Abstract
Objectives
To contrast the coverage of diseases between the Disease Ontology (DO) and SNOMED CT, and to compare the hierarchical structure of the two ontologies.
Methods
We establish a reference list of mappings. We characterize unmapped concepts in DO semantically and structurally. Finally, we compare the hierarchical structure between the two ontologies.
Results
Overall, 4478 (65%) the 6931 DO concepts are mapped to SNOMED CT. The cancer and neoplasm subtrees of DO account for many of the unmapped concepts. The most frequent differentiae in unmapped concepts include morphology (for cancers and neoplasms), specific subtypes (for rare genetic disorders), and anatomical subtypes. Unmapped concepts usually form subtrees, and less often correspond to isolated leaves or intermediary concepts.
Conclusion
This detailed analysis of the gaps in coverage and structural differences between DO and SNOMED CT contributes to the interoperability between these two ontologies and will guide further validation of the mapping.
Keywords: Biological Ontologies, Systematized Nomenclature of Medicine, Unified Medical Language System
Introduction
Different ontologies are used to represent disease concepts in biomedical research and in clinical settings. The Disease Ontology (DO) is widely used in the research community, especially in genomic and cancer research. SNOMED CT is primarily used in healthcare and clinical settings. Interoperability between these two important ontologies is critical for translational applications in biomedicine. For example, research findings about a disease (coded with DO) and clinical findings from EHR data about the same disease (coded with SNOMED CT) can be analyzed together only if the DO and SNOMED CT codes for the disease are mapped together.
In this paper, we investigate the coverage of disease concepts between DO and SNOMED CT. More specifically, we identify and characterize the concepts present in DO but not covered by SNOMED CT. We also analyze the differences in hierarchical structure between the two ontologies.
Background
Resources
Disease Ontology
The Disease Ontology (DO) [1] is part of the Open Biomedical Ontologies (OBO) [2] collection and is used in several research projects. The ontology is implemented using description logics (DL) and available in the Web Ontology Language (OWL) format. We worked with the August 2016 release of the Disease Ontology. This version has 6931 active disease concepts. Some concepts in DO have explicit cross-references (represented by “obo:hasDbXref” relations) to concepts from SNOMED CT and other bio-ontologies. In this paper, we refer to these cross-references as “mappings”.
SNOMED CT
The Systematized Nomenclature of Medicine - Clinical Terms (SNOMED CT) is the largest clinical terminology. We used the March 2016 release of SNOMED CT (US Edition), as this is the version cross-referenced by the August 2016 version of DO. SNOMED CT contains over 300,000 clinical concepts, with about 100,000 disease concepts. As for DO, SNOMED CT is developed using description logics. However, since SNOMED CT is distributed in a proprietary format, we converted it to OWL using the script provided as part of the release. We processed the OWL versions of DO and SNOMED CT using the Java OWL API.
UMLS
The Unified Medical Language System (UMLS) Metathesaurus [3], developed by the U.S. National Library of Medicine, provides mappings across concepts from various standard biomedical terminologies and ontologies, including SNOMED CT. However, the UMLS does not currently integrate the Disease Ontology. The UMLS provides a RESTful API to identify lexical matches among all the concepts from its sources. Each UMLS concept is linked to one of 15 Semantic Groups, including Disorders. Using UMLS allows us to leverage the rich synonymy across all its source vocabularies for mapping and the semantic characterization of its concepts for consistency checking. We used the 2016AA release of UMLS in this research as it contains the March 2016 release of SNOMED CT.
Related work
Kibbe et. al. [4] report on the overall coverage of the DO and its cross-references to other terminologies in their update on the Disease Ontology. In this study, we perform a deeper analysis of the cross-references to SNOMED CT specifically.
Previous work has investigated the coverage of concepts within specific subdomains of medicine [5]. It has been demonstrated that the UMLS semantics can be exploited successfully for mapping across vocabularies [6]. In previous work from our group, Dhombres et. al. [7] evaluated the coverage of phenotypes across the Human Phenotype Ontology (HPO) [8] and SNOMED CT. Fung et. al. [9] assessed coverage of rare diseases in ICD and SNOMED CT. In this paper, we use similar techniques for assessment of disease concepts between DO and SNOMED CT.
To the best of our knowledge, this is the first attempt to contrast the coverage and structure between DO and SNOMED CT.
Methods
Our approach to investigating the coverage and organization of disease concepts in DO and SNOMED CT can be summarized as follows. We first establish a reference list of mappings of DO concepts to SNOMED CT. We characterize the unmapped DO concepts semantically and structurally. Finally, we compare the hierarchical structure between the two ontologies.
Establishing a reference list of mappings
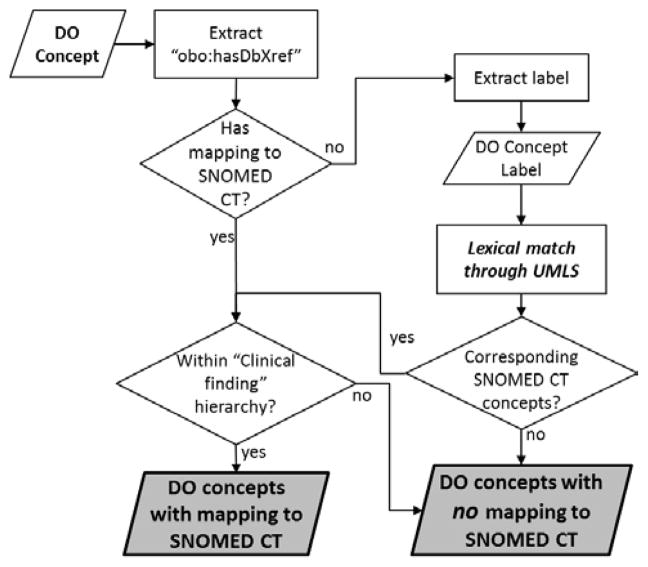
As shown in Figure 1, to establish a reference list of mappings, we start by updating the mappings provided by DO, from which we filter out semantically inconsistent mappings. We identify additional mappings lexically using the UMLS.
Figure 1.
Methodology for establishing a reference list of mappings
Updating and filtering mappings provided by DO
From the mappings (“cross-references”) to SNOMED CT provided by DO, we remove those mappings to retired SNOMED CT concepts and resolve the mappings to “moved” (remapped) concepts in SNOMED CT. Because DO concepts are expected to represent diseases, we consider semantically inconsistent and filter out those mappings to concepts outside the “Clinical finding” hierarchy of SNOMED CT, which contains all diseases, disorders and findings.
Finding additional lexical mappings
We leverage the UMLS in an attempt to identify lexical mappings for those DO concepts without any mappings to the “Clinical finding” hierarchy of SNOMED CT. More specifically, we first extract the labels for each DO concept, including preferred terms and synonyms. We take advantage of the rich set of synonyms provided by the UMLS to map these terms to UMLS concepts, using exact or normalized string matches. Finally, as we did for the mappings provided by DO, we select semantically consistent lexical mappings by restricting the mappings to the “Clinical finding” hierarchy of SNOMED CT.
All semantically consistent mappings (from DO cross-references or obtained lexically) form the reference list of mappings used in the rest of this investigation. All other DO concepts are considered unmapped.
Characterizing unmapped DO concepts
We characterize the unmapped DO concepts semantically and structurally, and analyze the differentia(e) between unmapped concepts and their parent(s).
Semantically
To identify whether coverage is better for some types of diseases than others, we compute the distribution of mapped and unmapped DO concepts with respect to the top-level subtrees of DO.
Structurally
To investigate whether unmapped DO concepts are isolated unmapped leaf concepts, subtrees of unmapped concepts, or unmapped intermediary concepts, we cluster them into groups of hierarchically related concepts (“connected components” in graph theory parlance).
Differentiae
Moreover, for isolated unmapped leaf concepts and subtrees of unmapped concepts, one author (OB) performed a manual review to identify the differentia(e) between each unmapped DO concept and its immediate parent concept(s). For example, the unmapped DO concept “ovarian germ cell teratoma” differs from its parent concept “ovarian germ cell cancer” by the addition of a morphology distinction (teratoma is a morphologic type of cancer).
Comparing hierarchical organization between DO and SNOMED CT
DO and SNOMED CT both represent disease concepts and share important classificatory principles (e.g., by localization, by etiology, by morphologic type). Therefore, we expect their hierarchical organization to be similar. In other words, we expect that most hierarchical relations present in ontology will also be present in the other. And we do not expect that two hierarchically related concepts in one ontology will have no hierarchical relation or will be considered the same concept in the other ontology.
In practice, we take each pair of hierarchically related concepts (direct parent-child pair) present in one ontology and examine the relations between the two concepts in the other ontology. As illustrated in Figure 2, we consider 4 patterns of hierarchical relations across ontologies.
Figure 2.
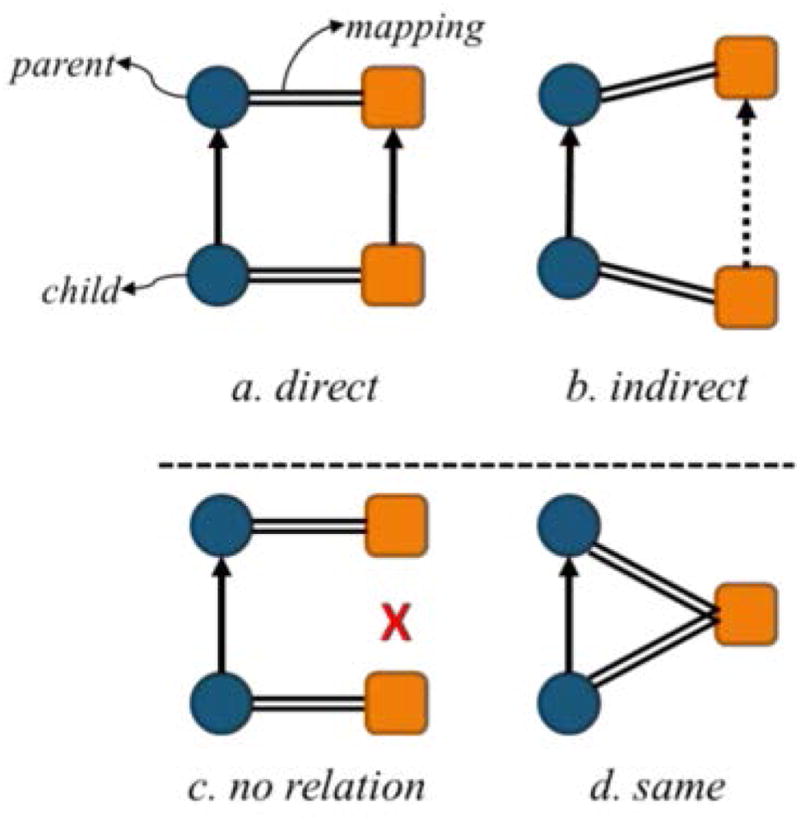
Types of possible hierarchical relations between corresponding mapped concepts
The two concepts are in the same direct parent-child relation in both ontologies.
The hierarchical relation is direct in one ontology but indirect in the other. The two ontologies are consistent, but the ontology with the indirect relation is finer grained than the other.
The hierarchical relation present in one ontology is missing from the other. The two ontologies are inconsistent.
The two hierarchically related concepts in one ontology map to the same concept in the other ontology. The two ontologies are inconsistent.
To make this comparison possible, we restrict it to pairs of concepts in which both concepts have a reference mapping to the other ontology. Since there may exist multiple mappings for an individual parent or child concept, we compare all possible parent-child pairs. We apply this method in both directions (i.e., both from DO to SNOMED CT concepts and from SNOMED CT to DO).
Results
Establishing a reference list of mappings
Updating and filtering mappings provided by DO
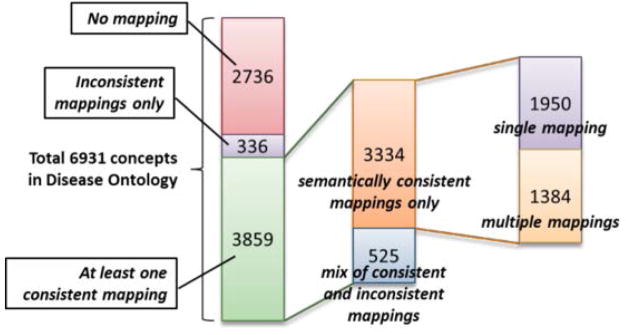
There were 12,470 mappings to SNOMED CT provided by DO as cross-references. We removed 224 mappings to retired SNOMED CT concepts and resolved 6552 mappings to moved concepts. A total 4195 DO concepts have one or more mappings to SNOMED CT (involving 8352 mappings).
Of the 8352 mappings, we filtered out 1184 semantically inconsistent mappings. After this filtering step, 336 DO concepts were left with no mapping. In most cases, a disease concept from DO was mapped to a concept in the “Morphologic abnormality” hierarchy of SNOMED CT. For example, “mixed cell type cancer” [DOID:154] mapped to “Mixed tumor, malignant (morphologic abnormality)” [SCTID:8145008].
Of the 3859 DO concepts with at least one semantically consistent mapping, 3334 had only semantically consistent mappings, while 525 had at least one inconsistent mapping.
Finally, of the 3334 DO concepts with only semantically consistent mappings, 1950 concepts had a single mapping (e.g., “Cycloplegia” [DOID:10033] mapped to “Cycloplegia (disorder)” [SCTID:68158006] only), while 1384 had multiple semantically consistent mappings. Of these 1384 concepts, 110 had a mapping to concepts in both the “Disease” and the “Clinical Findings” hierarchies, usually to a disease its associated finding. For example, “Mevalonic aciduria” [DOID:0050452] is mapped to “Mevalonic aciduria (disorder)” [SCTID:124327008] and “Hyperimmunoglobulin D with periodic fever (finding)” [SCTID:234538002].
Overall, as shown in Figure 3, of the 6931 disease concepts in DO, 3859 (56%) were mapped to SNOMED CT through at least one semantically consistent mapping provided by DO, 336 (5%) had only semantically inconsistent mappings, and 2736 (39%) were unmapped.
Figure 3.
Breakdown of explicit mappings of the DO concepts
Finding Additional Lexical Mappings
Leveraging lexical mapping through the UMLS, we identified a mapping to SNOMED CT for 619 (20%) of the 3072 DO concepts with no semantically consistent mapping.
Overall, our reference list of mappings includes 7949 semantically consistent mappings covering 4478 (65%) the 6931 DO concepts, mapped to 6440 unique SNOMED CT concepts.
Characterizing unmapped concepts
Semantically
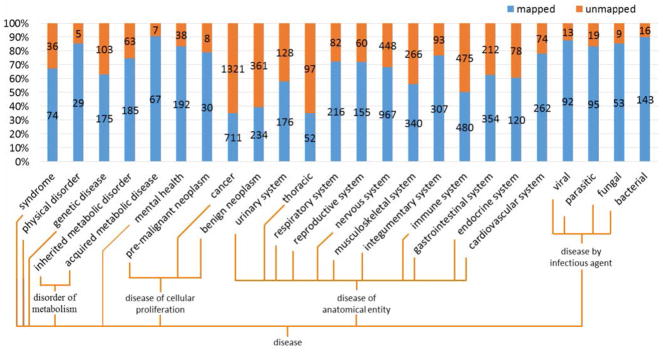
Of the 6931 DO concepts, 2453 (35%) remained unmapped to SNOMED CT. Figure 4 shows the distribution of mapped and unmapped concepts by the top-level subtrees of DO. Concepts belonging to multiple subtrees are counted multiple times. The top subtrees for unmapped concepts include cancers, neoplasms, diseases of the thoracic sys tem, immune system diseases, and nervous system diseases. In contrast, very few of infectious diseases remain unmapped.
Figure 4.
Distribution of mapped versus unmapped concepts within top level subtrees of the Disease Ontology. Numbers indicate the actual number of concepts in each subtree.
Structurally
The 2453 unmapped concepts can be grouped into 261 clusters of hierarchically related concepts (connected components). From a structural perspective, unmapped concepts fall under three possible categories: isolated unmapped leaf concepts, subtrees of unmapped concepts, and unmapped intermediary concepts.
We found 401 cases of isolated unmapped leaf concepts, namely 214 with a single parent and 187 with multiple parents. For example, “multiple mucosal neuroma” [DOID:5155] is the only unmapped child of “neuroma” [DOID:2001], which means that all of its siblings (e.g., “Neurilemmoma” [DOID:3192]), are mapped to SNOMED CT. Of note, 69 of these leaf concepts are the only child of their parent.
We found 1806 unmapped concepts in subtrees of unmapped concepts. These are clusters where none of the concepts is mapped, while they share a common mapped ancestor. For example, the subtree rooted at “chromosomal deletion syndrome” [DOID:0060388] contains 35 concepts, including “distal 10q deletion syndrome” [DOID:0060390] and “chromosome 15q11.2 deletion syndrome” [DOID: 0060393].
Finally, we found 246 cases of unmapped intermediary concepts (located between a mapped ancestor and a mapped descendant). 58 are intermediary “grouper” concepts in DO with all of its parents and children mapped to some SNOMED CT concept. 50 of these concepts sit between a single mapped parent and child. An example is “multifocal dystonia” [DOID:0050837]. This concept is unmapped to SNOMED CT, while its parent, “dystonia” [DOID:543], and child, “hemidystonia” [DOID:0050846], are mapped to SNOMED CT.
Differentiae
We examined all 2207 unmapped concepts that are isolated leaf or in subtrees, which represent the majority (90%) of the 2453 unmapped concepts, and analyzed how they differed from their parent concept(s). The most frequent differentiae, listed in Table 1 along with examples, include morphology (for cancers and neoplasms), specific subtypes (for rare genetic disorders), and anatomical subtypes. Of note, about a third of the unmapped concepts have more than one differentia. Typically these concepts have more than one parent. For example, “urethra adenocarcinoma” is a child of both “adenocarcinoma” (anatomical differentia) and “urethra cancer” (morphology differentia).
Table 1.
Characterization of differentiae added by unmapped DO concepts
| Type of differentiae | Count |
|---|---|
| Morphology (e.g. follicular dendritic cell sarcoma) | 831 |
| Morphology and anatomic site | 520 |
| Specific subtype (e.g. spinocerebellar ataxia type 1) | 253 |
| Anatomic site (e.g. intramuscular hemangioma) | 147 |
| Morphology and period of onset | 61 |
| Period of onset (e.g. pediatric osteosarcoma) | 45 |
| Chromosomal location and anomaly | 45 |
| Complex syndrome (e.g. agnathia-otocephaly complex) | 42 |
| Subtype | 42 |
| Organism (e.g. screw worm infectious disease) | 30 |
| Others | 191 |
Comparing hierarchical organization between DO and SNOMED CT
We found 4233 direct parent-child pairs among the mapped DO concepts and 5772 among the mapped SNOMED CT concepts. After classifying each pair of hierarchically related concepts into the four patterns of hierarchical relations across ontologies presented earlier, we established the distribution of patterns shown in Table 2.
Table 2.
Characterization of the mapped parent-child concepts in comparison to the relation between their corresponding mapped concepts. The types are illustrated in Figure 2 above.
| Mapping direction | Type (a) or (b) only | Type (c) or (d) only | (a or b) & (c or d) | Total pairs |
|---|---|---|---|---|
| DO to SNCT | 1198 [28%] | 1075 [25%] | 1978 [48%] | 4233 |
| SNCT to DO | 1842 [32%] | 2792 [48%] | 1138 [20%] | 5772 |
As mentioned earlier, in a given pair of hierarchically related concepts, each concept can be mapped to more than one concepts. Therefore, a pair of hierarchically related concepts can exhibit more than one pattern. To simplify the analysis, we distinguish between patterns indicative of semantic consistency (a and b) and patterns indicative of semantic inconsistency (c and d). Only about 30% of the hierarchical relations in DO and SNOMED CT are semantically consistent between the two ontologies (a/b only). The other hierarchical relations are either completely (c/d only) or partially inconsistent (a/b and c/d). This analysis reveals critical differences in hierarchical organization and concept orientation (i.e., whether two concepts correspond to the same entity) in the two ontologies.
Here are examples of relations for patterns. (The arrow, →, represents the “child of” relation).
For the DO relation “peliosis hepatis” [DOID:914] → “hepatic vascular disease” [DOID:272], there is a direct corresponding relation in SNOMED CT, “Peliosis hepatis (disorder)” [SCTID:58008004] → “Vascular disorder of liver (disorder)” [SCTID:235878005]. The two ontologies are perfectly aligned in this case.
For the DO relation “diphtheritic cystitis” [DOID:13306] → “cystitis” [DOID:1679], there is an indirect corresponding relation in SNOMED CT, “Diphtheritic cystitis (disorder)” [SCTID:48278001] → “Bacterial cystitis (disorder)” → “Infective cystitis (disorder)” → “Cystitis (disorder)” [SCTID:38822007]. The two ontologies are semantically consistent, but SNOMED CT is finer-grained in this case as its two intermediary concepts are missing from DO.
For the DO relation “portal hypertension” [DOID:10762] → “hepatic vascular disease” [DOID:272], there is no corresponding hierarchical relation in SNOMED CT between these two concepts. Instead, “Portal hypertension (disorder)” [SCTID:34742003] and “Vascular disorder of liver (disorder)” [SCTID:235878005] are in different parts of the “disorder of abdomen” hierarchy. In this case, the two ontologies are inconsistent.
Discussion
Pre- vs. post-coordination
Most biomedical terminologies, including DO, only consider pre-coordinated concepts. In other words, there is no built-in mechanism in DO to combine existing concepts to derive new concepts. As a result, only existing, pre-coordinated concepts are available to applications (e.g., for annotation purposes). In contrast, SNOMED CT supports post-coordination through a compositional grammar [10], which reflects semantic constraints expressed in the SNOMED CT concept model [11]. For this reason, SNOMED CT tends to adopt a parsimonious approach to pre-coordination, i.e., avoid pre-coordinating what can be expressed through post-coordination.
As shown in Table 1, the combination of differentiae morphology and anatomic site is the single most frequent combination. While pre-coordinated concepts are generally easier to use, the proliferation of pre-coordinated concepts may add unnecessary to the terminology.
Resolving multiple mappings
The mappings (cross-references) to SNOMED CT provided by DO frequently involve multiple SNOMED CT concepts (1-many mappings). Even after filtering out semantically inconsistent mappings (e.g., mapping of a disorder to a morphology concept), many multiple mappings remain.
In fact, in our reference mapping, 1384 (42%) of the 3334 DO concepts with mapping to SNOMED CT have multiple (semantically consistent) mappings to SNOMED CT. Of these, there are 110 concepts with a mapping to both a “Disease” concept and a “Clinical Findings” concept. In such cases, the “Disease” concept could be given precedence. The remaining 1274 DO concepts have multiple mappings within the same hierarchy. In this case, the structural analysis we performed can help guide the mapping. Mappings involved in semantically consistent patterns of hierarchical relations (namely type a and b) could be given precedence.
Conclusion
Overall, 4478 (65%) the 6931 DO concepts are mapped to SNOMED CT. The cancer and neoplasm subtrees of DO account for many of the unmapped concepts. The most frequent differentiae include morphology (for cancers and neoplasms), specific subtypes (for rare genetic disorders), and anatomical subtypes. Unmapped concepts usually form subtrees, and less often correspond to isolated leaf concepts or isolated intermediary concepts. This detailed analysis of the gaps in coverage and structural differences between DO and SNOMED CT contributes to the interoperability between these two ontologies and will guide further validation.
Acknowledgments
This work is supported in part by the Intramural Research Program of the U.S. National Library of Medicine (NLM) and an appointment to the NLM Research Participation Program administered by ORISE through an interagency agreement between the U.S Dept. of Energy and the NLM.
References
- 1.Schriml LM, Arze C, Nadendla S, Chang YW, Mazaitis M, Felix V, Feng G, Kibbe WA. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40:D940–946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S-A, Scheuermann RH, Shah N, Whetzel PL, Lewis S. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotech. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D, Parkinson H, Schriml LM. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015;43:D1071–1078. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chute CG, Cohn SP, Campbell KE, Oliver DE, Campbell JR. The content coverage of clinical classifications. For The Computer-Based Patient Record Institute’s Work Group on Codes & Structures. Journal of the American Medical Informatics Association. 1996;3:224–233. doi: 10.1136/jamia.1996.96310636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenreider O, Nelson SJ, Hole WT, Chang HF. Beyond synonymy: exploiting the UMLS semanticsin mapping vocabularies. Proceedings of the AMIA Symposium. 1998:815–819. [PMC free article] [PubMed] [Google Scholar]
- 7.Dhombres F, Winnenburg R, Case JT, Bodenreider O. Extending the coverage of phenotypes in SNOMED CT through post-coordination. Stud Health Technol Inform. 2015;216:795–799. [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditarydisease. The American Journal of Human Genetics. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung KW, Richesson R, Bodenreider O. Coverage of rare disease names in standard terminologies and implications for patients, providers, and research. AMIA Annu Symp Proc. 2014;(2014):564–572. [PMC free article] [PubMed] [Google Scholar]
- 10.IHTSDO. SNOMED CT Compositional Grammar Specification and Guide. 2016. [Google Scholar]
- 11.IHTSDO. SNOMED CT Starter Guide. 2016. SNOMED CT Concept Model. [Google Scholar]