Abstract
While the vast majority of cellular DNA in eukaryotes is contained in long linear strands in chromosomes, we have long recognized some exceptions like mitochondrial DNA, plasmids in yeasts and double-minutes in cancer cells where the DNA is present in extrachromosomal circles. In addition, specialized extrachromosomal circles of DNA (eccDNA) have been noted to arise from repetitive genomic sequences like telomeric DNA or ribosomal DNA. Recently eccDNA arising from unique (non-repetitive) DNA have been discovered in normal and malignant cells, raising interesting questions about their biogenesis, function and clinical utility. Here we will review recent results and future directions of inquiry on these new forms of eccDNA.
Keywords: eccDNA, DNA repair, microDNA
History of extrachromosomal circular DNA in eukaryotes
EccDNA was first found by Alix Bassel and Yasuo Hoota while investigating Franklin Stahl’s theory that chromosomes of higher organisms are made of a series of DNA circles [1] in 1964. They found DNA circles of various sizes, ranging from hundreds to thousands of base pairs within a preparation of mammalian DNA. These large DNA circles are referred to as Double Minutes (DMs). Another group verified the existence of DMs in human cancer cells by karyotype preparations and by CsCl gradient purification [2, 3]. CsCl gradient purification and EM imaging experiments were used to study DNA from a number of other organisms [4–10].
Southern blots determined that eccDNA molecules were homologous to genomic DNA. The majority of eccDNA were <500 base pairs and were named poly-disperse circular DNA (spcDNA) [11–15]. Although the vast majority appeared to come from repetitive sequences [11–15], a few spcDNA molecules hybridized to unique sequences [11, 13, 15, 16]. Some groups found that some non-repetitive spcDNA sequences were flanked on both sides by direct repeats of an average length of 9–11 bp [13–15]. This suggested that DNA repair pathways such as homologous recombination or microhomology mediated end joining between short repeats could generate the circles. However, later studies isolated and sequenced eccDNA with unique sequences that do not contain repetitive regions of any length within or flanking the DNA [16]. Around the same time, a group used exonuclease III to quantify eccDNA amounts and found that eccDNA levels vary between tissues in mice [17].
Using techniques that mostly studied eccDNA from repetitive sequences, a few groups attempted to determine which processes contribute to the formation of eccDNA. Cycloheximide, an inhibitor of protein synthesis, caused a 70-fold increase of eccDNA in murine cells [14]; increases were also seen with a carcinogen, 7,1-dimethylbenz[a]anthracene, and a DNA replication inhibitor, hydroxyurea [14]. Cells from patients with Fanconi Anemia, with defects in a specific DNA repair pathway, contained longer and more eccDNA molecules [12]. Two-dimensional gel electrophoresis expanded the exploration of the smaller eccDNA to show that carcinogens increase eccDNA levels [18] and that eccDNA production changes with developmental stage in frogs and flies [19, 20]. EccDNA could be formed by foreign DNA within a cell [21] and the organization of DNA sequences in tandem repeats predisposed the DNA for eccDNA formation [21–23]. Collectively, these results suggested that eccDNA formation is dependent on DNA sequence, organization and DNA damage repair.
As regards function of eccDNA, sequencing of larger eccDNA molecules, DMs, indicated their capacity to code for full genes, and indeed eccDNA molecules in cancer cells were found to contain amplified oncogenes and drug resistant genes [18, 24–26].
Recent advances in eccDNA
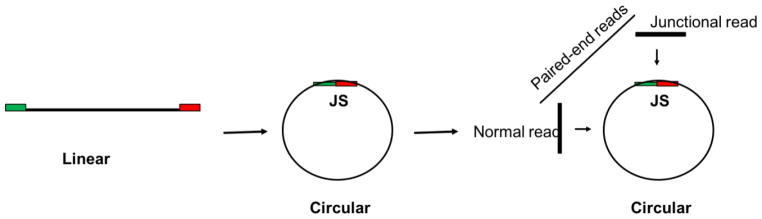
Recently, a paired-end high-throughput sequencing technology approach was used to sequence the full complement of eccDNA in mouse tissues and human cancer cell lines [27]. High-throughput sequencing on exonuclease-resistant, rolling circle amplified, extrachromosomal cellular DNA followed by a computational method to identify junctional sequences (Fig 1) allowed the characterization of eccDNA sequences at base pair resolution [27]. Unique (from non-repetitive DNA) eccDNA sizes peaked around 180 and 380 bp with 5% of the molecules extending to as long as 2–3 kb; the pattern was consistent between all mouse tissues and human cancer cell lines. These molecules were named microDNA. Longer eccDNA could have been under-represented in this study because rolling circle amplification will amplify smaller circles at a greater rate than larger circles, but electron microscopy of the circles also suggests that the majority of the circles are small [28]. The eccDNA mapped to over a hundred-thousand unique sites in the genome, and were enriched in specific regions, hotspots, including 5′UTR regions and CpG regions, areas with high GC content (60%), and transcriptionally active chromatin [27]. The genomic DNA flanking most microDNA contain 2–15 base direct repeats suggesting a microhomology-mediated mode of generation of the circles [27]. The chromosomal locations of the eccDNA sequences weakly clustered prostate and ovarian cancer cell-lines away from each other suggesting that sites of eccDNA formation could be correlated to cell lineage [28]. Further, deletion of MSH3, encoding a protein in the DNA mismatch repair pathway, caused eccDNA levels to decrease by 80% [28]. It is not yet clear whether these small eccDNAs replicate. A very rough estimate, probably an underestimate, of the abundance of the eccDNA counted by electron microscopy on preparations from defined numbers of cells suggest that there are at least 125–200 circles per DT40 cell [28].
Figure 1. Junctional tag.
Schematic representation of eccDNA and junctional sequence genesis from linear DNA. The two ends of linear DNA get ligated and ligation event creates a new junctional sequence which is not present in parent linear DNA; The sequencing reads partially mapping to the left and right side of ligation point are the junctional sequence. Because the junctional sequence is not present in linear DNA it acts as a discriminatory feature within computer algorithms to validate that the DNA fragment was within a circular DNA molecule [27, 28, 49]. The paired end reads pairs where one read completely maps inside the body of circle and other second reads maps on the junctional sequence are used for the final validation of circular nature of the starting DNA fragment.
A similar study done in Saccharomyces cerevisiae [29] identified about two-thousand extrachromosomal circles of DNA covering nearly a quarter of the yeast genome. The approach ignored potential eccDNAs less than 1kb in size and did not depend on the identification of junctional sequences to call a sequence an eccDNA. Thus the eccDNAs identified varied from 1–38 kb in length, with a significant enrichment of circles from repeated parts of the genome like transposons, ribosomal DNA circles, gene duplications etc, suggesting a homologous recombination mediated biogenesis of the circles. Nevertheless, nearly 60% of the eccDNA arose from unique sequences, and over 90% of the genomic sites revealed 7 base direct repeats that may lend them to a microhomology-mediated mechanism of circle formation.
A very recent study done in Caenorhabditis elegans and in human cell lines used an approach that relies on density gradient centrifugation (Cesium Chloride- Ethidium Bromide) followed by tagmentation and high-throughput sequencing [30]. The authors also report circles mapping on both coding and noncoding regions of the genome. The eccDNA frequently appeared to be derived from exons of protein coding genes like mucin and titin. The authors hypothesized that eccDNA may contribute to the expression of different isoforms of a gene by interfering with or promoting the transcription of specific exons [27].
Fluorescent microscopy has been used to quantify changes in large eccDNAs, DMs, between cancerous and normal cells [31, 32]. EGFR and myc genes were amplified in cancer cells after a few passages through the formation of eccDNA [32]. An image analysis software package combined with fluorescence imaging to quantify copies of oncogene, found MYC and EGFR to be amplified in ~40% of examined human cancer tissues whereas no enrichment was found in normal tissues [31]. The oncogenes were significantly more amplified through the mechanism of eccDNA formation than through chromosomal amplification [31]. The failure to detect eccDNA in normal tissues is probably due to the absence of an enrichment procedure for circles and because the small eccDNA (microDNA) do not bind enough dye to be detected under the microscope. Together these studies suggest that eccDNA could contribute to tumor heterogeneity and evolution of tumors by increasing the copy number of oncogenes [28]. These findings were validated by a study where an oncogene, MET, in glioblastoma cells was found to be amplified on eccDNA molecules as seen by FISH [33].
Yeast also contained an eccDNA carrying the GAP1 gene, encoding a protein involved in amino acid absorption, that was amplified after metabolite limitation [34]. The eccDNA was flanked by two long direct repeats and it was hypothesized that homologous recombination between the two repeat regions could form an eccDNA.
Clearly the eccDNA identified to date are distributed over a wide size range and can also be easily divided into those that arise from repetitive DNA versus unique DNA (Table 1). Thus it is likely that eccDNAs may arise by many different pathways and have different functions.
Table 1.
Size range of circular DNA in eukaryotes
| Name of circular DNA | Size range | Replication | References | Function (if any) |
|---|---|---|---|---|
| Double-minute chromosomes | 100kb – 3mb | Self | [31, 42–45] | Double minutes contain proto-oncogenes in cancers |
| Extra chromosomal rDNA circle | 19.3 and 40.4 kb | Self | [41, 55] | Accumulation associated with aging, suppress mitochondrial “cheats” in Yeast |
| Telomeric circle | Integral multiples of 738 bp | Self | [22] | Telomeric circles in a wide range of organisms provide telomeric repeat template for restoring telomere length by homologous recombination |
| Mitochondrial circular DNA | 16 kb | Self | [56] | Contains genes essential for mitochondria function |
| Chloroplast circular DNA | 120–160 kb | Self | [57] | Contains genes including essential genes for photosynthesis |
| Alpha satellite circle | 2–20 kb | Self | [22] | Centromere evolution? |
| MicroDNA | 100–1000 bases | ND | [27, 28, 49] | miRNA generation? |
| Kinetoplast maxicircles | 20–40 kb | Self | [58, 59] | Encode ribosomal RNAs and mitochondrial proteins |
| Kinetoplast minicircles | 0.5–1 kb | Self | [58, 59] | Produce guide RNA to decode maxicircle gene information for mitochondria |
Future Questions
Biogenesis
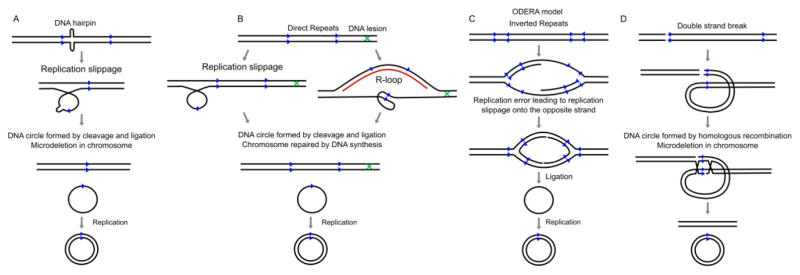
The mechanisms that lead to the biogenesis of eccDNA have yet to be fully discovered. A large percentage of eccDNA molecules contain or are proximal to short direct repeats, suggesting that some form of microhomology directed repair may form eccDNA [35, 36]. However, a significant portion of eccDNA contain no repeats and could not recombine with any nearby sequence [16]. EccDNA levels have been known to increase with the addition of carcinogens [16, 37]. The contribution of DNA replication in eccDNA production, however, is controversial where some labs find eccDNA levels increase when ongoing replication is blocked by replication inhibitors [14] while others find that eccDNA can be formed in the absence of any DNA replication [21]. Further, specific DNA repair proteins are necessary for eccDNA formation such as MSH3 (involved in mismatch repair) in DT40 cells [28]; or unnecessary for eccDNA formation, such as okra, mus309, and mei41 (involved in homologous recombination, non-homologous end joining, and DNA damage recognition respectively) in Drosophila melanogaster cells [20]. Homologous recombination could excise repetitive DNA sequences during early development to give rise to larger eccDNA. This would contribute to the heterogeneity of tandem repeat sequences between individuals, including genes organized in tandem arrays such as rDNA and tDNA [21, 22]. Another hypothesis is that DNA synthesis through regions rich in repeats would require increased recruitment of DNA repair proteins, which may cause part of the chromosome to be incompletely condensed prior to mitosis. This would cause breakage of large fragments, which are ligated into circles [38]. Finally, the enrichment of eccDNA in normal cells from GC rich, transcriptionally active areas of the genome suggest that R-loop formation and its repair may contribute to eccDNA formation [28]. In Figure 2 we have speculated on the different mechanism by which the eccDNAs may arise. Overall, it seems that eccDNAs arise from various processes and more research is needed to determine how the size, GC content, and repeat sequences of eccDNA are tied to pathways involving DNA metabolism.
Figure 2. Examples of how eccDNA is formed.
(A) Replication slippage creates a loop on the template strand through mis-priming of a dissociated polymerase at the wrong direct repeat. The loop is then excised and ligated into a circle, leaving a microdeletion on the chromosome. (B) Replication slippage creates a loop in the product strand which is then excised and ligated into a circle, but no microdeletion is left on the chromosome. An R-loop displaces the non-template strand and allows the direct repeats on the unpaired strand to form into a loop which is then excised and ligated into a circle. Alternatively (not shown), the RNA paired DNA strand could be excised, released from the RNA and ligated between direct repeats to form a circle. In either case, the gap in the chromosomal DNA is repaired by gap filling and leaves no deletions on the chromosome. (C) ODERA mechanism of eccDNA formation. Replication slippage on pairs of inverted repeats and ligation forms a single-strand circle, e.g. [54]. (D) Double strand break within a repeat region with a proximal homologous repeat sequence is repaired by homologous recombination. The small fragment forms a circle, while the chromosome suffers a microdeletion. An example of this is in [34].
When unique DNA is detected as an eccDNA, there is also the issue of whether the biogenesis arose from excision of the chromosomal DNA, leading to a corresponding deletion in the chromosomal sequence, or whether the circular DNA was produced by some kind of copying mechanism, as during DNA replication or repair, where there is no concurrent loss of chromosomal DNA. Indeed, in at least two hot-spots for microDNA production, a very high depth sequencing of the chromosomal DNA from adult mouse brain identified microdeletions present substoichiometrically (in a somatically mosaic pattern) [27]. The microdeletions were rare, occurring in 1 of 400–4000 alleles from the brain and would be missed if the genomic sequencing is not done at a very high depth. Nevertheless, the presence of eccDNA from over a hundred-thousand sites in mouse, human and chicken cells makes it unlikely that all of them are created by an excision event that leaves behind over a hundred-thousand somatically mosaic deletions on the chromosomes.
Circular DNA containing telomeric DNA (t-circles) and ribosomal sequences (rDNA) have been studied for many more years and many mechanisms of formation have been proposed, though none proven. These theories include genomic rearrangement, excision and ligation, recombination between tandem repeats, and reverse transcription of mRNA [39]. One protein complex, CTC1/STN1/TEN1, known to be involved in telomere maintenance has been shown to significantly contribute to t-circle formation [40]. Also, in yeast cells lacking SGS1, a protein known to be involved in DNA repair, there are significantly increased levels of eccDNA [41]. These results further suggest that multiple pathways contribute to eccDNA formation.
Functions
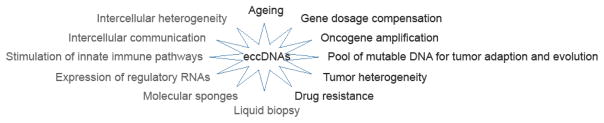
Known functions of eccDNA include contribution to intercellular genetic heterogeneity in tumors, particularly amplification of oncogenes and drug resistance genes, which presumes that the genes on the eccDNA are expressed. Theoretical functions of eccDNA can include expression of regulatory RNAs contributing to intercellular heterogeneity, sponging of transcription factors, production of a pool of mutable DNA for evolution of tumors, gene dose compensation, ageing, release from cells for intercellular communication, use in liquid biopsy and stimulation of innate immune pathways (Figure 3).
Figure 3. Functions of eccDNA in mammalian cells.
The known functions of eccDNA are listed in black text; the hypothesized roles of eccDNA are listed in grey text.
EccDNAs have been linked to cancers and drug resistance. Double minutes carry oncogenes or drug resistance genes and advance cancers by gene amplification [42–45]. Recent evidence suggests oncogene amplification mediated by eccDNAs is a much more common phenomenon than previously thought, and is critical for tumor heterogeneity and evolution. For example, eccDNAs were detected in nearly 40% of the tumor cell lines and nearly 90% of patient-derived brain tumor models [31]. This study also provides mathematical and experimental evidence that driver oncogene amplification and tumor heterogeneity may be significantly higher when the amplification occurs on eccDNA than when the amplification occurs within chromosomes. Due to the random distribution of eccDNAs to daughter cells, during each division one of the daughter cells may inherit a higher copy number of eccDNAs with a driver oncogene, and thus acquire a proliferative advantage [31]. The number of specific eccDNAs in cells may also change in response to environmental conditions thereby introducing an additional mechanism for tumor adaptation to challenging conditions. An example of this is in glioblastomas, where EGFR is frequently mutated and commonly gives rise to the EGFRvIII oncogenic variant. While EGFRvIII promotes tumor growth, it also makes the tumor cells sensitive to EGFR tyrosine kinase inhibitors [46]. It is found that resistance to EGFR tyrosine kinase inhibitors is developed by losing double minutes carrying the mutant EGFR [46]. It is also possible that driver mutations can occur extrachromosomally during the amplification of eccDNAs, and this will also be critical for tumor heterogeneity and evolution [47]. Overall, eccDNAs appear to be a common phenomenon in cancers contributing to tumor heterogeneity, adaptation, and evolution.
EccDNA has also been linked to aging. EccDNAs containing ribosomal RNA genes accumulate with time and contribute to the aging of yeast cells [41]. These extrachromosomal rDNAs have an ARS sequence (autonomously replicating sequence), and are able to replicate [41]. Moreover, in each cell division they are preferentially segregated to mother cells. These properties are responsible for an exponential increase of these eccDNAs in aging mother cells, while limiting the number of eccDNAs in daughter cells prolongs their lifespans. The exact mechanism how the eccDNAs trigger the senescence and eventual death of old cells is not clear. It was proposed that sheer abundance of these eccDNAs might titrate the components of replication and/or transcription machineries, and triggers the senescence and eventual death of old cells. Corroborating the titration hypothesis, ectopic expression of ARS plasmid is sufficient to trigger a final arrested state of old cells and eventual cell death [41]. Recent discoveries of eccDNA in normal cells and tissues raises the interesting question of whether eccDNA accumulation may contribute to ageing in higher eukaryotes.
EccDNAs may also play a role in gene compensation [48]. In S. cerevisiae, H2A and H2B histones are encoded by two gene pairs named as HTA1-HTB1 and HTA2-HTB2. When HTA1-HTB1 is deleted, dosage compensation occurs by gene amplification of HTA2-HTB2 via formation of a new eccDNA containing 39kb of chromosome II that includes HTA2-HTB2, the histone H3–H4 locus, a centromere and origins of replication [48]. This new eccDNA is created by recombination between two Ty1 retrotransposon elements that flank this region [48]. In HTA1-HTB1 deleted strains formation of the HTA2-HTB2 eccDNA is significantly elevated to compensate for the decrease in H2A and H2B [48].
In contrast to the DMs, the smaller type of eccDNAs are more widespread, but much less is known about their function in cell biology [27, 49]. They are too small to contain protein-coding genes, but long enough to code for regulatory short RNAs or fragments of genes. Another possibility for the microDNA function may be molecular sponging: they may function as sponges for transcription factors to control gene expression indirectly. Finally, it was recently found that microDNAs are present in serum and plasma of both mouse and humans as circulating DNA [49, 50]. Other than their potential as biomarkers in liquid biopsy experiments, if other cells can take in these microDNAs, this may be a novel way of communication between cells. This possibility is a speculation at this point, but is an important subject of future investigation.
Cells react to naked DNA in the cytoplasm by activation of the cGAS pathway that culminates in expression of interferon and stimulation of the immune system [51, 52]. This is part of the innate immune response that is used in response to foreign pathogens. One interesting possibility is that the eccDNAs are released to the cytoplasm during mitosis and are either degraded by enzymes like TREX1 or activate the cGAS pathway. Thus the eccDNA, especially if they are not protected by chromatin, may be an endogenous antigen that can activate autoimmune pathways.
Clinical utility
Tumor specific features of eccDNA could be important for the identification and prognosis of the disease and therefore it is important to find any differences in eccDNA between tumor and matched normal tissue. Human cancer cell lines were found to have a population of longer eccDNAs than that in normal mouse tissues [27]. MicroDNA was present in both tumor and normal lung tissue and most of the known properties of eccDNA were similar between the normal and tumor [49]. However, the eccDNA identified in human lung cancers were slightly longer on average compared to matched normal tissue from the same patients (3 out of 4 pairs of tumor and normal samples) [49]. It will, of course, be more important for the use of eccDNA as biomarkers for cancers, if their abundance or sequence of origin change in a predictable way when a normal cell is transformed into a cancer cell.
There are numerous reports using high-throughput sequencing technology to identify tumor-specific linear DNA fragments present in the serum or plasma, the so called liquid biopsy [53]. The presence of eccDNA in circulation has also recently been shown [49, 50]. The characteristics of the microDNA (higher GC content, 2–15 bases long direct repeat flanking their source genomic sites, genomic distribution etc.) of circulating eccDNA were very similar to that reported earlier in mouse tissue and mouse, chicken and human cell lines. Furthermore, the eccDNA arise from genic and intergenic regions. Consistent with the fact that microDNA from lung cancers was slightly longer than matched normal tissue (mentioned above), the circulating eccDNA in the patients before surgical resection of the tumor was generally longer compared to the circulating eccDNA in the same patients 6 weeks after surgery [49]. This may suggest that human cancers also release longer eccDNA into circulation than normal tissues. The eccDNAs are expected to be more stable compared to linear DNA and this may be an advantage for using eccDNA in blood for liquid biopsy experiments.
Concluding Remarks
The most important question to be resolved in order for eccDNA to be used as a biomarker for cancer is determine whether cell-free eccDNA arising from tumors can be differentiated from eccDNA arising from normal tissues. Further, because eccDNA has been shown to confer an ability to amplify oncogenes and drug resistant genes it is vital to determine what specific proteins are involved in eccDNA formation. The proteins involved in generating eccDNA from the genome could act as targets for therapy against this form of genomic plasticity. The functions of eccDNA needs to be further explored. The majority of eccDNA are 200–400 base pairs (microDNA, some of the spcDNA). However, most studies have focused on the gene amplification abilities of DMs, and it is possible that microDNA can contribute to similar amplification of small genes or regulatory RNAs. Lastly, the turnover of eccDNA in cells or in cell-free DNA in the circulation has yet to be determined, and the results will have implications for their intracellular functions and their utility for liquid biopsy, respectively. The half-life of these molecules inside the cells would be particularly interesting in proliferating cells where nuclear envelope breakdown in mitosis exposes them concurrently to cytoplasmic nucleases and to the cGAS DNA sensing system. Overall, research on eccDNA has increased the appreciation of the plasticity of genomic DNA and the dynamics of gene amplification and deletion and much more remains to be discovered regarding their biogenesis, function and possible clinical utility (See Outstanding Questions).
Outstanding Questions.
What repair pathways/proteins are involved in generating the different sizes of eccDNA?
Do eccDNA increase or decrease in specific diseases?
Do eccDNAs regulate cell function or play a part in inter-cellular communication?
Can eccDNA re-insert into genome and change the structure and function of genes?
Are eccDNAs passed on to the next generation of cells and organisms?
Trends box.
Genome sequencing, microscopic and biochemical methods have identified eccDNA in a wide range of species, tissues and cells.
eccDNA are present in mammalian tumor cells and larger eccDNA harbor oncogenes
eccDNA are heterogenous in size, ranging from few hundreds (microDNA) to few megabases (double minutes)
Some eccDNA have a role in conferring tumor drug resistance
eccDNA provide genetic diversity between cells even in normal tissues
Glossary
- Cycloheximide
protein synthesis inhibitor.
- Density gradient centrifugation
Is a technique to isolate closed circular DNA. The genomic DNA is mixed in a CsCl solution containing ethidium bromide (an intercalating agent) and subjected to ultracentrifugation to form a density gradient. Plasmid-DNA sample is run separately to locate where circular DNA will sediment. This location is used to isolate the eccDNA which is insufficient in abundance to produce a visible band.
- Double minutes
eccDNA first identified by karyotyping as existing independent of the chromosomes in cancer cells. They are long enough to have their own origins of replication and have been observed in tumor tissue where they most frequently harbor the oncogenes or genes involved in drug resistance in tumor.
- Homologous Recombination
A repair pathway that repairs double strand breaks by using the homologous sequence of a sister chromatid to align, fill in gaps, and then ligate the damaged chromosome.
- Junctional sequences
Nucleotide sequences created at the junction during circularization of the linear DNA. The junctional sequence does not exist in the linear chromosome.
- Junctional tags or reads
One of the ends of a paired-end read that cannot be mapped on to the genome and can only be explained as a junctional sequence produced by joining two shorter sequences that flank the location where the other end of the pair maps on the genome.
- Microhomology mediated end joining
A repair pathway which uses 5–25 bp of homology to align and then ligate the broken ends of a double strand break.
- Paired end reads
Reads from two ends of a defined size linear DNA fragment obtained by high-throughput sequencing.
- Rolling circle amplification
A DNA/RNA amplification process where tandem copies of a circular molecule are generated as linear DNA. The final linear DNA/RNA molecule has direct repeats of the original circular DNA/RNA molecule joined in tandem.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hotta Y, Bassel A. Molecular Size and Circularity of DNA in Cells of Mammals and Higher Plants. Proc Natl Acad Sci U S A. 1965;53:356–62. doi: 10.1073/pnas.53.2.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox D, et al. Minute Chromatin Bodies in Malignant Tumours of Childhood. Lancet. 1965;1(7402):55–8. doi: 10.1016/s0140-6736(65)90131-5. [DOI] [PubMed] [Google Scholar]
- 3.Radloff R, et al. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967;57(5):1514–21. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agsteribbe E, et al. Circular DNA from mitochondria of Neurospora crassa. Biochim Biophys Acta. 1972;269(2):299–303. doi: 10.1016/0005-2787(72)90439-x. [DOI] [PubMed] [Google Scholar]
- 5.Billheimer FE, Avers CJ. Nuclear and mitochondrial DNA from wild-type and petite yeast: circularity, length, and buoyant density. Proc Natl Acad Sci U S A. 1969;64(2):739–46. doi: 10.1073/pnas.64.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buongiorno-Nardelli M, et al. Electron microscope analysis of amplifying ribosomal DNA from Xenopus laevis. Exp Cell Res. 1976;98(1):95–103. doi: 10.1016/0014-4827(76)90467-5. [DOI] [PubMed] [Google Scholar]
- 7.Ono T, et al. Characterization of nuclear and satellite DNA from trypanosomes. Biken J. 1971;14(3):203–15. [PubMed] [Google Scholar]
- 8.Smith CA, Vinograd J. Small polydisperse circular DNA of HeLa cells. J Mol Biol. 1972;69(2):163–78. doi: 10.1016/0022-2836(72)90222-7. [DOI] [PubMed] [Google Scholar]
- 9.Stanfield S, Helinski DR. Small circular DNA in Drosophila melanogaster. Cell. 1976;9(2):333–45. doi: 10.1016/0092-8674(76)90123-9. [DOI] [PubMed] [Google Scholar]
- 10.Wong FY, Wildman SG. Simple procedure for isolation of satellite DNA’s from tobacco leaves in high yield and demonstration of minicircles. Biochim Biophys Acta. 1972;259(1):5–12. doi: 10.1016/0005-2787(72)90468-6. [DOI] [PubMed] [Google Scholar]
- 11.Bertelsen AH, et al. Molecular characterization of small polydisperse circular deoxyribonucleic acid from an African green monkey cell line. Biochemistry. 1982;21(9):2076–85. doi: 10.1021/bi00538a015. [DOI] [PubMed] [Google Scholar]
- 12.Motejlek K, et al. Increased amount and contour length distribution of small polydisperse circular DNA (spcDNA) in Fanconi anemia. Mutat Res. 1993;293(3):205–14. doi: 10.1016/0921-8777(93)90071-n. [DOI] [PubMed] [Google Scholar]
- 13.Stanfield SW, Helinski DR. Cloning and characterization of small circular DNA from Chinese hamster ovary cells. Mol Cell Biol. 1984;4(1):173–80. doi: 10.1128/mcb.4.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sunnerhagen P, et al. Molecular cloning and characterization of small polydisperse circular DNA from mouse 3T6 cells. Nucleic Acids Res. 1986;14(20):7823–38. doi: 10.1093/nar/14.20.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanfield SW, Lengyel JA. Small circular DNA of Drosophila melanogaster: chromosomal homology and kinetic complexity. Proc Natl Acad Sci U S A. 1979;76(12):6142–6. doi: 10.1073/pnas.76.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Loon N, et al. Formation of extrachromosomal circular DNA in HeLa cells by nonhomologous recombination. Nucleic Acids Res. 1994;22(13):2447–52. doi: 10.1093/nar/22.13.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaubatz JW, Flores SC. Purification of eucaryotic extrachromosomal circular DNAs using exonuclease III. Anal Biochem. 1990;184(2):305–10. doi: 10.1016/0003-2697(90)90685-3. [DOI] [PubMed] [Google Scholar]
- 18.Schneider SS, et al. Isolation and structural analysis of a 1.2-megabase N-myc amplicon from a human neuroblastoma. Mol Cell Biol. 1992;12(12):5563–70. doi: 10.1128/mcb.12.12.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen S, et al. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol Cell Biol. 1999;19(10):6682–9. doi: 10.1128/mcb.19.10.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen S, et al. Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 2003;13(6A):1133–45. doi: 10.1101/gr.907603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen S, Mechali M. A novel cell-free system reveals a mechanism of circular DNA formation from tandem repeats. Nucleic Acids Res. 2001;29(12):2542–8. doi: 10.1093/nar/29.12.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen S, et al. Extrachromosomal circles of satellite repeats and 5S ribosomal DNA in human cells. Mob DNA. 2010;1(1):11. doi: 10.1186/1759-8753-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navratilova A, et al. Survey of extrachromosomal circular DNA derived from plant satellite repeats. BMC Plant Biol. 2008;8:90. doi: 10.1186/1471-2229-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beland JL, et al. CpG island mapping of a mouse double-minute chromosome. Mol Cell Biol. 1993;13(8):4459–64. doi: 10.1128/mcb.13.8.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carroll SM, et al. Characterization of an episome produced in hamster cells that amplify a transfected CAD gene at high frequency: functional evidence for a mammalian replication origin. Mol Cell Biol. 1987;7(5):1740–50. doi: 10.1128/mcb.7.5.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl F, et al. Amplicon structure in multidrug-resistant murine cells: a nonrearranged region of genomic DNA corresponding to large circular DNA. Mol Cell Biol. 1992;12(3):1179–87. doi: 10.1128/mcb.12.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata Y, et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336(6077):82–6. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dillon LW, et al. Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity. Cell Rep. 2015;11(11):1749–59. doi: 10.1016/j.celrep.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller HD, et al. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci U S A. 2015;112(24):E3114–22. doi: 10.1073/pnas.1508825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoura MJ, et al. Intricate and Cell-type-specific Populations of Endogenous Circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens. G3 (Bethesda) 2017 doi: 10.1534/g3.117.300141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner KM, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543(7643):122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogt N, et al. Amplicon rearrangements during the extrachromosomal and intrachromosomal amplification process in a glioma. Nucleic Acids Res. 2014;42(21):13194–205. doi: 10.1093/nar/gku1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.deCarvalho AC, et al. Extrachromosomal DNA elements can drive disease evolution in glioblastoma. BioRxiv. 2017:1–13. [Google Scholar]
- 34.Gresham D, et al. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci U S A. 2010;107(43):18551–6. doi: 10.1073/pnas.1014023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misra R, et al. Recombination mediates production of an extrachromosomal circular DNA containing a transposon-like human element, THE-1. Nucleic Acids Res. 1989;17(20):8327–41. doi: 10.1093/nar/17.20.8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones RS, Potter SS. L1 sequences in HeLa extrachromosomal circular DNA: evidence for circularization by homologous recombination. Proc Natl Acad Sci U S A. 1985;82(7):1989–93. doi: 10.1073/pnas.82.7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen S, et al. Small polydispersed circular DNA (spcDNA) in human cells: association with genomic instability. Oncogene. 1997;14(8):977–85. doi: 10.1038/sj.onc.1200917. [DOI] [PubMed] [Google Scholar]
- 38.Hahn PJ. Molecular biology of double-minute chromosomes. Bioessays. 1993;15(7):477–84. doi: 10.1002/bies.950150707. [DOI] [PubMed] [Google Scholar]
- 39.Tomaska L, et al. Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol. 2009;16(10):1010–5. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang C, et al. The human CTC1/STN1/TEN1 complex regulates telomere maintenance in ALT cancer cells. Exp Cell Res. 2017;355(2):95–104. doi: 10.1016/j.yexcr.2017.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91(7):1033–42. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 42.Vogt N, et al. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci U S A. 2004;101(31):11368–73. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuberi L, et al. Rapid response to induction in a case of acute promyelocytic leukemia with MYC amplification on double minutes at diagnosis. Cancer Genet Cytogenet. 2010;198(2):170–2. doi: 10.1016/j.cancergencyto.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Storlazzi CT, et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20(9):1198–206. doi: 10.1101/gr.106252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del Rey J, et al. Centrosome clustering and cyclin D1 gene amplification in double minutes are common events in chromosomal unstable bladder tumors. BMC Cancer. 2010;10:280. doi: 10.1186/1471-2407-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nathanson DA, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343(6166):72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikolaev S, et al. Extrachromosomal driver mutations in glioblastoma and low-grade glioma. Nat Commun. 2014;5:5690. doi: 10.1038/ncomms6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Libuda DE, Winston F. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature. 2006;443(7114):1003–7. doi: 10.1038/nature05205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar P, et al. Normal and Cancerous Tissues Release Extrachromosomal Circular DNA (eccDNA) into the Circulation. Mol Cancer Res. 2017 doi: 10.1158/1541-7786.MCR-17-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, et al. Molecular characterization of cell-free eccDNAs in human plasma. Sci Rep. 2017;7(1):10968. doi: 10.1038/s41598-017-11368-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mackenzie KJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548(7668):461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Oliveira Mann CC, Kranzusch PJ. cGAS Conducts Micronuclei DNA Surveillance. Trends Cell Biol. 2017 doi: 10.1016/j.tcb.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Heitzer E, et al. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61(1):112–23. doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 54.Brewer BJ, et al. Origin-Dependent Inverted-Repeat Amplification: Tests of a Model for Inverted DNA Amplification. PLoS Genet. 2015;11(12):e1005699. doi: 10.1371/journal.pgen.1005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poole AM, et al. A positive role for yeast extrachromosomal rDNA circles? Extrachromosomal ribosomal DNA circle accumulation during the retrograde response may suppress mitochondrial cheats in yeast through the action of TAR1. Bioessays. 2012;34(9):725–9. doi: 10.1002/bies.201200037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410(2):103–23. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 57.Bendich AJ. Circular chloroplast chromosomes: the grand illusion. Plant Cell. 2004;16(7):1661–6. doi: 10.1105/tpc.160771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shlomai J. The structure and replication of kinetoplast DNA. Curr Mol Med. 2004;4(6):623–47. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- 59.Shapiro TA, Englund PT. The structure and replication of kinetoplast DNA. Annu Rev Microbiol. 1995;49:117–43. doi: 10.1146/annurev.mi.49.100195.001001. [DOI] [PubMed] [Google Scholar]