncRNAs often do not cause significant phenotypes[88]. It is likely that lncRNAs may function at specific stages of the developmental process or under specific conditions.
3.3 LncRNAs and Bone Remodeling
Despite the rapid accumulation of data about the role of lncRNAs in the epigenetic regulation of transcription and nuclear structure, their role in skeletal basic and clinical biology remains largely unknown. The delineation of specific miRs-targets regulating differentiation and function of bone cells and the role of lncRNAs in the process of miRs created an urgent need for greater understanding of the role of lncRNAs in bone metabolism.
In one of the earlier studies, a differential expression of an array of 116 lncRNAs was found during BMP-2 induced differentiation of C3H10T1/2 stem cells[99]. Among these, 59 were upregulated and 57 were downregulated in BMP-2 treated group. In addition, 24 cooperatively differentially expressed lncRNAs and nearby mRNA pairs were found. These observations indicate that lncRNA expression profiles are significantly altered in C3H10T1/2 undergoing early osteoblast differentiation and these results may provide insight into the mechanisms responsible for osteoblast differentiation.
A human study, using microarray analyses of monocytes in 73 Caucasian females with high or low hip BMD showed that 575 lncRNAs were differentially expressed between the two groups. In high BMD subjects, 309 lncRNAs were upregulated and 266 were downregulated. Analysis with genome browser showed that four genes and lncRNAs pairs were significantly correlated and cooperatively differentially expressed in high compared with low BMD subjects. In these four pairs, the coding genes, nuclear receptor subfamily 4 group A member 1(NR4A1) and ATP-binding cassette sub-family G member 1(ABCG1), seem to be important for monocytes differentiation and survival. These findings connected the lncRNAs profiles with osteoporosis and may suggest the regulatory mechanism between lncRNAs and protein coding genes in bone metabolism[100].
3.3.1 Specific roles
Specific lncRNAs are considered today to be important to bone formation (Table 2):
Table 2.
IncRNAs involved in Bone Metabolism.
| LncRNA | Function | Expression | Effects |
|---|---|---|---|
| HOTAIR | Protein recruiter | Adipogenic. Upregulated in osteosarcoma | Recruits EZH2 and SUZ12 to the HoxD cluster and silences the locus. β-catenin represses HOTAIR |
| DANCR | miRNA sponge | Adipogenic ↓ | May promote cell proliferation and differentiation toward the chondrocyte lineage |
| H19 | miRNA sponge miRNA precursor | Osteogenic ↑ Adipogenic ↓ |
Functions downstream of SOX9. H19 is processed into miR-675 that subsequently upregulates COL2A1. H19 is processed into miR-675, which targets TGFβ1 for degradation. H19 acts as a ceRNA, binding to miR-141 and miR-22, blocking binding |
| MEG3 | Protein recruiter | Osteogenic ↑ Adipogenic ↓ |
Upregulates BMP4 by disrupting the interaction between the SOX2 and BMP4 promoter |
↑upregulation; ↓downregulation.
Abbreviations: HOTAIR, HOX Transcript Antisense RNA; DANCR, Differentiation Antagonizing Non-Protein Coding RNA; MEG3, maternally expressed 3; EZH2, Enhancer of zeste homolog 2; SUZ12, Suppressor Of Zeste 12 Protein Homolog; SOX9, sex determining region Y)-box 9; miR, microRNA; TGFβ1, tumor growth factor β-1; BMP4, bone morphogenetic protein 4.
It has been shown that targeted disruption of the lncRNA for HOX transcript antisense RNA (HOTAIR) results in malformation of carpal and metacarpal bones and homeotic transformation of the spine[101]. HOTAIR is expressed in the posterior trunk and distal limb buds, as well as, in the mesenchymal cells of embryonic forelimbs[63]. HOTAIR binds to polycomb repressive complex 2 (PRC2) regulating the methylation of trimethylation of histone H3 at lysine 27 (H3K270, and in the formation of lysine (K)-specific demethylase 1A (Lsd1) complex, which demethylates H3K4. It has been proposed that HOTAIR creates a silent chromatin state and by repression of the expression of the Homeobox protein (HoxD) genes (Figure 4).
-
During osteogenesis it has been shown that lncRNA Differentiation Antagonizing Non-Protein Coding RNA (DANCR), recognized in the differentiation process of hFOB1.19 cells, may function as molecular switch regulating the commitment of MSCs[102]. DANCR associates with enhancer of zeste homolog 2 (EZH2), and this association results in the inhibition of Runx2 expression and subsequent osteoblast differentiation. DANCR-siRNA blocks the expression of endogenous DANCR, resulting in osteoblast differentiation, whereas DANCR overexpression is sufficient to inhibit osteoblast differentiation. These data suggested that DANCR is an essential participant of osteoblast commitment and differentiation.
Furthermore, DANCR inhibits adipogenic differentiation by acting as a sponge of miR-204[103]. Reduced expression of DANCR with the onset of adipogenic differentiation increases free-functioning miR-204 that downregulates the target genes Runx-2 and Sirtuin 1 (SIRT1)[103]. SIRT1 is an adipogenic inhibitor; therefore, downregulation of SIRT1 in C3H10T1/2 cells may promote adipogenic differentiation[104]. In addition, miR-204 suppresses Wnt/β-catenin signaling by modulating dishevelled homolog (DVL3) expression, which promotes adipogenic differentiation of human adipose-derived MSCs[105].
TGF-β and Wnt signaling pathways are involved in osteo-adipogenesis[106-109]. These signaling pathways are modulated by lncRNAs and associated regulators, such as miRs and other histone modifiers. The lncRNA H19 acts as a miR precursor of miR-675. Its expression increases during osteoblast differentiation but decreases during adipocyte differentiation in human MSCs and bone marrow MSCs[110]. ΜiR-675 may indirectly increase Runx2 expression and osteoblast differentiation in human MSCs[110]. Overexpression of miR-675 in human BMSCs inhibits adipogenic differentiation through the downregulation of class II HDACs[111,112]. H19 also acts as a miR sponge that captures miR-141, miR-22, miR-200a, and let-7 to inhibit their respective functions (Figure 5). Thus, the regulatory effects of H19 on osteo-adipogenesis are partially determined by its co-operating miRs.
The lncRN maternally expressed 3 (MEG3) is a maternally expressed, imprinted long non- coding RNA gene that has been shown to interact with the PRC2 complex and suppresses the expression of genes involved in the TGF-β pathway inhibiting the SMAD-dependent signaling pathway. In human BMSCs, increasing the expression of MEG3 activates BMP4 transcription and promotes osteogenic differentiation[113]. TGF-β1-mediated SMAD2/3 signaling negatively regulates the expression of miR-29[114,115]. In human osteoblasts, canonical Wnt signaling induces the expression of miR-29, as has been mentioned above which diminishes the effect of Wnt signaling by targeting the Wnt antagonists DKK-1, Kremen2, and SFRP2[27]. It has also been shown that miR-29 promotes osteoblast differentiation by downregulating anti-osteogenic factors, such as HDAC4, TGFB3, CTNNBIP1, activin receptor type-2A (ACVR2A), and dual specificity protein phosphatase 2 (DUSP2), in MC3T3-E1 cells[31]. Thus, the MEG3-miR-29 regulatory circuitry may promote osteoblast differentiation.
Figure 4.
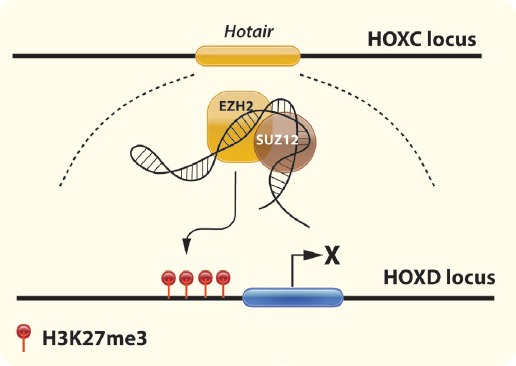
Regulation of HOXD gene expression by lncRNA HOTAIR. Transcribed from the HOXC locus, HOTAIR binds to SUZ12 and EZH2, part of the PRC2 complex. HOTAIR recruits the PRC2 complex and silences the HOXD locus through H3K27me3. (Abbreviations: HOXD, Homeobox D; lncRNA, long non coding RNA; HOTAIR, HOX transcript antisense RNA; SUZ12, SUZ12 Polycomb Repressive Complex 2 Subunit; EZH2, Enhancer of zeste homolog 2; PRC2, polycomb repressive complex 2).
Figure 5.
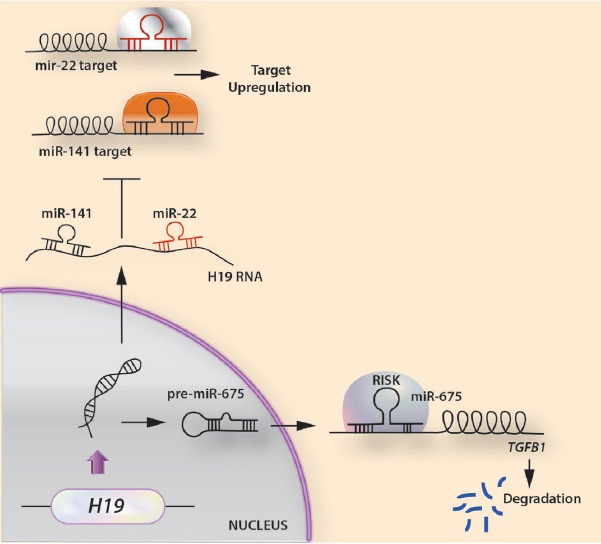
Modes of action of lncRNA H19 in osteogenesis. (A) H19 generates miR-675, which targets TGFβ1 leading it to degradation. (B) H19 acts as a sponge for removal of miR-141 and miR-22, thus leading to an up-regulation of their target mRNAs disrupting osteoblast differentiation.
4. Concluding Remarks
Non-coding RNAs were previously thought to arise as non-functional by-products in RNA splicing. Advancements in high-throughput sequencing technologies and bioinformatics, however, have changed our view regarding biogenesis and function of ncRNAs and their study has gradually become one of the most noticeable areas in the field of RNA biology. NcRNAs are an abundant, relatively stable, diverse and conserved class of RNA molecules, that act as competing endogenous nucleotides binding to RNAs and regulating their transcription or affect parental gene expression. The complex crosstalk among signalling molecules, lncRNAs, miRs, DNA methylases, and histone modifiers is crucial for achieving a balance between osteoblast and adipocyte lineage commitment and enchance bone remodeling. Αging is also characterized by epigenetic alterations that may induce trans-differentiation of osteoblasts into adipocytes, and explain age-associated marrow adipose tissue accumulation. Further functional studies of ncRNAs and their interacting partners during osteo-adipogenesis are necessary to clarify the specific roles of ncRNAs in bone metabolism and aging. The diverse functionality of the different classes of non-coding RNAs that participate in skeletal morphogenesis and development are leading the exploration of novel mechanisms that would explain bone metabolic diseases far beyond DNA mutational changes.
Studies identifying circulating small RNAs as potential “biomarkers” are emerging for cancer and other diseases. This would also be a future direction for skeletal disorders, as ncRNAs reflect changes in activity of bone cells, which may occur as an early sign of disease progression, as response to therapy or even lead to the development of specific molecular targets for therapeutic intervention. Up to now, the clinical utility of circulating ncRNAs in bone metabolic diseases has not been established, since study designs were not suited to identify which ncRNAs can give a prognosis for future risk of fragility fractures, or predict a treatment response. Despite the large gaps in our existing knowledge, however, these fascinating non-coding nucleotide sequences seem to act as critical regulators of normal development and disease and the greater understanding of their biogenesis and function will open a new era in the quest for novel biomarkers and target molecules for drug development.
Footnotes
The authors have no conflict of interest.
Edited by: P. Makras
References
- 1.Sandoval J, Peiro-Chova L, Pallardo FV, Garcia-Gimenez JL. Epigenetic biomarkers in laboratory diagnostics:emerging approaches and opportunities. Expert Rev Mol Diagn. 2013;13:457–71. doi: 10.1586/erm.13.37. [DOI] [PubMed] [Google Scholar]
- 2.Vrtacnik P, Marc J, Ostanek B. Epigenetic mechanisms in bone. Clin Chem Lab Med. 2014;52:589–608. doi: 10.1515/cclm-2013-0770. [DOI] [PubMed] [Google Scholar]
- 3.Sen P, Shah PP, Nativio R, Berger SL. Epigenetic Mechanisms of Longevity and Aging. Cell. 2016;166:822–39. doi: 10.1016/j.cell.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An JH, Ohn JH, Song JA, et al. Changes of microRNA profile and microRNA-mRNA regulatory network in bones of ovariectomized mice. J Bone Miner Res. 2014;29:644–56. doi: 10.1002/jbmr.2060. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs:target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther. 2011;13:220. doi: 10.1186/ar3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation:microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 9.Hackl M, Heilmeier U, Weilner S, Grillari J. Circulating microRNAs as novel biomarkers for bone diseases - Complex signatures for multifactorial diseases? Mol Cell Endocrinol. 2016;432:83–95. doi: 10.1016/j.mce.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–6. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 11.Aushev VN, Lee E, Zhu J, et al. Novel predictors of breast cancer survival derived from miRNA activity analysis. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-17-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer:biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–9. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Xie RL, Croce CM, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–8. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Hassan MQ, Volinia S, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906–11. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Xie RL, Gordon J, et al. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Biol Chem. 2012;287:21926–35. doi: 10.1074/jbc.M112.340398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–64. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu T, Zhou H, Hong Y, Li J, Jiang X, Huang H. miR-30 family members negatively regulate osteoblast differentiation. J Biol Chem. 2012;287:7503–11. doi: 10.1074/jbc.M111.292722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim EJ, Kang IH, Lee JW, Jang WG, Koh JT. MiR-433 mediates ERRgamma-suppressed osteoblast differentiation via direct targeting to Runx2 mRNA in C3H10T1/2 cells. Life Sci. 2013;92:562–8. doi: 10.1016/j.lfs.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Hassan MQ, Gordon JA, Beloti MM, et al. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107:19879–84. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tome M, Lopez-Romero P, Albo C, et al. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18:985–95. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao L, Yang X, Su X, et al. Redundant miR-3077-5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis. 2013;4:e600. doi: 10.1038/cddis.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu R, Liu W, Li H, et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J Biol Chem. 2011;286:12328–39. doi: 10.1074/jbc.M110.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Xie H, Liu W, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest. 2009;119:3666–77. doi: 10.1172/JCI39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, Cheng P, Chen C, et al. miR-93/Sp7 function loop mediates osteoblast mineralization. J Bone Miner Res. 2012;27:1598–606. doi: 10.1002/jbmr.1621. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Guo B, Li Q, et al. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 26.Yavropoulou MP, Yovos JG. The role of the Wnt signaling pathway in osteoblast commitment and differentiation. Hormones (Athens) 2007;6:279–94. doi: 10.14310/horm.2002.1111024. [DOI] [PubMed] [Google Scholar]
- 27.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 2010;285:25221–31. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassan MQ, Maeda Y, Taipaleenmaki H, et al. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem. 2012;287:42084–92. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Tu Q, Bonewald LF, et al. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26:1953–63. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, Xu Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun. 2010;402:186–9. doi: 10.1016/j.bbrc.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Hassan MQ, Jafferji M, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–84. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yavropoulou MP, Yovos JG. Osteoclastogenesis--current knowledge and future perspectives. J Musculoskelet Neuronal Interact. 2008;8:204–16. [PubMed] [Google Scholar]
- 33.Chen C, Cheng P, Xie H, et al. MiR-503 regulates osteoclastogenesis via targeting RANK. J Bone Miner Res. 2014;29:338–47. doi: 10.1002/jbmr.2032. [DOI] [PubMed] [Google Scholar]
- 34.Anastasilakis AD, Yavropoulou MP, Makras P, et al. Increased osteoclastogenesis in patients with vertebral fractures following discontinuation of denosumab treatment. Eur J Endocrinol. 2017;176:677–83. doi: 10.1530/EJE-16-1027. [DOI] [PubMed] [Google Scholar]
- 35.Pitari MR, Rossi M, Amodio N, et al. Inhibition of miR-21 restores RANKL/OPG ratio in multiple myeloma-derived bone marrow stromal cells and impairs the resorbing activity of mature osteoclasts. Oncotarget. 2015;6:27343–58. doi: 10.18632/oncotarget.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117:3648–57. doi: 10.1182/blood-2010-10-311415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 38.Sugatani T, Hruska KA. Down-regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J Cell Biochem. 2013;114:1217–22. doi: 10.1002/jcb.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu CH, Sui BD, Du FY, et al. miR-21 deficiency inhibits osteoclast function and prevents bone loss in mice. Sci Rep. 2017;7:43191. doi: 10.1038/srep43191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. 2009;284:4667–78. doi: 10.1074/jbc.M805777200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. 2007;101:996–9. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 42.Mann M, Barad O, Agami R, Geiger B, Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci U S A. 2010;107:15804–9. doi: 10.1073/pnas.0915022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng P, Chen C, He HB, et al. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res. 2013;28:1180–90. doi: 10.1002/jbmr.1845. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Li L, Moore BT, et al. MiR-133a in human circulating monocytes:a potential biomarker associated with postmenopausal osteoporosis. PLoS One. 2012;7:e34641. doi: 10.1371/journal.pone.0034641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeliger C, Karpinski K, Haug AT, et al. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res. 2014;29:1718–28. doi: 10.1002/jbmr.2175. [DOI] [PubMed] [Google Scholar]
- 46.Weilner S, Skalicky S, Salzer B, et al. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone. 2015;79:43–51. doi: 10.1016/j.bone.2015.05.027. [DOI] [PubMed] [Google Scholar]
- 47.Panach L, Mifsut D, Tarin JJ, Cano A, Garcia-Perez MA. Serum Circulating MicroRNAs as Biomarkers of Osteoporotic Fracture. Calcif Tissue Int. 2015;97:495–505. doi: 10.1007/s00223-015-0036-z. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Wang Z, Fu Q, Zhang J. Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers. 2014;19:553–6. doi: 10.3109/1354750X.2014.935957. [DOI] [PubMed] [Google Scholar]
- 49.You L, Pan L, Chen L, Gu W, Chen J. MiR-27a is Essential for the Shift from Osteogenic Differentiation to Adipogenic Differentiation of Mesenchymal Stem Cells in Postmenopausal Osteoporosis. Cell Physiol Biochem. 2016;39:253–65. doi: 10.1159/000445621. [DOI] [PubMed] [Google Scholar]
- 50.Heilmeier U, Hackl M, Skalicky S, et al. Serum miRNA Signatures Are Indicative of Skeletal Fractures in Postmenopausal Women With and Without Type 2 Diabetes and Influence Osteogenic and Adipogenic Differentiation of Adipose Tissue-Derived Mesenchymal Stem Cells In Vitro. J Bone Miner Res. 2016;31:2173–92. doi: 10.1002/jbmr.2897. [DOI] [PubMed] [Google Scholar]
- 51.Kocijan R, Muschitz C, Geiger E, et al. Circulating microRNA Signatures in Patients With Idiopathic and Postmenopausal Osteoporosis and Fragility Fractures. J Clin Endocrinol Metab. 2016;101:4125–34. doi: 10.1210/jc.2016-2365. [DOI] [PubMed] [Google Scholar]
- 52.Yavropoulou MP, Anastasilakis AD, Makras P, Tsalikakis DG, Grammatiki M, Yovos JG. Expression of microRNAs that regulate bone turnover in the serum of postmenopausal women with low bone mass and vertebral fractures. Eur J Endocrinol. 2017;176:169–76. doi: 10.1530/EJE-16-0583. [DOI] [PubMed] [Google Scholar]
- 53.Anastasilakis AD, Makras P, Pikilidou M, Tournis S, Makris K, Bisbinas I, Tsave O, Yovos JG, Yavropoulou MP. Changes of circulating micro-RNAs in response to treatment with teriparatide or denosumab in postmenopausal osteoporosis. J Clin Endocrinol Metab. doi: 10.1210/jc.2017-02406. doi.org/10.1210/jc.2017-02406. [DOI] [PubMed] [Google Scholar]
- 54.Lei SF, Papasian CJ, Deng HW. Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res. 2011;26:72–8. doi: 10.1002/jbmr.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs:insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 56.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clark MB, Amaral PP, Schlesinger FJ, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion:e1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs:lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith MA, Gesell T, Stadler PF, Mattick JS. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 2013;41:8220–36. doi: 10.1093/nar/gkt596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary conservation of long non-coding RNAs;sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063–71. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carninci P, Sandelin A, Lenhard B, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 63.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cawley S, Bekiranov S, Ng HH, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 65.Clark MB, Johnston RL, Inostroza-Ponta M, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 67.Ravasi T, Suzuki H, Pang KC, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–9. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chooniedass-Kothari S, Emberley E, Hamedani MK, et al. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–7. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- 69.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dinger ME, Gascoigne DK, Mattick JS. The evolution of RNAs with multiple functions. Biochimie. 2011;93:2013–8. doi: 10.1016/j.biochi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 71.Mercer TR, Wilhelm D, Dinger ME, et al. Expression of distinct RNAs from 3'untranslated regions. Nucleic Acids Res. 2011;39:2393–403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mercer TR, Dinger ME, Bracken CP, et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010;20:1639–50. doi: 10.1101/gr.112128.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dinger ME, Amaral PP, Mercer TR, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pang KC, Dinger ME, Mercer TR, et al. Genome-wide identification of long noncoding RNAs in CD8+T cells. J Immunol. 2009;182:7738–48. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 76.Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–91. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–33. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takeda K, Ichijo H, Fujii M, et al. Identification of a novel bone morphogenetic protein-responsive gene that may function as a noncoding RNA. J Biol Chem. 1998;273:17079–85. doi: 10.1074/jbc.273.27.17079. [DOI] [PubMed] [Google Scholar]
- 79.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 80.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson R, Teh CH, Jia H, et al. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA. 2009;15:85–96. doi: 10.1261/rna.1127009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 84.Nagano T, Mitchell JA, Sanz LA, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 85.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 87.Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–55. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 90.Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta. 2009;1790:936–47. doi: 10.1016/j.bbagen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 91.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs:functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Orom UA, Shiekhattar R. Noncoding RNAs and enhancers:complications of a long-distance relationship. Trends Genet. 2011;27:433–9. doi: 10.1016/j.tig.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14(Spec No 1):R121–32. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 94.Hung T, Chang HY. Long noncoding RNA in genome regulation:prospects and mechanisms. RNA Biol. 2010;7:582–5. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zuo C, Wang Z, Lu H, Dai Z, Liu X, Cui L. Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Mol Med Rep. 2013;8:463–7. doi: 10.3892/mmr.2013.1540. [DOI] [PubMed] [Google Scholar]
- 100.Zhou Ya DHW. Long non-coding RNA analyses for osteoporosis in Caucasian women. Transcriptomics-2015:Transcriptomics-2015. 2015:164. [Google Scholar]
- 101.Li L, Liu B, Wapinski OL, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–7. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 103.Li M, Sun X, Cai H, et al. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim Biophys Acta. 2016;1859:871–82. doi: 10.1016/j.bbagrm.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 104.Backesjo CM, Li Y, Lindgren U, Haldosen LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21:993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- 105.He H, Chen K, Wang F, et al. miR-204-5p promotes the adipogenic differentiation of human adipose-derived mesenchymal stem cells by modulating DVL3 expression and suppressing Wnt/beta-catenin signaling. Int J Mol Med. 2015;35:1587–95. doi: 10.3892/ijmm.2015.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rahimi N, Tremblay E, McAdam L, Roberts A, Elliott B. Autocrine secretion of TGF-beta 1 and TGF-beta 2 by pre-adipocytes and adipocytes:a potent negative regulator of adipocyte differentiation and proliferation of mammary carcinoma cells. In Vitro Cell Dev Biol Anim. 1998;34:412–20. doi: 10.1007/s11626-998-0023-z. [DOI] [PubMed] [Google Scholar]
- 107.Kanazawa A, Tsukada S, Kamiyama M, Yanagimoto T, Nakajima M, Maeda S. Wnt5b partially inhibits canonical Wnt/beta-catenin signaling pathway and promotes adipogenesis in 3T3-L1 preadipocytes. Biochem Biophys Res Commun. 2005;330:505–10. doi: 10.1016/j.bbrc.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 108.Kennell JA, MacDougald OA. Wnt signaling inhibits adipogenesis through beta-catenin-dependent and -independent mechanisms. J Biol Chem. 2005;280:24004–10. doi: 10.1074/jbc.M501080200. [DOI] [PubMed] [Google Scholar]
- 109.Wu M, Chen G, Li YP. TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang Y, Zheng Y, Jia L, Li W. Long Noncoding RNA H19 Promotes Osteoblast Differentiation Via TGF-beta1/Smad3/HDAC Signaling Pathway by Deriving miR-675. Stem Cells. 2015;33:3481–92. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 111.Nebbioso A, Dell'Aversana C, Bugge A, et al. HDACs class II-selective inhibition alters nuclear receptor-dependent differentiation. J Mol Endocrinol. 2010;45:219–28. doi: 10.1677/JME-10-0043. [DOI] [PubMed] [Google Scholar]
- 112.Huang Y, Zheng Y, Jin C, Li X, Jia L, Li W. Long Non-coding RNA H19 Inhibits Adipocyte Differentiation of Bone Marrow Mesenchymal Stem Cells through Epigenetic Modulation of Histone Deacetylases. Sci Rep. 2016;6:28897. doi: 10.1038/srep28897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhuang W, Ge X, Yang S, et al. Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells From Multiple Myeloma Patients By Targeting BMP4 Transcription. Stem Cells. 2015;33:1985–97. doi: 10.1002/stem.1989. [DOI] [PubMed] [Google Scholar]
- 114.Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS One. 2012;7:e33766. doi: 10.1371/journal.pone.0033766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kwon JJ, Nabinger SC, Vega Z, et al. Pathophysiological role of microRNA-29 in pancreatic cancer stroma. Sci Rep. 2015;5:11450. doi: 10.1038/srep11450. [DOI] [PMC free article] [PubMed] [Google Scholar]


