Abstract
Esophageal cancer ranks among the most aggressive malignant diseases. The limited improvements in treatment outcomes provided by conventional therapies have prompted us to seek innovative strategies for treating this cancer. More than 100 trillion microorganisms inhabit the human intestinal tract and play a crucial role in health and disease conditions, including cancer. The human intestinal microbiome is thought to influence tumor development and progression in the gastrointestinal tract by various mechanisms. For example, Fusobacterium nucleatum, which primarily inhabits the oral cavity and causes periodontal disease, might contribute to aggressive tumor behavior through activation of chemokines such as CCL20 in esophageal cancer tissue. Composition of the intestinal microbiota is influenced by diet, lifestyle, antibiotics, and pro‐ and prebiotics. Therefore, by better understanding how the bacterial microbiota contributes to esophageal carcinogenesis, we might develop novel cancer prevention and treatment strategies through targeting the gastrointestinal microflora. This review discusses the current knowledge, available data and information on the relationship of microbiota with esophagitis, Barrett's esophagus, esophageal adenocarcinoma and squamous cell carcinoma.
Keywords: esophagus, microbiome, microbiota
1. INTRODUCTION
Esophageal cancer is the sixth most common cause of cancer‐related death and the eighth most commonly diagnosed cancer worldwide.1 The predominant histological types of esophageal cancer are adenocarcinoma and squamous cell carcinoma.2 Adenocarcinoma of the distal esophagus predominates in the West, whereas squamous cell carcinoma, which tends to localize in the middle thoracic esophagus, predominates in the East. Molecular features also differ between adenocarcinoma and squamous cell carcinoma; for example, squamous cell carcinomas showed frequent genomic amplifications of CCND1 and SOX2 and/or TP63, whereas ERBB2, VEGFA and GATA4 and GATA6 were more commonly amplified in adenocarcinomas.3 Traditionally, both adenocarcinomas and squamous cell tumors have been treated by surgical resection.4 However, despite the development of multimodal therapies including surgery, chemotherapy, radiotherapy, and chemoradiotherapy, the prognosis remains poor even in patients who have undergone complete resection.5 Therefore, further studies are needed to clarify the pathogenesis of esophageal cancer and to explore new diagnostic and therapeutic possibilities.
Microbiome research is a rapidly advancing field in human cancers.6, 7, 8, 9, 10 More than 100 trillion bacteria inhabit the human body and form their own flora (ie microbiomes) in individual organs. The gut microbiota appears haphazard in infants, but begins resembling the adult microbiome by age 3 years. Nevertheless, the microbial distribution from the esophagus to the rectum varies spatially and temporally throughout the individual's lifespan. The normal gut microbiota carries out specific functions in host nutrient metabolism, xenobiotic and drug metabolism, structural integrity maintenance of the gut mucosal barrier, immunomodulation, and protection against pathogens.11, 12, 13, 14, 15 Recently, the gut microbiome has been shown to play a crucial role in health, as well as in diseases such as obesity,16 inflammatory bowel disease,17, 18 diabetes,19, 20 non‐alcoholic fatty liver disease,21, 22, 23 and several types of cancers.24, 25 Experimental evidence indicates that the human intestinal microbiome can influence tumor development and progression in the gastrointestinal tract by damaging DNA, activating oncogenic signaling pathways, producing tumor‐promoting metabolites, and suppressing the antitumor immune response.7, 25, 26, 27, 28, 29 As the gastrointestinal microbiota can be modified through the rational deployment of antibiotics, probiotics, and prebiotics,30, 31, 32 a better understanding of the relationship between human cancer and the microbiome may have clinical implications.
The present review discusses current knowledge on the relationship between the microbiome and esophageal cancer. Importantly, because two histological types (adenocarcinoma and squamous cell carcinoma) present as different diseases in terms of their epidemiology, pathogenesis, and tumor biology, the role of the microbiome is discussed separately for each histological type.
2. MICROBIOME OF THE NORMAL ESOPHAGUS
Distribution of the gut microbiota varies temporally and spatially at the genus level and higher. From the oral cavity, through the esophagus and distally to the rectum, the diversity and number of bacteria changes markedly, ranging from 101 per gram of contents in the esophagus and stomach to 1012 per gram of contents in the colon and distal gut.33 Importantly, the esophagus, unlike other luminal organs of the digestive system, does not retain food contents.
In the 1990s, microbiological studies depended mainly on conventional bacterial culture‐based methods. These studies demonstrated that the esophagus is either sterile or contains only a few transient microbes swallowed from the oropharynx or ejected from the stomach by gastroesophageal reflux.34 Gagliardi et al.34 revealed that Streptococcus viridans is the most frequent microorganism in both the normal esophagus and the oropharynx. These findings were consolidated by Norder Grusell et al.,35 who collected brush samples and biopsy samples from the esophagus, and reported the occurrence rate of Streptococcus viridans as 95‐98%. These studies support a possible correlation between the flora in the oropharynx and the esophagus. However, as most of the autochthonic esophageal microbiome is viable but non‐culturable, it will likely be missed by standard culturing methods. More recently, the diversity of the non‐culturable microbiota has been characterized by advanced approaches such as polymerase chain reaction (PCR) of 16S ribosomal RNA.36 Pei et al.37 examined the normal esophagus by broad‐range 16S rDNA PCR and identified 95 species in six phyla: Firmicutes (eg Streptococcus), Bacteroides (eg Prevotella), Actinobacteria (eg Rothia), Proteobacteria (eg Haemophilus), Fusobacteria (eg Fusobacterium), and TM7. Remarkably, the findings were similar across specimens, suggesting a stable esophageal biota that is distinct from the flora of the oropharynx, stomach, and food bolus in transit. Microscopic examination of the tissue confirmed a close association between the bacteria and the cell surfaces of the mucosal epithelium in situ, suggesting a residential, rather than a transient, biota.
Collectively, the normal esophagus has a distinct microbiome of predominantly oral flora. Members of the phylum Firmicutes as represented by Streptococcus viridans appear to be major components of the microbiota of the normal esophagus, although the presence of several other phyla (eg Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria, TM7) has also been reported.
3. MICROBIOME IN ESOPHAGITIS AND BARRETT'S ESOPHAGUS
Gastroesophageal reflux disease (GERD) is an important risk factor for esophageal adenocarcinoma. GERD can lead to erosive esophagitis and (after an aberrant healing process) to a metaplastic, specialized intestinal epithelium (ie Barrett's esophagus).38 Among the 6‐14% of GERD patients who develop Barrett's esophagus, 0.5‐1% will progress to adenocarcinoma.39 In a meta‐analysis of population‐based studies, weekly symptoms of GERD were estimated to increase the risk of esophageal adenocarcinoma approximately fivefold.40
Several studies have documented microbiome status in esophagitis and Barrett's esophagus. In a microscopic study of Barret's esophageal biopsy specimens, Osias et al.41 found that cultivable bacteria were closely associated with the mucosa of the specimens. They concluded that Barrett's mucosa are colonized rather than transiently visited by resident bacteria. Macfarlane et al.42 isolated a broader range of bacteria from patients with Barrett's esophagus than from individuals without Barrett's esophagus, suggesting a higher microbiological diversity in patients with Barrett's esophagus. Using 16S rDNA sequencing technology, Yang et al.43 characterized the diversity of the distal esophagus microbiota in individuals with normal esophagus, reflux esophagitis and Barrett's esophagus. They classified the esophageal microbiota into two types (Figure 1). Type I microbiome, mainly associated with normal esophagus, was predominated by Gram‐positive bacteria, primarily phylum Firmicutes. Type II microbiome contained a greater proportion of Gram‐negative anaerobes/microaerophiles (phyla Bacteroidetes, Proteobacteria, Fusobacteria, and Spirochaetes), and was primarily correlated with reflux esophagitis and Barrett's esophagus. As the microbiomes did not differ between GERD and Barrett's esophagus patients, the authors concluded that inflammation and intestinal metaplasia are associated with global alteration of the microbiome in the distal esophagus. Using culture‐independent techniques, Liu et al.44 examined the bacterial composition at the 16S rDNA gene site in subjects with a normal esophagus, reflux esophagitis, or Barrett's esophagus. Veillonella, Prevotella, Neisseria, and Fusobacterium prevailed in patients with reflux esophagitis and Barrett's esophagus, but were not detected in normal esophagus. More recently, Gall et al.45 showed that Streptococcus and Prevotella dominate the esophageal microbiota of Barrett's esophagus patients, with no substantial intraindividual differences between normal and metaplastic esophageal mucosa. They also found a significant association between the Streptococcus/Prevotella ratio and some important risk factors for Barrett's esophagus and esophageal adenocarcinoma (eg waist‐to‐hip ratio, hiatal hernia length). Overall, the esophageal bacteria differ among normal esophagus, reflux esophagitis and Barrett's esophagus, supporting that esophageal disease is related to the bacterial community profile, possibly through the innate immune system. Gram‐negative organisms, which predominate in reflux esophagitis and Barrett's esophagus, produce specific constituents such as lipopolysaccharide (LPS) that activate the innate immune responses.46 Either directly or indirectly, LPS may stimulate the innate immune system's Toll‐like receptor (TLR) 4 in the epithelial or inflammatory cells, leading to nuclear factor kappa B (NF‐κB) activation. Increased NF‐κB activation is associated with elevated levels of inflammatory cytokines.47 Activation of the NF‐κB pathway increases stepwise along the spectrum of reflux esophagitis, Barrett epithelium, and adenocarcinoma, paralleling the increases in interleukin (IL)‐1β, IL‐6, IL‐8, and tumor necrosis factor (TNF)‐α.48, 49, 50 Taken together, in reflux esophagitis and Barrett's esophagus, the change of microbiomes (ie increased Gram‐negative organisms) may contribute to esophageal adenocarcinoma by inducing chronic inflammation, triggering a cascade that leads to adenocarcinoma.46
Figure 1.
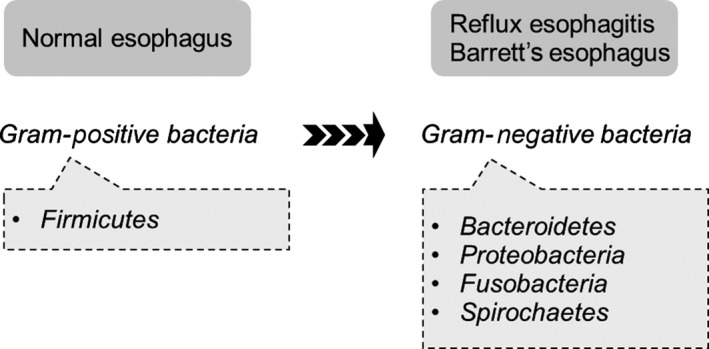
Microbiome status of normal esophagus, reflux esophagitis, and Barrett's esophagus. In the human distal esophagus, inflammation and intestinal metaplasia are associated with global alteration of the microbiome43
4. MICROBIOME AND ESOPHAGEAL ADENOCARCINOMA
Given the important role of the gut microbiome in human malignancies, a better understanding of the microbiome in esophageal cancer is increasingly important. In the 1980s, culture‐based methods and surgically resected specimens of esophageal adenocarcinoma and squamous cell carcinoma revealed the same microbiota in normal and cancerous tissues. However, these studies were focused on identifying pathogens related to postoperative infections, rather than comparing the non‐pathogenic bacteria in esophageal cancer cases and control cases.51, 52, 53 More recently, Narikiyo et al.54 characterized the microbiota of normal and cancerous esophageal tissue by 16S sequencing technology. Cancerous tissues were obtained from 20 patients undergoing surgical resection for esophageal cancer. Both microbiota were consistently dominated by the oral periodontopathic spirochete Treponema denticola, Streptococcus mitis, and Streptococcus anginosus, but the pathological subtypes of the tumors were not specified.54 Using a mixed culture‐dependent and culture‐independent approach, Blackett et al.55 compared the microbiota in reflux‐asymptomatic controls and in patients with GERD, Barrett's esophagus, and esophageal adenocarcinoma. Campylobacter were significantly more enriched in GERD and Barrett's esophagus than in the controls and esophageal adenocarcinoma. In addition, cytokines associated with carcinogenesis (eg IL‐18) were more highly expressed in the tissues colonized by Campylobacter.55 Given the potential human pathogenicity of Campylobacter species (which has been recently recognized),56 the role of Campylobacter in esophageal adenocarcinoma progression might mimic that of Helicobacter pylori in gastric cancer.
The relationship between the microbiome and esophageal adenocarcinoma development has been experimentally investigated. Sawada et al.57 investigated whether altering the microbiome with antibiotics affected the development of esophageal adenocarcinoma in a rat model with esophagojejunostomy. Terminal restriction fragment length polymorphism analysis showed that the esophageal microbiomes differed between the two groups; for instance, the proportions of Lactobacillales and Clostridium were reduced and elevated in the antibiotics group, respectively. However, the altered microbiome did not affect the incidence of esophageal adenocarcinoma. In a rat model with esophagojejunal anastomosis, Zaidi et al.58 revealed a prevalence of Escherichia coli in Barrett's esophagus and esophageal adenocarcinoma; moreover, TLR 1‐3, 6, 7 and 9 were significantly upregulated in esophageal adenocarcinoma compared with normal epithelium. This suggests an association between the TLR signaling pathway and E. coli, hinting that early molecular changes are mediated by microbes in the rat model of esophageal adenocarcinoma carcinogenesis.58 At this time, less conclusive information is available about the effects of the microbiome on esophageal adenocarcinoma. Nonetheless, alteration of microbiome status is potentially involved in the progression of GERD and Barrett's esophagus toward adenocarcinoma.
5. MICROBIOME AND ESOPHAGEAL SQUAMOUS CELL CARCINOMA
The microbiome is less well characterized in esophageal squamous cell carcinoma than in esophageal adenocarcinoma.59 Yu et al.60 observed a negative correlation between esophageal microbial richness and esophageal squamous dysplasia (the precursor lesion of esophageal squamous cell carcinoma) in a human oral microbe identification microarray. They suggested that individuals with lower esophageal microbial complexity are more prone to developing esophageal squamous dysplasia.60 Another study using 16S rDNA sequencing technology demonstrated that, relative to controls, the gastric corpus microbiota of patients affected by esophageal squamous dysplasia and esophageal squamous cell carcinoma are enriched in Clostridiales and Erysipelotrichales, suggesting that gastric dysbiosis is involved in the progression from esophageal squamous dysplasia to squamous cell carcinoma.61 Gao et al.62 revealed that a specific microbiome Porphyromonas gingivalis infects the cancerous and adjacent esophageal mucosa of esophageal squamous cell carcinoma patients but not the healthy mucosa of controls, supporting a pathogenesis role of this organism in esophageal squamous cell carcinoma. The presence of Porphyromonas gingivalis was also positively correlated with the severity (ie cancer cell differentiation and metastasis) of esophageal squamous cell carcinoma and with poor clinical outcome. Therefore, Porphyromonas gingivalis may serve as a biomarker of esophageal squamous cell carcinoma. According to Chen et al.,63 altered bacterial microbiota in the saliva is related to a higher risk of esophageal squamous cell carcinoma. The carriage of genera Lautropia, Bulleidia, Catonella, Corynebacterium, Moryella, Peptococcus and Cardiobacterium is lower in esophageal squamous cell carcinoma patients than in individuals without this cancer.
Given that poor oral health increases the risk of esophageal squamous cell carcinoma,64 these findings should be verified in prospective and long‐term cohort studies, along with functional studies. By establishing the association between the oral microbiome and risk of esophageal squamous cell carcinoma, we can better understand cancer etiology, and possibly develop a novel research paradigm for cancer chemoprevention.
Recently, we revealed that the prognosis of esophageal squamous cell carcinoma relates to the presence of Fusobacterium nucleatum, which primarily inhabits the oral cavity and causes periodontal disease.65 Fusobacterium nucleatum is frequently detected in colon cancer tissue, and may influence the development of colorectal cancer. Given the close proximity of the esophagus to the oral cavity, we suspect that Fusobacterium nucleatum also plays an important role in esophageal cancer. Using real‐time PCR analysis, we assessed DNA in the cancer tissues of 325 patients who underwent surgical removal of esophageal cancer. Seventy‐four out of 325 patients (nearly 23%) contained Fusobacterium nucleatum in their cancer tissues. Importantly, the presence of Fusobacterium nucleatum in cancer tissue was associated with significantly shorter survival time. Using microarray data, we also identified significant pathways in Fusobacterium nucleatum‐positive esophageal cancer tissues. The top‐ranked KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway in Fusobacterium nucleatum‐positive tissues was “Cytokine‐cytokine receptor interaction”. Detailed analysis of these data revealed that the genes of specific chemokines (ie CCL20) had increased in number, suggesting that Fusobacterium nucleatum contributes to the acquisition of aggressive tumor behavior by activating chemokines such as CCL20. Further analysis by more institutions, preferably worldwide, is desired because intestinal flora differ among individuals. Accumulating evidence suggests the crucial role of gut microbiota in the development and progression of esophageal squamous cell carcinoma. Further studies are needed to validate the previous findings and to elucidate the mechanism(s) whereby the gut microbiome affects tumor behavior.
6. FUTURE DIRECTIONS
By elucidating the mechanisms and microbiome contributions to the development and progression of esophageal cancer, we hope to develop novel therapeutics and strategies that treat or prevent esophageal cancer by modulating the microbiota (Figure 2). First, importantly, the composition of intestinal microbiota can be modified by antibiotics, probiotics, prebiotics or microbiota transplants. Limited‐spectrum and non‐absorbable antibiotics can remove or suppress unwanted components of the human microbiome. Probiotics can introduce missing microbial components with known beneficial functions for the human host. Prebiotics can maximize sustainable changes in the human microbiome by enhancing the proliferation of beneficial microbes or probiotics. Prebiotics or probiotics might target the microbiome for cancer prevention, especially in high‐risk populations. Second, the microbiota's potential ability to modulate the toxicity and efficacy of chemotherapy has also attracted interest.66 For example, the microbiota and immune system have reportedly enhanced the efficacy of oxaliplatin, a platinum‐based anticancer drug that treats esophageal cancer.67 Gut microbiota stimulate the production of reactive oxygen species (ROS) by immune cells. ROS enhance the DNA damage caused by oxaliplatin, blocking DNA replication and transcription and resulting in cell death.67 Third, given the intertwined nature of the microbiota and the immune system, microbiota likely influence their host's responsiveness to immunotherapy. Immunotherapy (eg antibodies to PD‐L1) ranks among the most exciting and successful developments in cancer care over the past decade. Antibiotic‐mediated disruption of the microbiota impaired the effectiveness of CpG oligonucleotide immunotherapy in mice with subcutaneous tumors.25, 67 Fourth, the microbiota is a potential biomarker of diagnosis or clinical outcome. If correct, the relationship between Fusobacterium nucleatum and poor clinical outcome identified in our previous work will have clinical implications.65
Figure 2.
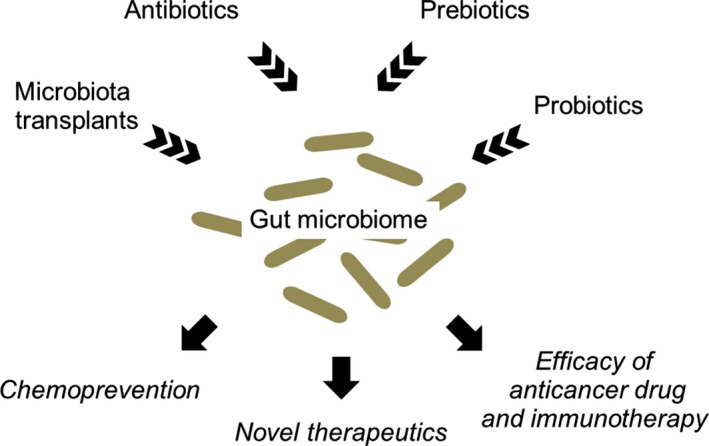
Clinical implication of the gut microbiome in human cancers
7. CONCLUSIONS
Accumulating evidence suggests that imbalanced gut microbiota induces changes in the enteric environment that lead to esophageal mucosal inflammation or tumorigenesis. Understanding the diverse ways that the bacterial microbiota contributes to esophageal carcinogenesis will open new possibilities for the diagnosis, prevention and treatment of esophageal cancer.
DISCLOSURE
Conflict of Interest: Authors declare no conflicts of interest for this article.
Baba Y, Iwatsuki M, Yoshida N, Watanabe M, Baba H. Review of the gut microbiome and esophageal cancer: Pathogenesis and potential clinical implications. Ann Gastroenterol Surg. 2017;1:99–104. https://doi.org/10.1002/ags3.12014
REFERENCES
- 1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 2. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–52. [DOI] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer Agency , et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba Y, Yoshida N, Shigaki H, et al. Prognostic impact of postoperative complications in 502 patients with surgically resected esophageal squamous cell carcinoma: a retrospective single‐institution study. Ann Surg. 2016;264:305–11. [DOI] [PubMed] [Google Scholar]
- 5. Baba Y, Watanabe M, Yoshida N, Kawanaka K, Yamashita Y, Baba H. Radiofrequency ablation for pulmonary metastases from gastrointestinal cancers. Ann Thorac Cardiovasc Surg. 2014;20:99–105. [DOI] [PubMed] [Google Scholar]
- 6. Arthur JC, Gharaibeh RZ, Muhlbauer M, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria‐induced colorectal cancer. Nat Commun. 2014;5:4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abreu MT, Peek RM Jr. Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146:1534–46. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Al‐Haddad S, El‐Zimaity H, Hafezi‐Bakhtiari S, et al. Infection and esophageal cancer. Ann NY Acad Sci. 2014;1325:187–96. [DOI] [PubMed] [Google Scholar]
- 10. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–72. [DOI] [PubMed] [Google Scholar]
- 11. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. [DOI] [PubMed] [Google Scholar]
- 14. Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9. [DOI] [PubMed] [Google Scholar]
- 15. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. [DOI] [PubMed] [Google Scholar]
- 17. Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–7. [DOI] [PubMed] [Google Scholar]
- 20. Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low‐grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3:279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoneda M, Naka S, Nakano K, et al. Involvement of a periodontal pathogen, Porphyromonas gingivalis on the pathogenesis of non‐alcoholic fatty liver disease. BMC Gastroenterol. 2012;12:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Furusho H, Miyauchi M, Hyogo H, et al. Dental infection of Porphyromonas gingivalis exacerbates high fat diet‐induced steatohepatitis in mice. J Gastroenterol. 2013;48:1259–70. [DOI] [PubMed] [Google Scholar]
- 23. Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut. 2016;65:2035–44. [DOI] [PubMed] [Google Scholar]
- 24. Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br J Cancer. 2016;114:237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garrett WS. Cancer and the microbiota. Science. 2015;348:80–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and anticancer immunosurveillance. Cell. 2016;165:276–87. [DOI] [PubMed] [Google Scholar]
- 27. Johnson CH, Spilker ME, Goetz L, Peterson SN, Siuzdak G. Metabolite and microbiome interplay in cancer immunotherapy. Can Res. 2016;76:6146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation‐induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–71. [DOI] [PubMed] [Google Scholar]
- 29. Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur J Immunol. 2015;45:17–31. [DOI] [PubMed] [Google Scholar]
- 30. Becattini S, Taur Y, Pamer EG. Antibiotic‐induced changes in the intestinal microbiota and disease. Trends Mol Med. 2016;22:458–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic‐resistant pathogens. Science. 2016;352:535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65:1906–15. [DOI] [PubMed] [Google Scholar]
- 33. O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gagliardi D, Makihara S, Corsi PR, et al. Microbial flora of the normal esophagus. Dis Esophagus. 1998;11:248–50. [DOI] [PubMed] [Google Scholar]
- 35. Norder Grusell E, Dahlen G, Ruth M, et al. Bacterial flora of the human oral cavity, and the upper and lower esophagus. Dis Esophagus. 2013;26:84–90. [DOI] [PubMed] [Google Scholar]
- 36. Morgan XC, Huttenhower C. Meta'omic analytic techniques for studying the intestinal microbiome. Gastroenterology. 2014;146:1437–48. e1. [DOI] [PubMed] [Google Scholar]
- 37. Pei Z, Yang L, Peek RM Jr, Levine SM, Pride DT, Blaser MJ. Bacterial biota in reflux esophagitis and Barrett's esophagus. World J Gastroenterol. 2005;11:7277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rubenstein JH, Shaheen NJ. Epidemiology, diagnosis, and management of esophageal adenocarcinoma. Gastroenterology. 2015;149:302–17. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wheeler JB, Reed CE. Epidemiology of esophageal cancer. Surg Clin N Am. 2012;92:1077–87. [DOI] [PubMed] [Google Scholar]
- 40. Rubenstein JH, Taylor JB. Meta‐analysis: the association of oesophageal adenocarcinoma with symptoms of gastro‐oesophageal reflux. Aliment Pharmacol Ther. 2010;32:1222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Osias GL, Bromer MQ, Thomas RM, et al. Esophageal bacteria and Barrett's esophagus: a preliminary report. Dig Dis Sci. 2004;49:228–36. [DOI] [PubMed] [Google Scholar]
- 42. Macfarlane S, Furrie E, Macfarlane GT, Dillon JF. Microbial colonization of the upper gastrointestinal tract in patients with Barrett's esophagus. Clin Infect Dis. 2007;45:29–38. [DOI] [PubMed] [Google Scholar]
- 43. Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137:588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu N, Ando T, Ishiguro K, et al. Characterization of bacterial biota in the distal esophagus of Japanese patients with reflux esophagitis and Barrett's esophagus. BMC Infect Dis. 2013;13:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gall A, Fero J, McCoy C, et al. Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett's esophagus cohort. PLoS One. 2015;10:e0129055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang L, Francois F, Pei Z. Molecular pathways: pathogenesis and clinical implications of microbiome alteration in esophagitis and Barrett esophagus. Clin Cancer Res. 2012;18:2138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pikarsky E, Porat RM, Stein I, et al. NF‐kappaB functions as a tumour promoter in inflammation‐associated cancer. Nature. 2004;431:461–6. [DOI] [PubMed] [Google Scholar]
- 48. Konturek PC, Nikiforuk A, Kania J, Raithel M, Hahn EG, Muhldorfer S. Activation of NFkappaB represents the central event in the neoplastic progression associated with Barrett's esophagus: a possible link to the inflammation and overexpression of COX‐2, PPARgamma and growth factors. Dig Dis Sci. 2004;49:1075–83. [DOI] [PubMed] [Google Scholar]
- 49. O'Riordan JM, Abdel‐latif MM, Ravi N, et al. Proinflammatory cytokine and nuclear factor kappa‐B expression along the inflammation‐metaplasia‐dysplasia‐adenocarcinoma sequence in the esophagus. Am J Gastroenterol. 2005;100:1257–64. [DOI] [PubMed] [Google Scholar]
- 50. Abdel‐Latif MM, Kelleher D, Reynolds JV. Potential role of NF‐kappaB in esophageal adenocarcinoma: as an emerging molecular target. J Surg Res. 2009;153:172–80. [DOI] [PubMed] [Google Scholar]
- 51. Lau WF, Wong J, Lam KH, Ong GB. Oesophageal microbial flora in carcinoma of the oesophagus. Aust NZ J Surg. 1981;51:52–5. [DOI] [PubMed] [Google Scholar]
- 52. Finlay IG, Wright PA, Menzies T, McArdle CS. Microbial flora in carcinoma of oesophagus. Thorax. 1982;37:181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mannell A, Plant M, Frolich J. The microflora of the oesophagus. Ann R Coll Surg Engl. 1983;65:152–4. [PMC free article] [PubMed] [Google Scholar]
- 54. Narikiyo M, Tanabe C, Yamada Y, et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 2004;95:569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blackett KL, Siddhi SS, Cleary S, et al. Oesophageal bacterial biofilm changes in gastro‐oesophageal reflux disease, Barrett's and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther. 2013;37:1084–92. [DOI] [PubMed] [Google Scholar]
- 56. Man SM. The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol. 2011;8:669–85. [DOI] [PubMed] [Google Scholar]
- 57. Sawada A, Fujiwara Y, Nagami Y, et al. Alteration of esophageal microbiome by antibiotic treatment does not affect incidence of rat esophageal adenocarcinoma. Dig Dis Sci. 2016;61:3161–8. [DOI] [PubMed] [Google Scholar]
- 58. Zaidi AH, Kelly LA, Kreft RE, et al. Associations of microbiota and toll‐like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer. 2016;16:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Di Pilato V, Freschi G, Ringressi MN, Pallecchi L, Rossolini GM, Bechi P. The esophageal microbiota in health and disease. Ann NY Acad Sci. 2016;1381:21–33. [DOI] [PubMed] [Google Scholar]
- 60. Yu G, Gail MH, Shi J, et al. Association between upper digestive tract microbiota and cancer‐predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev. 2014;23:735–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nasrollahzadeh D, Malekzadeh R, Ploner A, et al. Variations of gastric corpus microbiota are associated with early esophageal squamous cell carcinoma and squamous dysplasia. Sci Rep. 2015;5:8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gao S, Li S, Ma Z, et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer. 2016;11:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen X, Winckler B, Lu M, et al. Oral microbiota and risk for esophageal squamous cell carcinoma in a high‐risk area of China. PLoS One. 2015;10:e0143603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen X, Yuan Z, Lu M, Zhang Y, Jin L, Ye W. Poor oral health is associated with an increased risk of esophageal squamous cell carcinoma ‐ a population‐based case‐control study in China. Int J Cancer. 2017;140:626–35. [DOI] [PubMed] [Google Scholar]
- 65. Yamamura K, Baba Y, Nakagawa S, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22:5574–81. [DOI] [PubMed] [Google Scholar]
- 66. Carmody RN, Turnbaugh PJ. Host‐microbial interactions in the metabolism of therapeutic and diet‐derived xenobiotics. J Clin Investig. 2014;124:4173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]


