Abstract
The Arabidopsis LEAFY COTYLEDON2 (LEC2) gene is a central embryonic regulator that serves critical roles both early and late during embryo development. LEC2 is required for the maintenance of suspensor morphology, specification of cotyledon identity, progression through the maturation phase, and suppression of premature germination. We cloned the LEC2 gene on the basis of its chromosomal position and showed that the predicted polypeptide contains a B3 domain, a DNA-binding motif unique to plants that is characteristic of several transcription factors. We showed that LEC2 RNA accumulates primarily during seed development, consistent with our finding that LEC2 shares greatest similarity with the B3 domain transcription factors that act primarily in developing seeds, VIVIPAROUS1/ABA INSENSITIVE3 and FUSCA3. Ectopic, postembryonic expression of LEC2 in transgenic plants induces the formation of somatic embryos and other organ-like structures and often confers embryonic characteristics to seedlings. Together, these results suggest that LEC2 is a transcriptional regulator that establishes a cellular environment sufficient to initiate embryo development.
Embryogenesis in flowering plants begins with the double fertilization event in which the zygote and endosperm are formed after fusion of sperm cells with the egg cell and central cell of the female gametophyte, respectively. The endosperm initially undergoes syncytial development with formation of nuclear-cytoplasmic domains, but later cellularizes (1). The developing embryo is nourished by the endosperm and, in many plants, only the peripheral layer of the endosperm remains in the mature seed. Development of the zygote into the mature embryo can be divided conceptually into two distinct phases. During the early morphogenesis phase, the basic body plan of the plant is established with expression of polarity as a shoot–root axis, specification of morphological domains within the embryo, and formation of embryonic tissue and organ systems (2–4). The morphogenesis phase is followed by a period of maturation in which processes critical for seed formation occur (5, 6). During this late phase, reserves such as storage proteins and lipids are synthesized at high rates and accumulate in the seed. It is also during the maturation phase that the embryo acquires the ability to withstand desiccation at the final stage of seed development. At the end of embryogenesis, the seed consists of a mature, desiccated embryo that is quiescent metabolically. Although many aspects of embryogenesis have been characterized extensively, little is known at a mechanistic level of the processes that initiate embryo development.
The Arabidopsis LEAFY COTYLEDON (LEC) genes, LEC1, LEC2, and FUSCA3 (FUS3), play key roles in controlling embryo development (7). Unlike most other embryonic regulators that function during specific stages of embryogenesis (8–11), LEC genes are unique in that they are required for normal development during both the morphogenesis and maturation phases. Early in embryogenesis, LEC genes are required to specify suspensor cell fate and cotyledon identity (12–16). Late in embryogenesis, LEC genes are needed during the maturation phase for the acquisition of desiccation tolerance and the expression of many maturation-specific genes (13–17). Consistent with the finding that conditions that promote maturation suppress germination (6), lec mutant embryos prematurely activate the postgermination program (13, 15, 16, 18). Thus, LEC genes play a central role in controlling many aspects of embryogenesis, and they are candidates as regulators that coordinate the morphogenesis and maturation phases.
Identification and analysis of two LEC genes confirmed their regulatory role in embryogenesis and provided insight into their functions. LEC1 shares extensive sequence similarity with the HAP3 subunit of CCAAT-binding transcription factor, implicating LEC1 as a transcriptional regulator (12). Ectopic expression of LEC1 confers embryonic characteristics to seedlings and results in the formation of embryo-like structures on the surfaces of leaves, suggesting that the gene plays a role in conferring embryogenic competence to cells (12). Thus, we hypothesized that LEC1 establishes a cellular environment that promotes embryo development and that this environment coordinates the morphogenesis and maturation phases. FUS3 also encodes a regulatory protein: a B3 domain transcription factor that accumulates primarily during seed development (19). Transient assays showed that FUS3 is sufficient to activate genes usually expressed during maturation (20). Thus, two LEC genes seem to be involved in controlling embryo development by regulating transcription of other genes.
In this article, we focus on the LEC2 gene to determine its role in embryo development. Because genetic studies suggest that LEC1 and LEC2 may have partially redundant functions (12, 15), it is possible that LEC2 also functions in the initiation and coordination of embryo development. We cloned the LEC2 gene and showed that it is expressed preferentially during embryogenesis and encodes a protein with similarity to other seed-specific transcription factors. Significant insight into the role of the gene was obtained by showing that transgenic plants expressing the LEC2 gene ectopically form somatic embryos. Together, these results indicate that LEC2 is sufficient to induce embryogenic competence.
Materials and Methods
Plant Material.
Arabidopsis thaliana (L.) Heynh. was grown as described (16). lec2-1 mutant in ecotype Ws-0 was provided by David Meinke (Oklahoma State Univ., Stillwater). Three new lec2 mutant alleles were identified from a Ws-4 ecotype population mutagenized with T-DNA (21). Genetic tests showed that all three mutants failed to complement the original lec2-1 mutation. Mutant lines formerly designated CPT9, CUC3, and DLM1 were renamed lec2-3, lec2-4, and lec2-5, respectively. Homozygous lec2 mutant lines were maintained by immature seed rescue as described (16).
Gene Cloning.
To enrich for recombination breakpoints near the LEC2 gene, we crossed homozygous lec2-1 mutants (Ws-0 ecotype) with either homozygous distorted trichomes1-1 (dis1-1) or dis2-1 mutants (Ler ecotype). Among F2 progeny, we selected 381 recombinants homozygous for the dis1 mutation and heterozygous for lec2 and 173 that were homozygous for dis2 and heterozygous for lec2. Polymorphisms that distinguish Ws-0 and Ler chromosomes were identified by amplifying chromosomal regions with primers designed from restriction fragment length polymorphism and bacterial artificial chromosome (BAC) clones and sequencing the products. Polymorphic sequences were used to generate PCR-based markers for genetic mapping (22–24).
Cosmid clones for transgene complementation experiments were constructed by inserting DNA fragments from BAC clone F1A10 (GenBank accession no. AL080330) into the plant transformation vector pOCA28 (25). Cosmid clones containing BAC DNA that spanned the LEC2 gene were transferred into homozygous lec2-1 mutants as described (12). Clones that suppressed the mutation were identified by using progeny segregation tests and genotyping with PCR-based markers.
RNA Analysis.
Total RNA was isolated by the method of Wilkins and Smart (26). LEC2 RNA was detected in tissues by using PCR amplification of reverse transcription products with a primer containing the translation initiation codon, 5′-AAATGGATAACTTCTTACCCTTTCC-3′, and another immediately upstream of the translation termination codon, 5′-CGGATGAACCCACGTACG-3′ (27). Potential genomic DNA contaminants of RNA samples were eliminated by digestion with DNase I, MseI, and DdeI before the reverse transcription step.
In situ hybridization experiments were done and cruciferin and oleosin RNA probes were prepared as described (16, 28). To increase the hybridization specificity, LEC2 antisense RNA from bases −36 to +446 relative to the translation initiation site was used as a probe and RNase was used at a concentration of 100 μg/ml.
35S∷LEC2 Plants.
A LEC2 cDNA clone containing the complete protein-coding region was constructed, starting with a cDNA clone consisting of LEC2 sequences from 14 bases downstream of the translation initiation codon to the poly(A) tail. Amplification products of the LEC2 gene region were fused with this cDNA clone to add 5′ sequences to position −36. The sequence of this cDNA corresponded with that of amplified LEC2 reverse transcription products. This LEC2 cDNA was inserted between the cauliflower mosaic virus 35S promoter (−1329 to +7 relative to the transcription start site) and the octopine synthase 3′ terminator (29), and the 35S∷LEC2∷ocs construct was then transferred into pBJ49. lec2-1 and lec2-5 homozygous mutant plants and wild-type Ws-0 plants were transformed by standard methods (21). Transgenic seedlings were identified by their resistance to hygromycin and by the presence of LEC2 transgene-specific sequences as verified by PCR.
Results
LEC2 Gene Isolation.
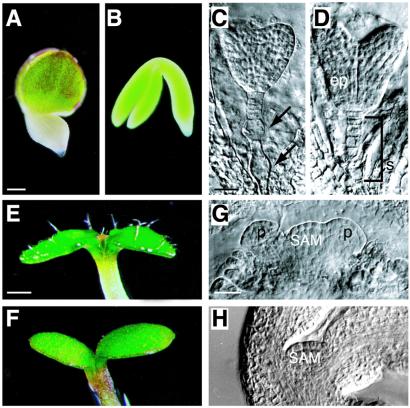
We identified three new mutant alleles of the LEC2 gene from T-DNA insertion lines (21) that were designated lec2-3, lec2-4, and lec2-5. Approximately 25% of late-stage embryos from plants heterozygous for these lec2 mutations were indistinguishable from the original lec2-1 mutant (15) and differed morphologically from wild-type and lec1 and fus3 mutant embryos (Fig. 1 A and B; data not shown). By contrast to wild type, mutant embryos from all lines had abnormal suspensors (Fig. 1 C and D), produced trichomes on cotyledon surfaces (Fig. 1 E and F), and possessed precociously activated shoot apical meristems (Fig. 1 G and H). Unlike lec1 and fus3 mutant embryos that die because of their intolerance to desiccation, 50–80% of embryos homozygous for the newly identified lec2 mutations and the original lec2-1 mutation were able to germinate from freshly dried seeds. Of those that germinated, 93–97% displayed defective cotyledons in which the distal regions had degenerated to different extents (data not shown; ref. 15). The variable sensitivity of lec2 mutants to desiccation suggests defects in maturation processes in distal regions of cotyledons.
Figure 1.
Morphological phenotype of lec2 mutants. (A) lec2-5, (C) lec2-3, and (E, G) lec2-4 mutant embryos. (B, D, F, H) Wild-type embryo. (A and B) Whole-mount photographs of maturing embryos. (C and D) Embryonic suspensors as viewed by using differential interference contrast (DIC) microscopy of cleared seeds. Arrows point to abnormal suspensor cells in lec2 mutants. (E and F) Cotyledons of seedlings grown for 4–5 days. A lec2 mutant seedling germinated before desiccation possessed trichomes on the adaxial surface of cotyledons. (G and H) Shoot apices of curled cotyledon-stage embryos seen with DIC optics. The shoot apical meristem of lec2 mutants is domed and possesses leaf primordia in contrast to the unactivated meristem of wild types. ep, Embryo proper; p, leaf primordium; SAM, shoot apical meristem; s, suspensor. [Bars = 100 μm (A), 20 μm (C, G), 300 μm (E).]
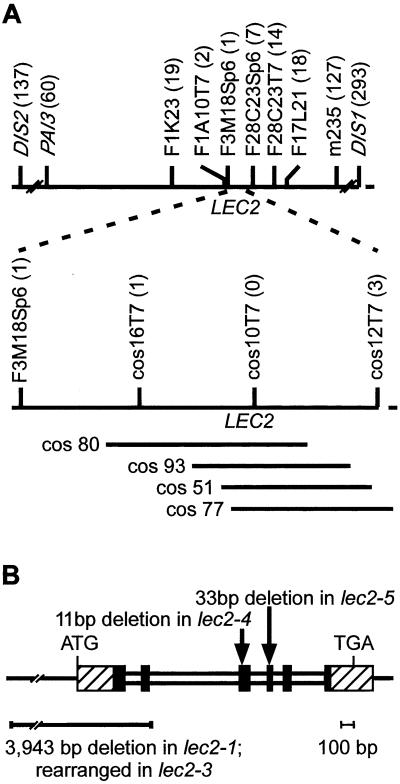
Although all four lec2 mutant alleles were derived from plants mutagenized with T-DNA, none of the mutations resulted from an insertion into the gene. Therefore, we cloned the LEC2 gene based on its position in the genome. LEC2 was mapped initially between two visual markers, dis2 and dis1, and the molecular markers PAI3 and m235 on chromosome 1 (30, 31). As shown diagrammatically in Fig. 2A, markers corresponding to the ends of BAC clones that lie between PAI3 and m235 were mapped by using plants selected for recombination breakpoints between LEC2 and DIS2 and between LEC2 and DIS1. Cosmid clones were prepared from the BAC F1A10 predicted to span the LEC2 gene. As detailed in Fig. 2A, genetic mapping of selected cosmid clone ends and transgene complementation experiments localized the LEC2 gene to an 11-kb region equivalent to positions 40,250–51,455 of BAC F3H9 (GenBank accession no. AC021044).
Figure 2.
Genetic mapping and positional cloning of the LEC2 gene. (A) Diagrammatic representation of the interval on chromosome 1 between DIS1 and DIS2. The position of the LEC2 gene relative to genetic markers, including the ends of BAC and cosmid clones, are indicated. Numbers in parentheses show recombinant breakpoints observed between the indicated marker and LEC2. Positions of cosmid clones that suppress the lec2 mutation are shown. (B) Representation of the LEC2 gene. Hatched and shaded boxes indicate exons, and narrow boxes represent introns. Shaded boxes depict the gene region encoding the B3 domain. The position of mutations in specific mutant alleles are indicated. Rearrangement of lec2-3 was assessed with DNA gel blot hybridization studies.
Nucleotide sequences of restriction fragments from the complementing cosmid clones combined with the release of the partial sequence of BAC F3H9 identified a putative gene specifying a single polypeptide in this 11-kb region. Analysis of a near-full-length cDNA clone and amplified reverse transcription products showed that the putative LEC2 gene consists of six exons as shown diagrammatically in Fig. 2B.
In addition to the transgene complementation experiments, analyses of mutant alleles provided support for the identity of the LEC2 gene. As indicated in Fig. 2B, the lec2-4 mutation is a deletion of bases 573–583 (11 bp) relative to the translation start codon that creates a truncated protein. The lec2-5 allele lacks bases 723–755 (33 bp). Both deletions disrupt a part of the encoded protein conserved with other proteins as discussed below. lec2-1 possesses a deletion that spans 3,048 bp and 895 bp, respectively, upstream and downstream of the translation start codon. DNA gel blot experiments showed that lec2-3 has a rearrangement or deletion in the 5′ region of the gene (data not shown). Thus, all four lec2 mutant alleles have defects in the same gene. Together, the transgene complementation experiments and analyses of lec2 mutant alleles demonstrate that we have isolated the LEC2 gene.
Predicted LEC2 Protein Shares Sequence Similarity with Plant Transcription Factors.
The predicted LEC2 polypeptide consists of 363 amino acid residues with an estimated molecular mass of 41,708 Da. A central region of the protein, whose coding region is indicated diagrammatically in Fig. 2B, shares extensive sequence similarity with the B3 domain, a DNA-binding region found in several plant transcription factors (10, 19, 32–35). The amino- and carboxyl-terminal regions do not share significant similarity with other proteins. LEC2 B3 domain is most similar to the B3 domains of ABA INSENSITIVE3 (ABI3)/VIVIPAROUS1 (VP1) and the LEC protein, FUS3, sharing 50% and 43% amino acid sequence identity, respectively, with ABI3 and FUS3 (Fig. 3; refs. 10, 19, 33). Because all three proteins are transcription factors that function primarily in seeds, we hypothesize that LEC2 may also serve as a transcriptional regulator of seed development.
Figure 3.
LEC2 contains a B3 domain. Amino acid alignment of residues from the B3 domains of LEC2, FUS3, ABI3, and VP1. Residues in black boxes are identical in at least two of the four proteins, and those in shaded boxes share similarity with conserved residues. Numbers in the right column indicate residue numbers in the predicted polypeptides.
LEC2 Is Expressed Primarily During Embryo Development.
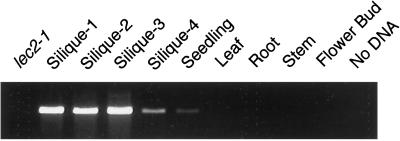
The effects of the lec2 mutation are limited to embryogenesis. Therefore, we asked whether LEC2 RNA accumulates specifically during seed development. Primers with sequences near the predicted LEC2 translation start and stop codons were used to amplify cDNA from wild-type and mutant siliques, seedlings, leaves, roots, stems, and flower buds. As shown in Fig. 4, amplification products were detected primarily in early- and middle-stage siliques and occasionally but not consistently in seedlings, leaves, roots, and stems, suggesting that the RNA may be present at very low levels in vegetative organs. This result suggests that LEC2 is expressed primarily during seed development.
Figure 4.
LEC2 RNA accumulates primarily during seed development. LEC2 RNA was detected at the indicated stages by amplifying reverse transcription products. Silique stages 1 through 4, respectively, are from siliques containing zygote to early globular-stage embryos, globular-stage to heart-stage embryos, torpedo-stage to curled cotyledon-stage embryos, and mature green embryos. Control experiments showed that a ribosomal protein RNA was amplified with similar efficiency from each reverse transcription reaction.
Ectopic LEC2 Expression Induces Somatic Embryo Formation.
Because LEC2 RNA accumulates primarily during seed development, we expressed the gene postembryonically to obtain additional clues about its role in embryo development. We transferred a LEC2 cDNA clone under the transcriptional control of the cauliflower mosaic virus 35S promoter into lec2 mutant plants. Unlike lec2 mutants with defective cotyledons, transgenic seedlings had complete cotyledon margins, indicating that the transgene had suppressed the mutant phenotypes. However, transgenic seedlings exhibited a range of morphological phenotypes.
At one extreme, represented in Fig. 5A, transgenic seedlings possessed embryonic characteristics as exemplified by their small and fleshy cotyledons, short hypocotyls, and unextended roots. Callus-like growth formed primarily on adaxial cotyledon surfaces of seedlings cultured on medium. Remarkably, somatic embryo-like structures, as shown in Figs. 5 A and B and 6A, developed from these regions, although cotyledon-like and leaf-like structures and shoots also were observed (data not shown). Occasionally, somatic embryo-like structures emerged over the entire surface of an embryo-like seedling (Fig. 5D). Similar results were obtained with wild-type seedlings containing the 35S∷LEC2 transgene.
Figure 5.
LEC2 induces somatic embryo development. Seeds from lec2-1 and lec2-5 plants transformed with the 35S∷LEC2 gene were germinated, and their subsequent development was monitored. (A) Formation of somatic embryo-like clusters (arrows) on the cotyledons of an embryo-like 35S∷LEC2 seedling. (B) Somatic embryo-like structures emerging from the cotyledon (arrowhead) of a 35S∷LEC2 seedling. (C) Scanning electron microscopy (SEM) photograph of a wild-type linear cotyledon-stage zygotic embryo. (D) A mass of somatic embryo-like structures covering a 35S∷LEC2 seedling as observed by SEM. (E) SEM photograph of somatic embryo-like structures on a 35S∷LEC2 seedling. (F) Seedling resulting from the germination of a 35S∷LEC2 somatic embryo formed on a wild-type transgenic seedling. (G) A 35S∷LEC2 seedling with a wild-type phenotype. Arrowhead shows that the distal region of the cotyledon is not defective. (H) A short, bushy plant grown on soil that developed from a 35S∷LEC2 seedling such as that shown in G. (I) Somatic embryos (arrows) emerging from leaf-like organ dissected from a plantlet mass. (J) A mass of plantlets formed from a single 35S∷LEC2 seedling such as that shown in G. a, Embryonic axis; al, embryonic axis-like; c, cotyledon; cl, cotyledon-like. [Bars = 1 mm (A, B, D, F, G), 100 μm (C, E), 500 μm (I), 1 cm (J), and 5 cm (H).]
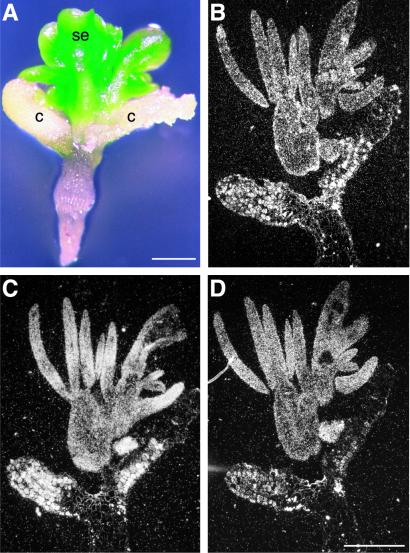
Figure 6.
LEC2-induced somatic embryos express embryo-specific genes. (A) Whole-mount photograph of a 35S∷LEC2 seedling with somatic embryo-like structures on its cotyledons. Sections of the same seedling were hybridized with antisense probes for LEC2 RNA (B), cruciferin A storage protein RNA (C), and oleosin lipid body protein RNA (D). After autoradiography, sections were photographed with use of dark-field optics. A sense-strand probe did not hybridize appreciably with the sections (data not shown). Regions of the seedlings in B–D that did not hybridize lacked cellular contents and were likely dead. c, Cotyledon; se, somatic embryo. (Bars = 0.5 mm.)
We believe that these structures are somatic embryos for several reasons. First, Fig. 5E shows that somatic embryo-like structures share morphological similarities with zygotic embryos, possessing cotyledon-like and embryonic axis-like organ systems typical of wild-type linear cotyledon-stage zygotic embryos (Fig. 5C). Structures similar in appearance to mature green-stage zygotic embryos were observed occasionally (Fig. 5B). Second, analyses of tissue sections suggest that somatic embryo-like structures possess axes in that both shoot and root apical meristems were formed (data not shown). Roots of somatic embryos “germinated” readily on medium or attached to plants, indicating that the root apical meristem was functional (Fig. 5F). Finally, somatic embryo-like structures expressed embryo-specific genes. Fig. 6 A, C, and D shows that RNAs encoding the storage protein cruciferin A and the lipid body protein oleosin accumulate in the somatic embryos and in regions of the underlying cotyledons of embryo-like seedlings. Significantly, LEC2 RNA was distributed similarly with the embryo-specific RNAs in seedlings giving rise to somatic embryos (Fig. 6B), indicating a correspondence between LEC2 gene expression and development of embryonic characteristics. Although LEC2-induced somatic embryos were larger than zygotic embryos, similar size differences are observed with Arabidopsis somatic embryos generated by other methods (36).
As shown in Fig. 5G, seedlings at the other morphological extreme had expanded cotyledons and resembled wild type (compare Fig. 1F). Seedlings of this type with extended roots typically developed into small, bushy plants when grown on soil, as shown in Fig. 5H. When 35S∷LEC2 seedlings were cultured on medium, we were initially surprised to find that these seedlings gave rise to masses of vegetatively growing plantlets (Fig. 5J), given the apparent role of LEC2 in promoting embryo development. However, further analysis showed that plantlets seem to have developed from somatic embryos, as shown in Fig. 5I. Taken together, these results suggest that LEC2 is sufficient to induce somatic embryo development.
Discussion
The pleiotropic effects of lec mutations, represented in Fig. 1, suggest that the LEC genes serve as central regulators of embryo and seed development. This prediction was confirmed by showing that LEC1 and FUS3 encode putative transcription factors that regulate critical embryonic processes (12, 19, 20). Here we present evidence suggesting that LEC2 likely encodes a transcriptional regulator with a significant role in embryogenesis.
LEC2 Possesses a B3 Domain Characteristic of Transcription Factors.
Our findings that the LEC2 gene encodes a B3 domain protein (Fig. 3) and is expressed primarily during embryogenesis (Fig. 4) suggest that it, like LEC1 and FUS3, is a transcriptional regulator of seed development. The B3 domain is an ≈120-amino acid residue region defined originally as the third basic region of maize VP1, which shares the largest contiguous block of sequence identity with its ortholog, Arabidopsis ABI3 (10). Subsequently, this domain was identified in other plant proteins, such as AUXIN RESPONSE FACTOR1 (34) and RELATED TO ABI3/VP1 (32). Many proteins containing B3 domains, including ABI3/VP1, FUS3, and AUXIN RESPONSE FACTOR1, function as transcription factors (20, 33, 34). The B3 domain is responsible, at least in part, for the DNA-binding activity of ABI3/VP1 and RELATED TO ABI3/VP1 (32, 37, 38). To our knowledge, the B3 DNA-binding domain is unique to plants.
Maximum parsimony analysis (39) of the B3 domains of 48 predicted Arabidopsis proteins suggests that LEC2, FUS3, and ABI3 constitute one class, perhaps indicating their common origin (S.L.S. and J.J.H., unpublished results). All three genes are expressed primarily during embryogenesis and are required genetically for the completion of normal seed development. Although they all play roles in seed development, their specific functions have diverged. ABI3 is a transcription factor that operates primarily during the maturation phase, whereas LEC2 and FUS3 are LEAFY COTYLEDON proteins required for processes during both the morphogenesis and maturation phases.
Role of LEC2 in Seed Development.
Two consequences of expressing the LEC2 gene postembryonically provide insight into its role in embryo development and strongly support the conclusion that LEC2 is a key regulator of embryogenesis. First, although seedlings expressing the 35S∷LEC2 gene exhibited a range of morphologies (Fig. 5), approximately one-third possessed embryonic rather than postgerminative characteristics. Cotyledons of these seedlings remained fleshy and did not expand, and their roots and hypocotyls did not extend, suggesting that ectopic LEC2 expression can extend embryonic development. 35S∷LEC2 seedlings at the other end of the phenotypic spectrum initially resembled wild type. The reason for this range of phenotypes is not clear, but one possibility is that it relates to differences in transgene expression levels. Second, regardless of their initial morphology, 35S∷LEC2 seedlings gave rise to somatic embryos as indicated by their morphological similarities with zygotic embryos, their expression of embryo-specific genes, and their establishment of functional apical meristems (Figs. 5 and 6). Together, these results suggest that ectopic LEC2 expression is sufficient to establish an embryonic environment that promotes somatic embryo formation. In this regard, it is possible that LEC2 RNA accumulation is regulated posttranscriptionally, thereby accounting for the ability of somatic embryos to “germinate” and, in some cases, give rise to masses of vegetatively growing plantlets.
Transgenic seedlings ectopically expressing a different LEC gene, LEC1, have embryonic characteristics that are substantially similar to those of 35S∷LEC2 embryo-like seedlings (12). Moreover, postembryonic expression of LEC1 is sufficient to induce somatic embryogenesis in transgenic plants as occurs with 35S∷LEC2 seedlings, although somatic embryo formation is more robust with LEC2. Recessive mutations in the Arabidopsis PICKLE (PKL) gene also induce somatic embryogenesis in postgerminative roots cultured on hormone-free medium (40). However, LEC1 is expressed in the roots of pkl mutant but not wild-type seedlings (41). This result suggests that the chromodomain protein PKL acts normally to repress LEC1 in postgerminative roots, and therefore that LEC1, at least in part, mediates somatic embryo formation in pkl mutants. Given our results, we hypothesize that LEC2 may also be derepressed in pkl mutants.
The ability of LEC2 and LEC1 to induce somatic embryogenesis suggests a role for the genes in the establishment of embryogenic competence. Somatic plant cells generally must be induced to become competent for somatic embryogenesis (42–44). Embryogenic competence is often induced by culturing cells with the hormone auxin and, sometimes cytokinin, although completion of somatic embryogenesis usually requires removal of the hormone(s). LEC2 and LEC1 obviate the need for hormone treatments in the acquisition of embryogenic competence, suggesting the two LEC transcription factors activate genes that play roles in the initiation of somatic embryogenesis. Our finding that both LEC2 and LEC1 RNAs are detected at the earliest embryonic stages tested (Fig. 4; ref. 12) opens the possibility that both genes are involved in establishing embryogenic competence during zygotic embryogenesis. Thus, the role of LEC2 and LEC1 in somatic embryogenesis may reflect their function in zygotic embryogenesis. Similarities in the expression patterns and overexpression phenotypes of LEC2 and LEC1 suggest that they may have partially overlapping roles early in zygotic embryogenesis to induce embryo formation. This interpretation is consistent with analyses showing that lec1 lec2 double mutants arrest at an earlier embryonic stage than either single mutant, which indicates partial genetic redundancy (12, 15).
Although LEC1 and LEC2 are each sufficient to induce embryogenic competence in somatic cells, they have similar but not identical functions. Mutations in each gene result in distinct phenotypes, and the double mutant displays a synergistic phenotype (12, 15, 16). Furthermore, the vast majority of 35S∷LEC1 seedlings arrest as embryo-like seedlings and fail to develop further, although cotyledon-like organs sometimes form in place of the first true leaves (12). By contrast, 35S∷LEC2 embryo-like seedlings continued to proliferate, producing callus, cotyledon-like and leaf-like organs in addition to somatic embryos. Thus, LEC1 and LEC2 may have complementary but partially redundant functions in embryo formation. The precise roles of LEC1 and LEC2 in embryo development remain to be determined.
Acknowledgments
We thank David Meinke for providing the lec2-1 mutant, the Arabidopsis Biological Resource Center for BAC clones and mutant lines, Sara Wortley and Julia Little for technical assistance, and Chuck Gasser and John Bowman and members of their labs for plasmids and advice. This work was supported by grants from Ceres, Inc., and the Department of Energy (to J.J.H.).
Abbreviations
- ABI3
ABA INSENSITIVE3
- BAC
bacterial artificial chromosome
- DIS
distorted trichomes
- FUS3
FUSCA3
- LEC
LEAFY COTYLEDON
- PKL
PICKLE
- VP1
VIVIPAROUS1
Footnotes
References
- 1.Olsen O A. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:241–267. doi: 10.1146/annurev.arplant.52.1.233. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg R B, de Paiva G, Yadegari R. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- 3.West M A, Harada J J. Plant Cell. 1993;5:1361–1369. doi: 10.1105/tpc.5.10.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurgens G. EMBO J. 2001;20:3609–3616. doi: 10.1093/emboj/20.14.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bewley J D. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harada J J. In: Advances in Cellular and Molecular Biology of Plants. Larkins B A, Vasil I K, editors. Vol. 4. Dordrecht, The Netherlands: Kluwer; 1997. pp. 545–592. [Google Scholar]
- 7.Harada J J. J Plant Physiol. 2001;158:405–409. [Google Scholar]
- 8.Di Laurenzio L, Wysocka-Diller J, Malamy J E, Pysh L, Helariutta Y, Freshour G, Hahn M G, Feldmann K A, Benfey P N. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 9.Long J A, Moan E I, Medford J I, Barton M K. Nature (London) 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 10.Giraudat J, Hauge B M, Valon C, Smalle J, Parcy F, Goodman H M. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelstein R R, Lynch T J. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotan T, Ohto M, Matsudaira Yee K, West M A L, Lo R, Kwong R W, Yamagishi K, Fischer R L, Goldberg R B, Harada J J. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 13.Keith K, Kraml M, Dengler N G, McCourt P. Plant Cell. 1994;6:589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meinke D W. Science. 1992;258:1647–1650. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- 15.Meinke D W, Franzmann L H, Nickle T C, Yeung E C. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West M A L, Matsudaira Yee K, Danao J, Zimmerman J L, Fischer R L, Goldberg R B, Harada J J. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumlein H, Misera S, Luerben H, Kolle K, Horstmann C, Wobus U, Muller A J. Plant J. 1994;6:379–387. [Google Scholar]
- 18.Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S. Dev Biol. 2000;220:412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- 19.Luerben H, Kirik V, Herrmann P, Misera S. Plant J. 1998;15:755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- 20.Reidt W, Wohlfarth T, Ellerstroem M, Czihal A, Tewes A, Ezcurra I, Rask L, Baumlein H. Plant J. 2000;21:401–408. doi: 10.1046/j.1365-313x.2000.00686.x. [DOI] [PubMed] [Google Scholar]
- 21.Bechtold N, Ellis J, Pelletier G. C R Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- 22.Neff M M, Neff J D, Chory J, Pepper A E. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 23.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 24.Bell C, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 25.Olszewski N E, Martin F B, Ausubel F M. Nucleic Acids Res. 1988;16:10765–10782. doi: 10.1093/nar/16.22.10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkins T A, Smart L B. In: A Laboratory Guide to RNA: Isolation, Analysis and Synthesis. Krieg P, editor. New York: Wiley–Liss; 1996. pp. 21–41. [Google Scholar]
- 27.Kawasaki E S. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, editor. San Diego: Academic; 1990. pp. 21–27. [Google Scholar]
- 28.Dietrich R A, Maslyar D J, Heupel R C, Harada J J. Plant Cell. 1989;1:73–80. doi: 10.1105/tpc.1.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gleave A P. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 30.Feenstra W J. Arabidopsis Inf Serv. 1978;15:35–38. [Google Scholar]
- 31.Bender J, Fink G R. Cell. 1995;83:725–734. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 32.Kagaya Y, Ohmiya K, Hattori T. Nucleic Acids Res. 1999;27:470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarty D R, Hattori T, Carson C B, Vasil V, Lazar M, Vasil I K. Cell. 1991;66:895–906. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 34.Ulmasov T, Hagen G, Guilfoyle T J. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 35.Hardtke C S, Berleth T. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mordhorst A P, Voerman K J, Hartog M V, Meijer E A, van Went J, Koornneef M, de Vries S C. Genetics. 1998;149:549–563. doi: 10.1093/genetics/149.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ezcurra I, Wycliffe P, Nehlin L, Ellerstrom M, Rask L. Plant J. 2000;24:57–66. doi: 10.1046/j.1365-313x.2000.00857.x. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki M, Kao C Y, McCarty D R. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hershkovitz M A, Leipe D D. In: Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins. Baxevanis A D, editor. New York: Wiley–Liss; 1998. pp. 189–230. [Google Scholar]
- 40.Ogas J, Cheng J-C, Sung Z R, Somerville C. Science. 1997;277:91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- 41.Ogas J, Kaufmann S, Henderson J, Somerville C. Proc Natl Acad Sci USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mordhorst A P, Toonen M A J, de Vries S C. Crit Rev Plant Sci. 1997;16:535–576. [Google Scholar]
- 43.Zimmerman J L. Plant Cell. 1993;5:1411–1423. doi: 10.1105/tpc.5.10.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dudits D, Bogre L, Gyorgyey J. J Cell Sci. 1991;99:475–484. [Google Scholar]